Association of the p75NTR Ser205Leu Polymorphism with Asymptomatic HTLV-1 Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Characterization and Sample Collection
2.2. DNA Extraction
2.3. Genotyping of NGF -198C/T, NGF Ala35Val, and p75NTR Ser205Leu
2.4. Quantification of HTLV-1 Proviral Load
2.5. Plasma Cytokine Level
2.6. Statistical Analysis
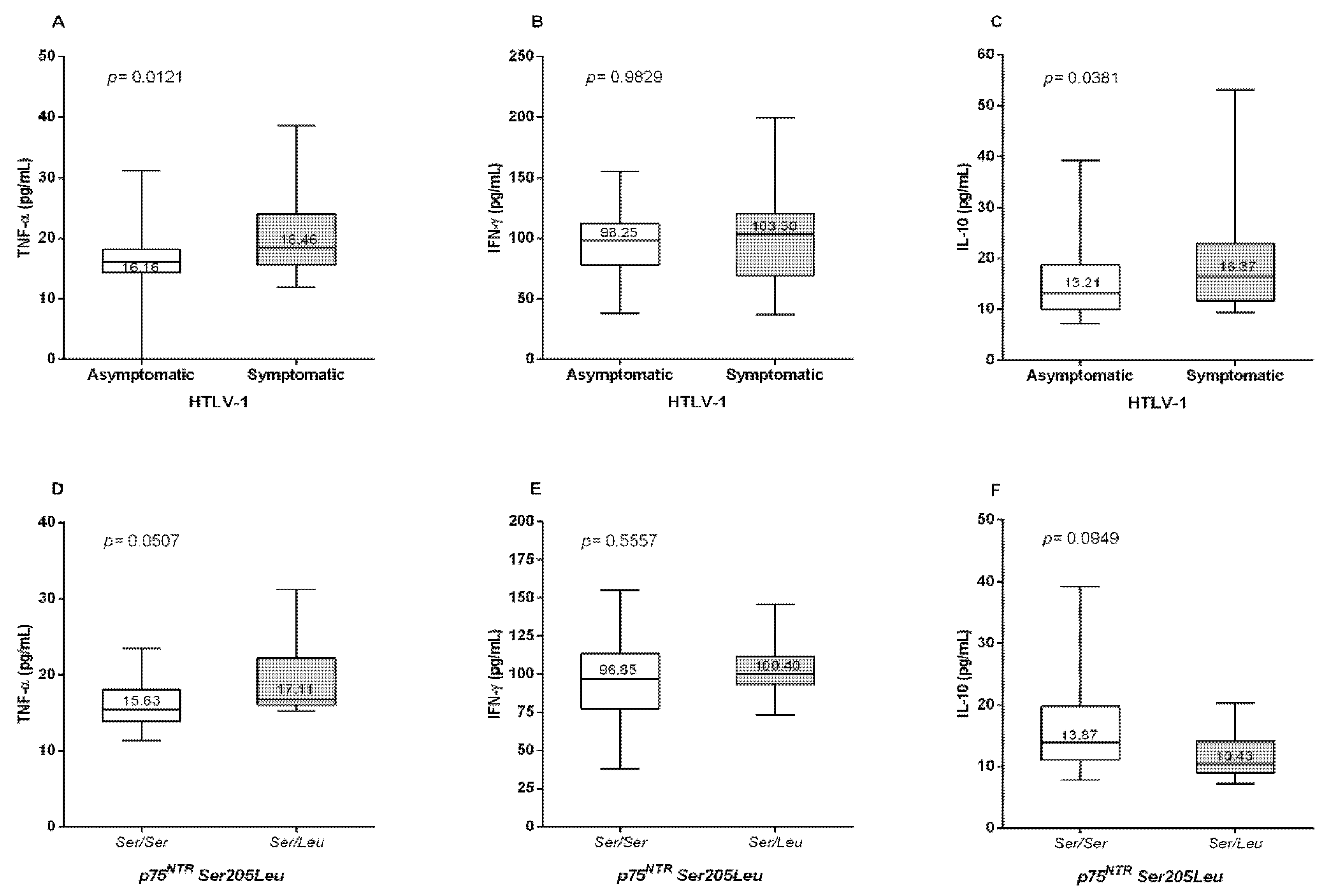
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kalyanaraman, V.S.; Sarngadharan, M.G.; Nakao, Y.; Ito, Y.; Aoki, T.; Gallo, R.C. Natural antibodies to the structural core protein (p24) of the human T-cell leucemia (lymphoma) retrovírus found in sera of leucemia in Japan. Proc. Natl. Acad. Sci. USA 1982, 79, 1653–1657. [Google Scholar] [CrossRef] [PubMed]
- Osame, M.; Usuku, K.; Izumo, S. HTLV-I associated myelopathy, a new clinical entity. Lancet 1986, 1, 1031–1032. [Google Scholar] [CrossRef]
- Quaresma, J.A.; Yoshikawa, G.T.; Koyama, R.V.; Dias, G.A.; Fujihara, S.; Fuzii, H.T. HTLV-1, Immune Response and Autoimmunity. Viruses 2015, 8, 5. [Google Scholar] [CrossRef] [PubMed]
- Poiesz, B.J.; Ruscetti, F.W.; Gazdar, A.F.; Bunn, P.A.; Minna, J.D.; Gallo, R.C. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. USA 1980, 77, 7415–7419. [Google Scholar] [CrossRef]
- Cassar, O.; Gessain, A. Serological and Molecular Methods to Study Epidemiological Aspects of human T-cell lymphotropi vírus type 1 infection. Methods Mol. Biol. 2017, 1582, 3–24. [Google Scholar]
- Martin, F.; Tagaya, Y.; Gallo, R. Time to eradicate HTLV-1: An open letter to WHO. Lancet 2018, 391, 1893–1894. [Google Scholar] [CrossRef]
- Rosadas, C.; Assone, T.; Yamashita, M.; Adonis, A.; Puccioni-Sohler, M.; Santos, M.; Paiva, A.; Casseb, J.; Oliveira, A.C.P.; Taylor, G.P. Health state utility values in people living with HTLV-1 and in patients with HAM/TSP: The impact of a neglected disease on the quality of life. PLoS Negl. Trop. Dis. 2020, 14, e0008761. [Google Scholar] [CrossRef]
- Rosadas, C.; Taylor, G.P. Health inequities and HTLV-1. Lancet Microbe 2021. [Google Scholar] [CrossRef]
- Assone, T.; de Souza, F.V.; Gaester, K.O.; Fonseca, L.A.; Luiz Odo, C.; Malta, F.; Pinho, J.R.; de Gonçalves, F.T.; Duarte, A.J.; de Oliveira, A.C.; et al. IL28B gene polymorphism SNP rs8099917 genotype GG is associated with HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP) in HTLV-1 carriers. PLoS Negl. Trop. Dis. 2014, 8, e3199. [Google Scholar] [CrossRef]
- Rosado, J.; Morales, S.; López, G.; Clark, D.; Verdonck, K.; Gotuzzo, E.; Van Camp, G.; Talledo, M. The FAS-670 AA genotype is associated with high proviral load in peruvian HAM/TSP patients. J. Med. Virol. 2017, 89, 726–731. [Google Scholar] [CrossRef]
- Queiroz, M.A.F.; Azevedo, V.N.; Amoras, E.S.G.; Moura, T.C.F.; Guimarães Ishak, M.O.; Ishak, R.; Vallinoto, A.C.R.; Martins Feitosa, R.N. IFNG +874A/T Polymorphism Among Asymptomatic HTLV-1-Infected Individuals Is Potentially Related to a Worse Prognosis. Front. Microbiol. 2018, 9, 795. [Google Scholar] [CrossRef] [PubMed]
- Vallinoto, A.C.R.; Cayres-Vallinoto, I.; Freitas Queiroz, M.A.; Ishak, M.O.G.; Ishak, R. Influence of Immunogenetic Biomarkers in the Clinical Outcome of HTLV-1 Infected Persons. Viruses 2019, 11, 974. [Google Scholar] [CrossRef] [PubMed]
- Silva, D.C.; Amoras, E.S.G.; Moura, T.C.F.; Lopes, F.T.; Gomes, S.T.M.; Costa, C.A.D.; Sousa, M.S.; Ishak, R.; Vallinoto, A.C.R.; Queiroz, M.A.F. TREX1 531C>T Polymorphism is Associated with High Proviral Load Levels in HTLV-1-Infected Persons. Viruses 2019, 12, 7. [Google Scholar] [CrossRef] [PubMed]
- Queiroz, M.A.F.; Amoras, E.S.G.; Moura, T.C.F.; da Costa, C.A.; de Sousa, M.S.; Lima, S.S.; Ishak, R.; Vallinoto, A.C.R. The SAMHD1 rs6029941 (A/G) Polymorphism Seems to Influence the HTLV-1 Proviral Load and IFN-Alpha Levels. Front. Cell Infect. Microbiol. 2020, 10, 246. [Google Scholar] [CrossRef] [PubMed]
- Levi-Montalcini, R. The nerve growth factor 35 years later. Science 1987, 237, 1154–1162. [Google Scholar] [CrossRef]
- Bracci-Laudiero, L.; De Stefano, M.E. NGF in Early Embryogenesis, Differentiation, and Pathology in the Nervous and Immune Systems. Curr. Top. Behav. Neurosci. 2016, 29, 125–152. [Google Scholar]
- Minnone, G.; De Benedetti, F.; Bracci-Laudiero, L. NGF and Its Receptors in the Regulation of Inflammatory Response. Int J. Mol. Sci. 2017, 18, 1028. [Google Scholar] [CrossRef]
- Roux, P.P.; Barker, P.A. Neurotrophin signaling through the p75 neurotrophin receptor. Prog. Neurobiol. 2002, 67, 203–233. [Google Scholar] [CrossRef]
- Alam, M.; Pravica, V.; Fryer, A.A.; Hawkins, C.P.; Hutchinson, I.V. Novel polymorphism in the promoter region of the human nerve growth-factor gene. Int J. Immunogenet. 2005, 32, 379–382. [Google Scholar] [CrossRef]
- Akkad, D.A.; Kruse, N.; Arning, L.; Gold, R.; Epplen, J.T. Genomic NGFB variation and multiple sclerosis in a case control study. BMC Med. Genet. 2008, 9, 107. [Google Scholar] [CrossRef]
- Cozza, A.; Melissari, E.; Iacopetti, P.; Mariotti, V.; Tedde, A.; Nacmias, B.; Conte, A.; Sorbi, S.; Pellegrini, S. SNPs in neurotrophin system genes and Alzheimer’s disease in an Italian population. J. Alzheimers Dis. 2008, 15, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Di Maria, E.; Giorgio, E.; Uliana, V.; Bonvicini, C.; Faravelli, F.; Cammarata, S.; Novello, M.C.; Galimberti, D.; Scarpini, E.; Zanetti, O.; et al. Possible influence of a non-synonymous polymorphism located in the NGF precursor on susceptibility to late-onset Alzheimer’s disease and mild cognitive impairment. J. Alzheimers Dis. 2012, 29, 699–705. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.T.; Lu, X.M.; Shu, Y.H.; Xiao, L.; Chen, K.T. Selection of human p75NTR tag SNPs and its biological significance for clinical association studies. Biomed. Mater. Eng. 2014, 24, 3833–3839. [Google Scholar] [CrossRef] [PubMed]
- Kunugi, H.; Hashimoto, R.; Yoshida, M.; Tatsumi, M.; Kamijima, K. A missense polymorphism (S205L) of the low-affinity neurotrophin receptor p75NTR gene is associated with depressive disorder and attempted suicide. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2004, 129B, 44–46. [Google Scholar] [CrossRef] [PubMed]
- Nagata, T.; Shinagawa, S.; Nukariya, K.; Nakayama, R.; Nakayama, K.; Yamada, H. Association between nerve growth factor gene polymorphism and executive dysfunction in Japanese patients with early-stage Alzheimer’s disease and amnestic mild cognitive impairment. Dement. Geriatr. Cogn. Disord. 2011, 32, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Pereira, L.M.S.; da Silva Graça Amoras, E.; da Silva Conde, S.R.S.; Demachki, S.; Dos Santos, E.J.M.; Lima, S.S.; Ishak, R.; Rosário Vallinoto, A.C. NGF (-198C > T, Ala35Val) and p75NTR (Ser205Leu) gene mutations are associated with liver function in different histopathological profiles of the patients with chronic viral hepatitis in the Brazilian Amazon. Mol. Med. 2020, 26, 12. [Google Scholar] [CrossRef]
- Tamegão-Lopes, B.P.; Rezende, P.R.; Maradei-Pereira, L.M.C.; Lemos, J.A.R. HTLV-1 and HTLV-2 proviral load: A simple method using quantitative real-time PCR. Rev. Soc. Bras. Med. Trop. 2006, 39, 548–552. [Google Scholar] [CrossRef]
- Enose-Akahata, Y.; Vellucci, A.; Jacobson, S. Role of HTLV-1 Tax and HBZ in the Pathogenesis of HAM/TSP. Front. Microbiol. 2017, 8, 2563. [Google Scholar] [CrossRef]
- Fuzii, H.T.; da Silva Dias, G.A.; de Barros, R.J.; Falcão, L.F.; Quaresma, J.A. Immunopathogenesis of HTLV-1-assoaciated myelopathy/tropical spastic paraparesis (HAM/TSP). Life Sci. 2014, 104, 9–14. [Google Scholar] [CrossRef]
- Saito, M. Association Between HTLV-1 Genotypes and Risk of HAM/TSP. Front. Microbiol. 2019, 10, 1101. [Google Scholar] [CrossRef]
- Yamauchi, J.; Araya, N.; Yagishita, N.; Sato, T.; Yamano, Y. An update on human T-cell leukemia virus type I (HTLV-1)-associated myelopathy/tropical spastic paraparesis (HAM/TSP) focusing on clinical and laboratory biomarkers. Pharmacol. Ther. 2021, 218, 107669. [Google Scholar] [CrossRef]
- Cartier, L.M.; Cea, J.G.; Vergara, C.; Araya, F.; Born, P. Clinical and neuropathological study of six patients with spastic paraparesis associated with HTLV-I: An axomyelinic degeneration of the central nervous system. J. Neuropathol. Exp. Neurol. 1997, 56, 403–413. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Albrecht, D.; García, L.; Cartier, L.; Kettlun, A.M.; Vergara, C.; Collados, L.; Valenzuela, M.A. Trophic factors in cerebrospinal fluid and spinal cord of patients with tropical spastic paraparesis, HIV, and Creutzfeldt-Jakob disease. AIDS Res. Hum. Retrovir. 2006, 248–254. [Google Scholar] [CrossRef]
- Keefe, K.M.; Sheikh, I.S.; Smith, G.M. Targeting Neurotrophins to Specific Populations of Neurons: NGF, BDNF, and NT-3 and Their Relevance for Treatment of Spinal Cord Injury. Int J. Mol. Sci. 2017, 18, 548. [Google Scholar] [CrossRef] [PubMed]
- Hempstead, B.L.; Martin-Zanca, D.; Kaplan, D.R.; Parada, L.F.; Chao, M.V. High-affinity NGF binding requires coexpression of the trk proto-oncogene and the low-affinity NGF receptor. Nature 1991, 350, 678–683. [Google Scholar] [CrossRef] [PubMed]
- Mahadeo, D.; Kaplan, L.; Chao, M.V.; Hempstead, B.L. High affinity nerve growth factor binding displays a faster rate of association than p140trk binding. Implications for multi-subunit polypeptide receptors. J. Biol. Chem. 1994, 269, 6884–6891. [Google Scholar] [CrossRef]
- Amoras, E.S.G.; Gomes, S.T.; Freitas, F.B.; Santana, B.B.; Ishak, G.; de Araújo, M.T.; Demachki, S.; da Silva Conde, S.R.; de Oliveira Guimarães Ishak, M.; Ishak, R.; et al. NGF and P75NTR gene expression is associated with the hepatic fibrosis stage due to viral and non-viral causes. PLoS ONE 2015, 10, e0121754. [Google Scholar]
- Otten, U.; Gadient, R.A. Neurotrophins and cytokines--intermediaries between the immune and nervous systems. Int. J. Dev. Neurosci. 1995, 13, 147–151. [Google Scholar] [CrossRef]
- Kuno, R.; Yoshida, Y.; Nitta, A.; Nabeshima, T.; Wang, J.; Sonobe, Y.; Kawanokuchi, J.; Takeuchi, H.; Mizuno, T.; Suzumura, A. The role of TNF-alpha and its receptors in the production of NGF and GDNF by astrocytes. Brain Res. 2006, 1116, 12–18. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, G.; Zheng, Y.; Lu, L.; Wu, C.; Zhang, Y.; Liu, Q.; Cao, X. TLR4 signaling induces functional nerve growth factor receptor p75NTR on mouse dendritic cells via p38MAPK and NF-kappa B pathways. Mol. Immunol. 2008, 45, 1557–1566. [Google Scholar] [CrossRef]
- Paiva, A.M.; Assone, T.; Haziot, M.E.J.; Smid, J.; Fonseca, L.A.M.; Luiz, O.D.C.; de Oliveira, A.C.P.; Casseb, J. Risk factors associated with HTLV-1 vertical transmission in Brazil: Longer breastfeeding, higher maternal proviral load and previous HTLV-1-infected offspring. Sci. Rep. 2018, 8, 7742. [Google Scholar] [CrossRef] [PubMed]

| Genotypes and Alleles | HTLV-1 n = 166 n (%) | Control n = 200 n (%) | p |
|---|---|---|---|
| NGF -198C/T rs11102930 | |||
| CC | 53 (31.9) | 67 (33.5) | 0.9358 * |
| CT | 83 (50.0) | 99 (49.5) | |
| TT | 30 (18.1) | 34 (17.0) | |
| * C | 0.57 | 0.58 | 1.0000 ** |
| * T | 0.43 | 0.42 | |
| NGF Ala35Val rs6330 | |||
| Ala/Ala | 77 (46.4) | 90 (45.0) | 0.2389 * |
| Ala/Val | 79 (47.6) | 88 (44.0) | |
| Val/Val | 10 (6.0) | 22 (11.0) | |
| Ala | 0.70 | 0.67 | 0.6519 ** |
| Val | 0.30 | 0.33 | |
| p75NTR Ser205Leu rs2072446 | |||
| Ser/Ser | 155 (93.4) | 173 (86.5) | 0.0385 ** |
| Ser/Leu | 11 (6.6) | 27 (13.5) | |
| Ser | 0.97 | 0.93 | 0.3311 ** |
| Leu | 0.03 | 0.07 |
| Genotypes and Alleles | Asymptomatic n = 107 n (%) | Symptomatic n = 59 n (%) | p |
|---|---|---|---|
| NGF -198C/T rs11102930 | |||
| CC | 31 (29.0) | 22 (37.3) | 0.3415 * |
| CT | 58 (54.2) | 25 (42.4) | |
| TT | 18 (16.8) | 12 (20.3) | |
| * C | 0.56 | 0.58 | 0.8895 ** |
| * T | 0.44 | 0.42 | |
| NGF Ala35Val rs6330 | |||
| Ala/Ala | 53 (49.6) | 24 (40.7) | 0.4550 *** |
| Ala/Val | 47 (43.9) | 32 (54.2) | |
| Val/Val | 7 (6.5) | 3 (5.1) | |
| Ala | 0.71 | 0.68 | 0.649 ** |
| Val | 0.29 | 0.32 | |
| p75NTR Ser205Leu rs2072446 | |||
| Ser/Ser | 96 (89.7) | 59 (100) | 0.0082 ** |
| Ser/Leu | 11 (10.3) | 0 | |
| Ser | 0.94 | 1 | 0.0289 ** |
| Leu | 0.06 | 0 |
| Genotypes and Alleles | Asymptomatic n = 107 n (%) | HAM n = 38 n (%) | p |
|---|---|---|---|
| NGF -198C/T rs11102930 | |||
| CC | 31 (29.0) | 13 (34.2) | 0.8332 * |
| CT | 58 (54.2) | 19 (50.0) | |
| TT | 18 (16.8) | 6 (15.8) | |
| * C | 0.56 | 0.59 | 0.7749 ** |
| * T | 0.44 | 0.41 | |
| NGF Ala35Val rs6330 | |||
| Ala/Ala | 53 (49.6) | 16 (42.1) | 0.3899 *** |
| Ala/Val | 47 (43.9) | 21 (55.3) | |
| Val/Val | 7 (6.5) | 1 (2.6) | |
| Ala | 0.71 | 0.69 | 0.8775 ** |
| Val | 0.29 | 0.31 | |
| p75NTR Ser205Leu rs2072446 | |||
| Ser/Ser | 96 (89.7) | 38 (100) | 0.0381 ** |
| Ser/Leu | 11 (10.3) | 0 | |
| Ser | 0.94 | 1 | 0.0289 ** |
| Leu | 0.06 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Queiroz, M.A.F.; Lopes, F.T.; Botelho, B.J.S.; Torres, M.K.d.S.; Amoras, E.d.G.S.; Costa, C.A.d.; Sousa, M.S.; Ishak, R.; Vallinoto, A.C.R. Association of the p75NTR Ser205Leu Polymorphism with Asymptomatic HTLV-1 Infection. Viruses 2022, 14, 1175. https://doi.org/10.3390/v14061175
Queiroz MAF, Lopes FT, Botelho BJS, Torres MKdS, Amoras EdGS, Costa CAd, Sousa MS, Ishak R, Vallinoto ACR. Association of the p75NTR Ser205Leu Polymorphism with Asymptomatic HTLV-1 Infection. Viruses. 2022; 14(6):1175. https://doi.org/10.3390/v14061175
Chicago/Turabian StyleQueiroz, Maria Alice Freitas, Felipe Teixeira Lopes, Bruno José Sarmento Botelho, Maria Karoliny da Silva Torres, Ednelza da Graça Silva Amoras, Carlos A. da Costa, Maísa Silva Sousa, Ricardo Ishak, and Antonio Carlos Rosário Vallinoto. 2022. "Association of the p75NTR Ser205Leu Polymorphism with Asymptomatic HTLV-1 Infection" Viruses 14, no. 6: 1175. https://doi.org/10.3390/v14061175
APA StyleQueiroz, M. A. F., Lopes, F. T., Botelho, B. J. S., Torres, M. K. d. S., Amoras, E. d. G. S., Costa, C. A. d., Sousa, M. S., Ishak, R., & Vallinoto, A. C. R. (2022). Association of the p75NTR Ser205Leu Polymorphism with Asymptomatic HTLV-1 Infection. Viruses, 14(6), 1175. https://doi.org/10.3390/v14061175







