Drug Interactions in Lenacapavir-Based Long-Acting Antiviral Combinations
Abstract
:1. Introduction
2. Materials and Methods
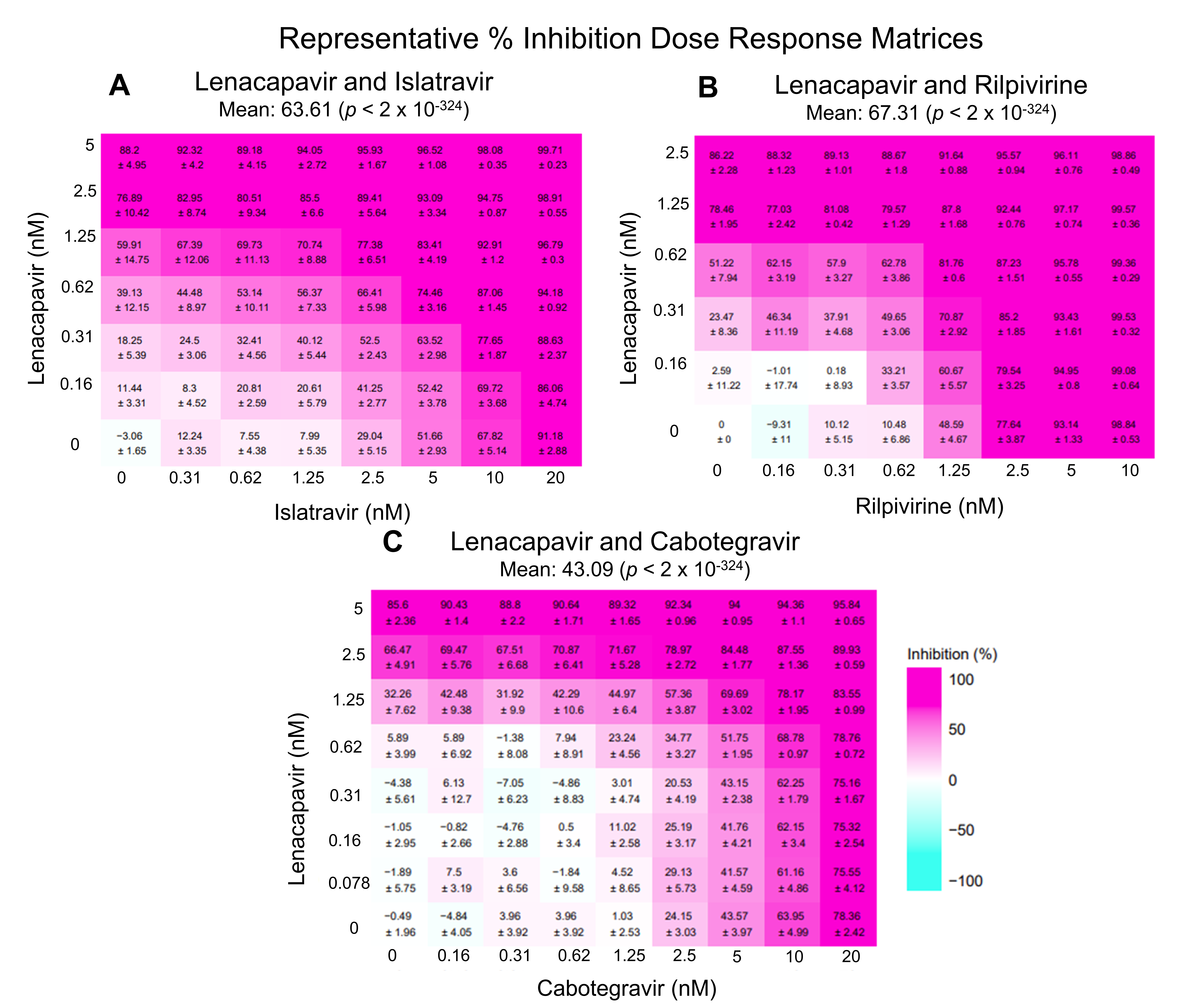
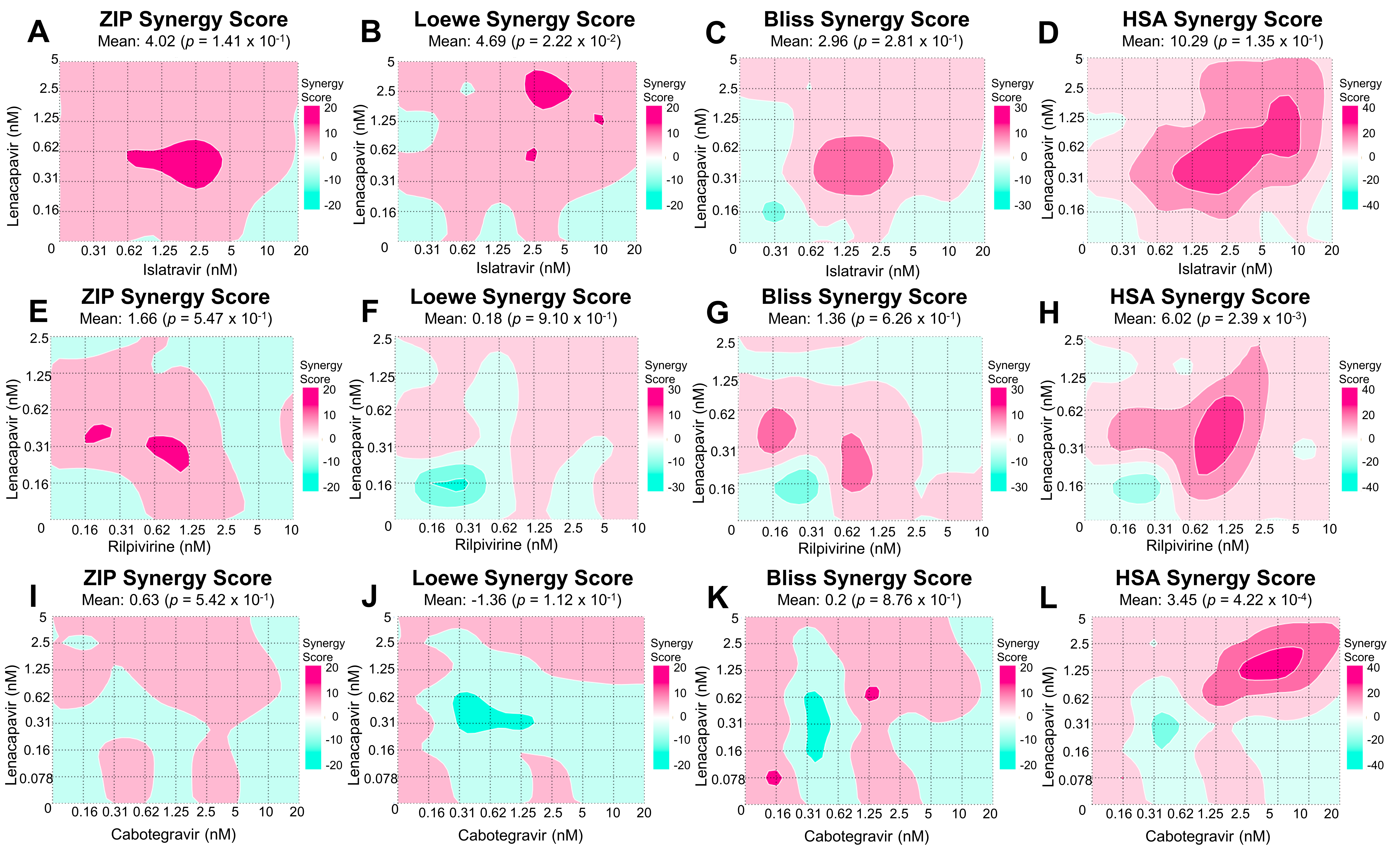
3. Results
3.1. LEN-ISL
3.2. LEN-RPV
3.3. LEN-CAB
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- UNIAIDS, Global HIV & AIDS Statistics—Fact Sheet. 2020. Available online: https://www.unaids.org/en/resources/fact-sheet (accessed on 20 March 2022).
- Riddell, J.; Amico, K.R.; Mayer, K.H. HIV Preexposure Prophylaxis: A Review. JAMA 2018, 319, 1261–1268. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.Y.; Wu, H.Y.; Yarla, N.S.; Xu, B.; Ding, J.; Lu, T.R. HAART in HIV/AIDS Treatments: Future Trends. Infect. Disord. Drug Targets 2018, 18, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Eggleton, J.S.; Nagalli, S. Highly Active Antiretroviral Therapy (HAART). In StatPearls, Treasure Island (FL); StatPearls Publishing LLC: Tampa, FL, USA, 2021. [Google Scholar]
- Markham, A. Cabotegravir Plus Rilpivirine: First Approval. Drugs 2020, 80, 915–922. [Google Scholar] [CrossRef] [PubMed]
- Thornhill, J.; Orkin, C. Long-acting injectable HIV therapies: The next frontier. Curr. Opin. Infect. Dis. 2021, 34, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Markowitz, M.; Sarafianos, S.G. 4′-Ethynyl-2-fluoro-2′-deoxyadenosine, MK-8591: A novel HIV-1 reverse transcriptase translocation inhibitor. Curr. Opin. HIV AIDS 2018, 13, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Pear, W.S.; Nolan, G.P.; Scott, M.L.; Baltimore, D. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 1993, 90, 8392–8396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeuchi, Y.; McClure, M.O.; Pizzato, M. Identification of gammaretroviruses constitutively released from cell lines used for human immunodeficiency virus research. J. Virol. 2008, 82, 12585–12588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, S.; Wang, W.; Aldahdooh, J.; Malyutina, A.; Shadbahr, T.; Tanoli, Z.; Pessia, A.; Tang, J. SynergyFinder Plus: Toward Better Interpretation and Annotation of Drug Combination Screening Datasets. Genom. Proteom. Bioinform. 2022. [Google Scholar] [CrossRef] [PubMed]
- Malyutina, A.; Majumder, M.M.; Wang, W.; Pessia, A.; Heckman, C.A.; Tang, J. Drug combination sensitivity scoring facilitates the discovery of synergistic and efficacious drug combinations in cancer. PLoS Comput. Biol. 2019, 15, e1006752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, B.; Wennerberg, K.; Aittokallio, T.; Tang, J. Searching for Drug Synergy in Complex Dose-Response Landscapes Using an Interaction Potency Model. Comput. Struct. Biotechnol. J. 2015, 13, 504–513. [Google Scholar] [CrossRef] [Green Version]
- Bliss, C.I. The toxicity of poisons applied jointly. Ann. Appl. Biol. 1939, 26, 585–615. [Google Scholar] [CrossRef]
- Loewe, S. The problem of synergism and antagonism of combined drugs. Arzneimittelforschung 1953, 3, 285–290. [Google Scholar]
- Berenbaum, M.C. What is synergy? Pharmacol. Rev. 1989, 41, 93–141. [Google Scholar] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cilento, M.E.; Ong, Y.T.; Tedbury, P.R.; Sarafianos, S.G. Drug Interactions in Lenacapavir-Based Long-Acting Antiviral Combinations. Viruses 2022, 14, 1202. https://doi.org/10.3390/v14061202
Cilento ME, Ong YT, Tedbury PR, Sarafianos SG. Drug Interactions in Lenacapavir-Based Long-Acting Antiviral Combinations. Viruses. 2022; 14(6):1202. https://doi.org/10.3390/v14061202
Chicago/Turabian StyleCilento, Maria E., Yee Tsuey Ong, Philip R. Tedbury, and Stefan G. Sarafianos. 2022. "Drug Interactions in Lenacapavir-Based Long-Acting Antiviral Combinations" Viruses 14, no. 6: 1202. https://doi.org/10.3390/v14061202
APA StyleCilento, M. E., Ong, Y. T., Tedbury, P. R., & Sarafianos, S. G. (2022). Drug Interactions in Lenacapavir-Based Long-Acting Antiviral Combinations. Viruses, 14(6), 1202. https://doi.org/10.3390/v14061202





