γδ T Cells in Emerging Viral Infection: An Overview
Abstract
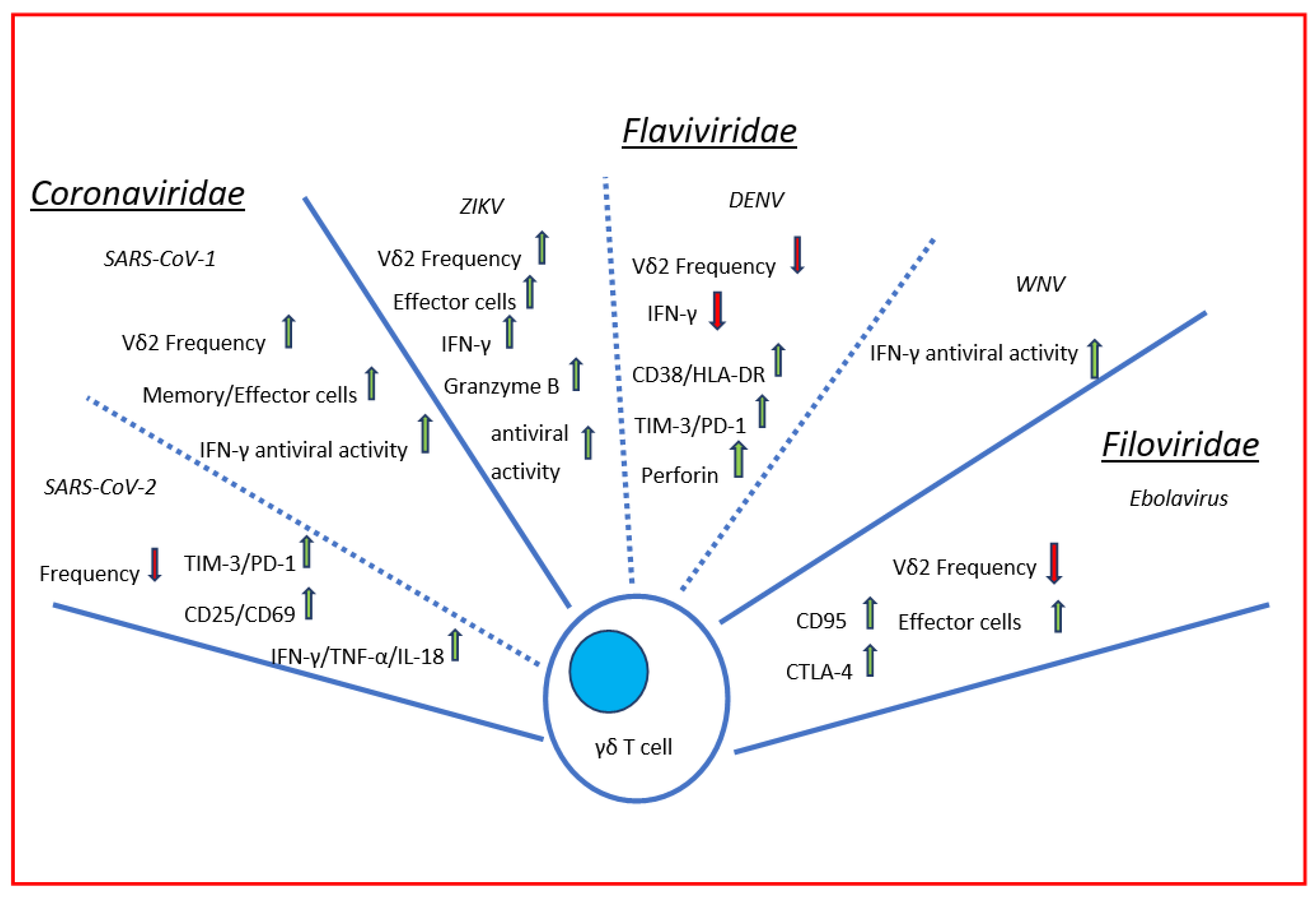
:1. Introduction
2. Coronaviruses
2.1. Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV)
2.2. Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2)
3. Flavivirus
3.1. Dengue Viruses
3.2. Zika Virus
3.3. West Nile Virus (WNV)
4. Filovirus
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ryu, W.-S. Chapter 21 New Emerging Viruses. Mol. Virol. Hum. Pathog. Viruses 2016, 289–302. [Google Scholar] [CrossRef]
- Hayday, A.C. γδ T Cell Update: Adaptate Orchestrators of Immune Surveillance. J. Immunol. 2019, 203, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Holtmeier, W.; Kabelitz, D. γδ T Cells Link Innate and Adaptive Immune Responses. Mech. Epithel. Def. 2005, 86, 151–183. [Google Scholar] [CrossRef]
- Giri, S.; Lal, G. Differentiation and functional plasticity of gamma-delta (γδ) T cells under homeostatic and disease conditions. Mol. Immunol. 2021, 136, 138–149. [Google Scholar] [CrossRef]
- Jin, Y.; Xia, M.; Saylor, C.M.; Narayan, K.; Kang, J.; Wiest, D.L.; Wang, Y.; Xiong, N. Cutting edge: Intrinsic programming of thymicgd T cells for specific peripheral tissue localization. J. Immunol. 2010, 185, 7156–7160. [Google Scholar] [CrossRef]
- Papadopoulou, M.; Sanchez, G.S.; Vermijlen, D. Innate and adaptive γδ T cells: How, when, and why. Immunol. Rev. 2020, 298, 99–116. [Google Scholar] [CrossRef]
- Hayday, A.C.; Vantourout, P. The Innate Biologies of Adaptive Antigen Receptors. Annu. Rev. Immunol. 2020, 38, 487–510. [Google Scholar] [CrossRef] [Green Version]
- Chennupati, V.; Worbs, T.; Liu, X.; Malinarich, F.H.; Schmitz, S.; Haas, J.D.; Malissen, B.; Förster, R.; Prinz, I. Intra- and Intercompartmental Movement of γδ T Cells: Intestinal Intraepithelial and Peripheral γδ T Cells Represent Exclusive Nonoverlapping Populations with Distinct Migration Characteristics. J. Immunol. 2010, 185, 5160–5168. [Google Scholar] [CrossRef] [Green Version]
- Castro, C.D.; Boughter, C.T.; Broughton, A.E.; Ramesh, A.; Adams, E.J. Diversity in recognition and function of human γδ T cells. Immunol. Rev. 2020, 298, 134–152. [Google Scholar] [CrossRef]
- Deseke, M.; Prinz, I. Ligand recognition by the γδ TCR and discrimination between homeostasis and stress conditions. Cell. Mol. Immunol. 2020, 17, 914–924. [Google Scholar] [CrossRef]
- Groh, V.; Steinle, A.; Bauer, S.; Spies, T. Recognition of Stress-Induced MHC Molecules by Intestinal Epithelial γδ T Cells. Science 1998, 279, 1737–1740. [Google Scholar] [CrossRef] [PubMed]
- Kabelitz, D. Editorial: “Recent advances in gamma/delta T cell biology: New ligands, new functions, and new translational perspectives”. Front. Immunol. 2015, 6, 371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herrmann, T.; Fichtner, A.S.; Karunakaran, M.M. An Update on the Molecular Basis of Phosphoantigen Recognition by Vγ9Vδ2 T Cells. Cells 2020, 9, 1433. [Google Scholar] [CrossRef] [PubMed]
- Barros, R.D.M.; Roberts, N.A.; Dart, R.J.; Vantourout, P.; Jandke, A.; Nussbaumer, O.; Deban, L.; Cipolat, S.; Hart, R.; Iannitto, M.L.; et al. Epithelia Use Butyrophilin-like Molecules to Shape Organ-Specific γδ T Cell Compartments. Cell 2016, 167, 203–218.e17. [Google Scholar] [CrossRef] [Green Version]
- Jandke, A.; Melandri, D.; Monin, L.; Ushakov, D.S.; Laing, A.G.; Vantourout, P.; East, P.; Nitta, T.; Narita, T.; Takayanagi, H.; et al. Butyrophilin-like proteins display combinatorial diversity in selecting and maintaining signature intraepithelial γδ T cell compartments. Nat. Commun. 2020, 11, 3769. [Google Scholar] [CrossRef]
- Poccia, F.; Agrati, C.; Martini, F.; Capobianchi, M.R.; Wallace, M.; Malkovsky, M. Antiviral reactivities of γδ T cells. Microbes Infect. 2005, 7, 518–528. [Google Scholar] [CrossRef]
- Caron, J.; Ridgley, L.A.; Bodman-Smith, M. How to Train Your Dragon: Harnessing Gamma Delta T Cells Antiviral Functions and Trained Immunity in a Pandemic Era. Front. Immunol. 2021, 12, 983. [Google Scholar] [CrossRef]
- Chien, Y.; Meyer, C.; Bonneville, M. gd T cells: First line of defense and beyond. Annu. Rev. Immunol. 2014, 32, 121–155. [Google Scholar] [CrossRef]
- Yazdanifar, M.; Barbarito, G.; Bertaina, A.; Airoldi, I. γδ T Cells: The Ideal Tool for Cancer Immunotherapy. Cells 2020, 9, 1305. [Google Scholar] [CrossRef]
- Zhao, Y.; Lin, L.; Xiao, Z.; Li, M.; Wu, X.; Li, W.; Li, X.; Zhao, Q.; Wu, Y.; Zhang, H.; et al. Protective Role of γδ T Cells in Different Pathogen Infections and Its Potential Clinical Application. J. Immunol. Res. 2018, 2018, 5081634. [Google Scholar] [CrossRef] [Green Version]
- Silva-Santos, B.; Mensurado, S.; Coffelt, S.B. γδ T cells: Pleiotropic immune effectors with therapeutic potential in cancer. Nat. Rev. Cancer 2019, 19, 392–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sebestyen, Z.; Prinz, I.; Déchanet-Merville, J.; Silva-Santos, B.; Kuball, J. Translating gammadelta (γδ) T cells and their receptors into cancer cell therapies. Nat. Rev. Drug Discov. 2019, 19, 169–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Payne, S. Family Coronaviridae. Viruses 2017, 149–158. [Google Scholar] [CrossRef]
- Stadler, K.; Masignani, V.; Eickmann, M.; Becker, S.; Abrignani, S.; Klenk, H.-D.; Rappuoli, R. SARS—Beginning to understand a new virus. Nat. Rev. Genet. 2003, 1, 209–218. [Google Scholar] [CrossRef]
- WHO. Available online: http://www.who.int/csr/sars/country/2003_08_15/en/ (accessed on 28 March 2022).
- Lam, W.K.; Zhong, N.S.; Tan, W.C. Overview on SARS in Asia and the World. Respirology 2003, 8, S2–S5. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Hui, D.; Wu, A.; Chan, P.; Cameron, P.; Joynt, G.M.; Ahuja, A.; Yung, M.Y.; Leung, C.; To, K.; et al. A Major Outbreak of Severe Acute Respiratory Syndrome in Hong Kong. N. Engl. J. Med. 2003, 348, 1986–1994. [Google Scholar] [CrossRef]
- Ho, A.S.; Sung, J.J.; Chan-Yeung, M. An outbreak of severe acute respiratory syndrome among hospital workers in a community hospital in Hong Kong. Ann. Intern. Med. 2003, 139, 564–567. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Gao, X. Immunological responses against SARS-coronavirus infection in humans. Cell MolImmunol. 2004, 1, 119–122. [Google Scholar] [PubMed]
- Li, C.K.-F.; Xu, X. Host Immune Responses to SARS Coronavirus in Humans. Mol. Biol. SARS-Coronavirus 2009, 22, 259–278. [Google Scholar] [CrossRef] [Green Version]
- Poccia, F.; Agrati, C.; Castilletti, C.; Bordi, L.; Gioia, C.; Horejsh, D.; Ippolito, G.; Chan, P.; Hui, D.; Sung, J.J.Y.; et al. Anti–Severe Acute Respiratory Syndrome Coronavirus Immune Responses: The Role Played by Vγ9Vδ2 T Cells. J. Infect. Dis. 2006, 193, 1244–1249. [Google Scholar] [CrossRef]
- Ren, L.-L.; Wang, Y.-M.; Wu, Z.-Q.; Xiang, Z.-C.; Guo, L.; Xu, T.; Jiang, Y.-Z.; Xiong, Y.; Li, Y.-J.; Li, X.-W.; et al. Identification of a novel coronavirus causing severe pneumonia in human: A descriptive study. Chin. Med. J. 2020, 133, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Giamarellos-Bourboulis, E.J.; Netea, M.G.; Rovina, N.; Akinosoglou, K.; Antoniadou, A.; Antonakos, N.; Damoraki, G.; Gkavogianni, T.; Adami, M.-E.; Katsaounou, P.; et al. Complex Immune Dysregulation in COVID-19 Patients with Severe Respiratory Failure. Cell Host Microbe 2020, 27, 992–1000.e3. [Google Scholar] [CrossRef] [PubMed]
- Matricardi, P.M.; Dal Negro, R.W.; Nisini, R. The first, holistic immunological model of COVID-19: Implications for preven-tion, diagnosis, and public health measures. Pediatr. Allergy Immunol. 2020, 31, 454–470. [Google Scholar] [CrossRef] [PubMed]
- Rokni, M.; Ghasemi, V.; Tavakoli, Z. Immune responses and pathogenesis ofSARS-CoV-2 during an outbreak in Iran: Comparison withSARSandMERS. Rev. Med. Virol. 2020, 30, e2107. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.; Cai, W.; Cheng, A.; Wang, M.; Yin, Z.; Jia, R. Flaviviruses: Innate Immunity, Inflammasome Activation, Inflammatory Cell Death, and Cytokines. Front. Immunol. 2022, 13, 829433. [Google Scholar] [CrossRef]
- Bordoni, V.; Sacchi, A.; Cimini, E.; Notari, S.; Grassi, G.; Tartaglia, E.; Casetti, R.; Giancola, M.L.; Bevilacqua, N.; Maeurer, M.; et al. An Inflammatory Profile Correlates with Decreased Frequency of Cytotoxic Cells in Coronavirus Disease 2019. Clin. Infect. Dis. 2020, 71, 2272–2275. [Google Scholar] [CrossRef]
- Sacchi, A.; Grassi, G.; Bordoni, V.; Lorenzini, P.; Cimini, E.; Casetti, R.; Tartaglia, E.; Marchioni, L.; Petrosillo, N.; Palmieri, F.; et al. Early expansion of myeloid-derived suppressor cells inhibits SARS-CoV-2 specific T-cell response and may predict fatal COVID-19 outcome. Cell Death Dis. 2020, 11, 921. [Google Scholar] [CrossRef]
- Rijkers, G.; Vervenne, T.; Van Der Pol, P. More bricks in the wall against SARS-CoV-2 infection: Involvement of γ9δ2 T cells. Cell. Mol. Immunol. 2020, 17, 771–772. [Google Scholar] [CrossRef]
- Wilk, A.J.; Rustagi, A.; Zhao, N.Q.; Roque, J.; Martínez-Colón, G.J.; McKechnie, J.L.; Ivison, G.T.; Ranganath, T.; Vergara, R.; Hollis, T.; et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020, 26, 1070–1076. [Google Scholar] [CrossRef]
- Odak, I.; Barros-Martins, J.; Bošnjak, B.; Stahl, K.; David, S.; Wiesner, O.; Busch, M.; Hoeper, M.M.; Pink, I.; Welte, T.; et al. Reappearance of effector T cells is associated with recovery from COVID-19. EBioMedicine 2020, 57, 102885. [Google Scholar] [CrossRef]
- Saris, A.; Reijnders, T.D.Y.; Nossent, E.J.; Schuurman, A.R.; Verhoeff, J.; van Asten, S.; Bontkes, H.; Blok, S.; Duitman, J.; Bogaard, H.-J.; et al. Distinct cellular immune profiles in the airways and blood of critically ill patients with COVID-19. Thorax 2021, 76, 1010–1019. [Google Scholar] [CrossRef] [PubMed]
- Liao, M.; Liu, Y.; Yuan, J.; Wen, Y.; Xu, G.; Zhao, J.; Cheng, L.; Li, J.; Wang, X.; Wang, F.; et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020, 26, 842–844. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.; Qian, H.; Yang, X.; Zhang, X.; Zhang, D.; Dai, T.; Guo, R.; Shi, L.; Cheng, Y.; Zhang, B.; et al. The phenotypic changes of γδ T cells in COVID-19 patients. J. Cell. Mol. Med. 2020, 24, 11603–11606. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, S.F.; Ramsdell, F.; Alderson, M.R. The activation antigen CD69. Stem Cells 1994, 12, 456–465. [Google Scholar] [CrossRef] [PubMed]
- Jouan, Y.; Guillon, A.; Gonzalez, L.; Perez, Y.; Boisseau, C.; Ehrmann, S.; Ferreira, M.; Daix, T.; Jeannet, R.; François, B.; et al. Phenotypical and functional alteration of unconventional T cells in severe COVID-19 patients. J. Exp. Med. 2020, 217, e20200872. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.-Y.; Liong, K.H.; Gunalan, M.G.; Li, N.; Liang Lim, D.S.; Dale, A.; Fisher, P.; MacAry, A.; Leo, Y.S.; Wong, S.-W.; et al. Type I IFNs and IL-18 Regulate the Antiviral Response of Primary Human gd T Cells against Dendritic Cells Infected with Dengue virus. J. Immunol. 2015, 194, 3890–3900. [Google Scholar] [CrossRef]
- Flament, H.; Rouland, M.; Beaudoin, L.; Toubal, A.; Bertrand, L.; Lebourgeois, S.; Rousseau, C.; Soulard, P.; Gouda, Z.; Cagninacci, L.; et al. Outcome of SARS-CoV-2 infection is linked to MAIT cell activation and cytotoxicity. Nat. Immunol. 2021, 22, 322–335. [Google Scholar] [CrossRef]
- Laing, A.G.; Lorenc, A.; del Barrio, I.D.M.; Das, A.; Fish, M.; Monin, L.; Muñoz-Ruiz, M.; McKenzie, D.R.; Hayday, T.S.; Francos-Quijorna, I.; et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020, 26, 1623–1635. [Google Scholar] [CrossRef]
- Carissimo, G.; Xu, W.; Kwok, I.; Abdad, M.Y.; Chan, Y.-H.; Fong, S.-W.; Puan, K.J.; Lee, C.Y.-P.; Yeo, N.K.-W.; Amrun, S.N.; et al. Whole blood immunophenotyping uncovers immature neutrophil-to-VD2 T-cell ratio as an early marker for severe COVID-19. Nat. Commun. 2020, 11, 5243. [Google Scholar] [CrossRef]
- Zhang, J.-Y.; Wang, X.-M.; Xing, X.; Xu, Z.; Zhang, C.; Song, J.-W.; Fan, X.; Xia, P.; Fu, J.-L.; Wang, S.-Y.; et al. Single-cell landscape of immunological responses in patients with COVID-19. Nat. Immunol. 2020, 21, 1107–1118. [Google Scholar] [CrossRef]
- Carter, M.J.; Fish, M.; Jennings, A.; Doores, K.J.; Wellman, P.; Seow, J.; Acors, S.; Graham, C.; Timms, E.; Kenny, J.; et al. Peripheral immunophenotypes in children with multisystem inflammatory syndrome associated with SARS-CoV-2 infection. Nat. Med. 2020, 26, 1701–1707. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, D.L.; Haque, S.F.Y.; Silberzahn, T.; Hamilton, K.; Langford, C.; Ellis, P.; Carr, R.; Hayday, A.C. Neonates harbour highly active γδ T cells with selective impairments in preterm infants. Eur. J. Immunol. 2009, 39, 1794–1806. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Mo, X.; Hu, Y.; Qi, X.; Jiang, F.; Jiang, Z.; Tong, S. Epidemiology of COVID-19 Among Children in China. Pediatrics 2020, 145, e20200702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerapio, J.P.; Perrier, M.; Pont, F.; Tosolini, M.; Laurent, C.; Bertani, S.; Fournie, J.-J. Single-Cell RNAseq Profiling of Human γδ T Lymphocytes in Virus-Related Cancers and COVID-19 Disease. Viruses 2021, 13, 2212. [Google Scholar] [CrossRef] [PubMed]
- Kuno, G.; Chang, G.-J.J.; Tsuchiya, K.R.; Karabatsos, N.; Cropp, C.B. Phylogeny of the Genus Flavivirus. J. Virol. 1998, 72, 73–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazeaud, C.; Freppel, W.; Chatel-Chaix, L. The Multiples Fates of the Flavivirus RNA Genome During Pathogenesis. Front. Genet. 2018, 9, 595. [Google Scholar] [CrossRef]
- Rathore, A.P.S.; St John, A.L. Cross-Reactive Immunity Among Flaviviruses. Front. Immunol. 2020, 11, 334. [Google Scholar] [CrossRef] [Green Version]
- Saron, W.A.A.; Rathore, A.P.S.; Ting, L.; Ooi, E.E.; Low, J.; Abraham, S.N.; John, A.L.S. Flavivirus serocomplex cross-reactive immunity is protective by activating heterologous memory CD4 T cells. Sci. Adv. 2018, 4, eaar4297. [Google Scholar] [CrossRef] [Green Version]
- Dos Santos Franco, L.; Gushi, L.T.; Luiz, W.B.; Amorim, J.H. Seeking Flavivirus Cross-Protective Immunity. Front. Immunol. 2019, 10, 2260. [Google Scholar] [CrossRef]
- Slon-Campos, J.L.; Mongkolsapaya, J.; Screaton, G.R. The immune response against flaviviruses. Nat. Immunol. 2018, 19, 1189–1198. [Google Scholar] [CrossRef]
- Suthar, M.S.; Aguirre, S.; Fernandez-Sesma, A. Innate Immune Sensing of Flaviviruses. PLoS Pathog. 2013, 9, e1003541. [Google Scholar] [CrossRef] [PubMed]
- Cimini, E.; Bordoni, V.; Sacchi, A.; Visco-Comandini, U.; Montalbano, M.; Taibi, C.; Casetti, R.; Lalle, E.; D’Offizi, G.; Capobianchi, M.; et al. Intrahepatic Vγ9Vδ2 T-cells from HCV-infected patients show an exhausted phenotype but can inhibit HCV replication. Virus Res. 2018, 243, 31–35. [Google Scholar] [CrossRef]
- Cimini, E.; Bonnafous, C.; Sicard, H.; Vlassi, C.; D’Offizi, G.; Capobianchi, M.R.; Martini, F.; Agrati, C. In VivoInterferon-Alpha/Ribavirin Treatment Modulates Vγ9Vδ2 T-Cell Function During Chronic HCV Infection. J. Interf. Cytokine Res. 2013, 33, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Cimini, E.; Bonnafous, C.; Bordoni, V.; Lalle, E.; Sicard, H.; Sacchi, A.; Berno, G.; Gioia, C.; D’Offizi, G.; Comandini, U.V.; et al. Interferon-α Improves Phosphoantigen-Induced Vγ9Vδ2 T-Cells Interferon-γ Production during Chronic HCV Infection. PLoS ONE 2012, 7, e37014. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue (accessed on 24 March 2022).
- World Health Organization. Dengue and Severe Dengue. April 2017. Available online: http://www.who.int/mediacentre/factsheets/fs117/en/ (accessed on 24 March 2022).
- Districy of Columbia Depatment of Health. Dengue Fever Factsheet. Available online: https://doh.dc.gov/sites/default/files/dc/sites/doh/publication/attachments/Dengue_Fever.pdf (accessed on 24 March 2022).
- Centers for Disease Control and Prevention. Epidemiology. Available online: https://www.cdc.gov/dengue/epidemiology/ (accessed on 24 March 2022).
- Centers for Disease Control and Prevention. Dengue. Available online: https://wwwnc.cdc.gov/travel/yellowbook/2016/infectious-diseases-related-to-travel/dengue (accessed on 24 March 2022).
- Nanaware, N.; Banerjee, A.; Bagchi, S.M.; Bagchi, P.; Mukherjee, A. Dengue Virus Infection: A Tale of Viral Exploitations and Host Responses. Viruses 2021, 13, 1967. [Google Scholar] [CrossRef] [PubMed]
- King, C.A.; Wegman, A.D.; Endy, T.P. Mobilization and Activation of the Innate Immune Response to Dengue Virus. Front. Cell. Infect. Microbiol. 2020, 10, 574417. [Google Scholar] [CrossRef]
- Mantri, C.K.; St. John, A.L. Immune synapses between mast cells and γδ T cells limit viral infection. J. Clin. Investig. 2019, 129, 1094–1108. [Google Scholar] [CrossRef] [Green Version]
- Cimini, E.; Grassi, G.; Beccacece, A.; Casetti, R.; Castilletti, C.; Capobianchi, M.R.; Nicastri, E.; Agrati, C. In Acute Dengue Infection, High TIM-3 Expression May Contribute to the Impairment of IFNγ Production by Circulating Vδ2 T Cells. Viruses 2022, 14, 130. [Google Scholar] [CrossRef]
- Azeredo, E.L.; De Oliveira-Pinto, L.M.; Zagne, S.M.; Cerqueira, D.I.S.; Nogueira, R.M.R.; Kubelka, C.F. NK cells, displaying early activation, cytotoxicity and adhesion molecules, are associated with mild dengue disease. Clin. Exp. Immunol. 2005, 143, 345–356. [Google Scholar] [CrossRef]
- Dick, G.W.A.; Kitchen, S.F.; Haddow, A.J. Zika Virus (I). Isolations and serological specificity. Trans. R. Soc. Trop. Med. Hyg. 1952, 46, 509–520. [Google Scholar] [CrossRef]
- Maurice, J. The Zika virus public health emergency: 6 months on. Lancet 2016, 388, 449–450. [Google Scholar] [CrossRef]
- Carteaux, G.; Maquart, M.; Bedet, A.; Contou, D.; Brugières, P.; Fourati, S.; de Langavant, L.C.; De Broucker, T.; Brun-Buisson, C.; Leparc-Goffart, I.; et al. Zika Virus Associated with Meningoencephalitis. N. Engl. J. Med. 2016, 374, 1595–1596. [Google Scholar] [CrossRef] [PubMed]
- Galliez, R.M.; Spitz, M.; Rafful, P.P.; Cagy, M.; Escosteguy, C.; Germano, C.S.B.; Sasse, E.; Gonçalves, A.L.; Silveira, P.P.; Pezzuto, P.; et al. Zika Virus Causing Encephalomyelitis Associated with Immunoactivation. Open Forum Infect. Dis. 2016, 3, ofw203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mécharles, S.; Herrmann, C.; Poullain, P.; Tran, T.-H.; Deschamps, N.; Mathon, G.; Landais, A.; Breurec, S.; Lannuzel, A. Acute myelitis due to Zika virus infection. Lancet 2016, 387, 1481. [Google Scholar] [CrossRef] [Green Version]
- Zucker, J.; Neu, N.; Chiriboga, C.A.; Hinton, V.J.; Leonardo, M.; Sheikh, A.; Thakur, K. Zika Virus–Associated Cognitive Impairment in Adolescent, 2016. Emerg. Infect. Dis. 2017, 23, 1047–1048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brasil, P.; Pereira, J.P., Jr.; Moreira, M.E.; Nogueira, R.M.R.; Damasceno, L.; Wakimoto, M.; Rabello, R.S.; Valderramos, S.G.; Halai, U.-A.; Salles, T.S.; et al. Zika Virus Infection in Pregnant Women in Rio de Janeiro-Preliminary report. N. Engl. J. Med. 2016, 375, 2321–2334. [Google Scholar] [CrossRef] [PubMed]
- Freitas, B.D.P.; Dias, J.R.D.O.; Prazeres, J.; Sacramento, G.A.; Ko, A.I.; Maia, M.; Belfort, R. Ocular Findings in Infants with Microcephaly Associated with Presumed Zika Virus Congenital Infection in Salvador, Brazil. JAMA Ophthalmol. 2016, 134, 529–535. [Google Scholar] [CrossRef] [Green Version]
- Vouga, M.; Baud, D. Imaging of congenital Zika virus infection: The route to identification of prognostic factors. Prenat. Diagn. 2016, 36, 799–811. [Google Scholar] [CrossRef]
- Rasmussen, S.A.; Jamieson, D.J.; Honein, M.A.; Petersen, L.R. Zika Virus and Birth Defects—Reviewing the Evidence for Causality. N. Engl. J. Med. 2016, 374, 1981–1987. [Google Scholar] [CrossRef]
- Lalle, E.; Colavita, F.; Iannetta, M.; Tekle, S.G.; Carletti, F.; Scorzolini, L.; Bordi, L.; Vincenti, D.; Castilletti, C.; Ippolito, G.; et al. Prolonged detection of dengue virus RNA in the semen of a man returning from Thailand to Italy, January 2018. Eurosurveillance 2018, 23, 18–00197. [Google Scholar] [CrossRef]
- Epelboin, S.; Dulioust, E.; Epelboin, L.; Benachi, A.; Merlet, F.; Patrat, C. Zika virus and reproduction: Facts, questions and current management. Hum. Reprod. Updat. 2017, 23, 629–645. [Google Scholar] [CrossRef] [PubMed]
- Musso, D.; Nhan, T.; Robin, E.; Roche, C.; Bierlaire, D.; Zisou, K.; Shan Yan, A.; Cao-Lormeau, V.M.; Broult, J. Potential for Zika virus transmission through blood transfusion demonstrated during an outbreak in French Polynesia, November 2013 to February 2014. Eurosurveillance 2014, 19, 20761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Available online: https://www.who.int/news-room/fact-sheets/detail/zika-virus (accessed on 24 March 2022).
- Lee, L.J.; Komarasamy, T.V.; Adnan, N.A.A.; James, W.; Balasubramaniam, V.R. Hide and Seek: The Interplay Between Zika Virus and the Host Immune Response. Front. Immunol. 2021, 12, 4379. [Google Scholar] [CrossRef] [PubMed]
- Pardy, R.D.; Rajah, M.M.; Condotta, S.A.; Taylor, N.G.; Sagan, S.M.; Richer, M.J. Analysis of the T Cell Response to Zika Virus and Identification of a Novel CD8+ T Cell Epitope in Immunocompetent Mice. PLoS Pathog. 2017, 13, e1006184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ngono, A.E.; Shresta, S. Immune Response to Dengue and Zika. Annu. Rev. Immunol. 2018, 36, 279–308. [Google Scholar] [CrossRef] [Green Version]
- Maucourant, C.; Petitdemange, C.; Yssel, H.; Vieillard, V. Control of Acute Arboviral Infection by Natural Killer Cells. Viruses 2019, 11, 131. [Google Scholar] [CrossRef] [Green Version]
- Cimini, E.; Castilletti, C.; Sacchi, A.; Casetti, R.; Bordoni, V.; Romanelli, A.; Turchi, F.; Martini, F.; Tumino, N.; Nicastri, E.; et al. Human Zika infection induces a reduction of IFN-γ producing CD4 T-cells and a parallel expansion of effector Vδ2 T-cells. Sci. Rep. 2017, 7, 6313. [Google Scholar] [CrossRef]
- Cimini, E.; Sacchi, A.; De Minicis, S.; Bordoni, V.; Casetti, R.; Grassi, G.; Colavita, F.; Castilletti, C.; Capobianchi, M.R.; Ippolito, G.; et al. Vδ2 T-Cells Kill ZIKV-Infected Cells by NKG2D-Mediated Cytotoxicity. Microorganisms 2019, 7, 350. [Google Scholar] [CrossRef] [Green Version]
- Available online: https://www.who.int/news-room/fact-sheets/detail/west-nile-virus (accessed on 7 March 2022).
- Saiz, J.-C.; Martín-Acebes, M.A.; Blázquez, A.B.; Escribano-Romero, E.; Poderoso, T.; de Oya, N.J. Pathogenicity and virulence of West Nile virus revisited eight decades after its first isolation. Virulence 2021, 12, 1145–1173. [Google Scholar] [CrossRef]
- Fang, H.; Welte, T.; Zheng, X.; Chang, G.-J.J.; Holbrook, M.R.; Soong, L.; Wang, T. γδd T cells promote the maturation of dendritic cells during West Nile virus infection. FEMS Immunol. Med. Microbiol. 2010, 59, 71–80. [Google Scholar] [CrossRef]
- Agrati, C.; Castilletti, C.; Cimini, E.; Romanelli, A.; Lapa, D.; Quartu, S.; Martini, F.; Capobianchi, M.R. Antiviral activity of human Vd2 T-cells against WNV includes both cytolytic and non-cytolytic mechanisms. FEMS Immunol. Med. Microbiol. 2016, 39, 139–142. [Google Scholar] [PubMed]
- Feldmann, H.; Jones, S.; Klenk, H.-D.; Schnittler, H.-J. Ebola virus: From discovery to vaccine. Nat. Rev. Immunol. 2003, 3, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, J.H.; Adachi, T.; Adhikari, N.K.J.; Arribas, J.R.; Bah, I.E.; Bausch, D.G.; Bhadelia, N.; Borchert, M.; Brantsaeter, A.B.; Brett-Major, D.M.; et al. New filovirus disease classification and nomenclature. Nat. Rev. Genet. 2019, 17, 261–263. [Google Scholar] [CrossRef]
- Available online: https://www.who.int/health-topics/ebola#tab=tab_1 (accessed on 7 March 2022).
- Available online: https://www.who.int/teams/blueprint/ebola-marburg (accessed on 7 March 2022).
- World Health Organization (WHO). Ebola Virus Disease. Available online: https://www.who.int/news-room/fact-sheets/detail/ebola-virus-disease (accessed on 28 December 2020).
- Ohimain, E.I.; Silas-Olu, D. The 2013–2016 Ebola virus disease outbreak in West Africa. Curr. Opin. Pharmacol. 2021, 60, 360–365. [Google Scholar] [CrossRef] [PubMed]
- Agrati, C.; Castilletti, C.; Casetti, R.; Sacchi, A.; Falasca, L.; Turchi, F.; Tumino, N.; Bordoni, V.; Cimini, E.; Viola, D.; et al. Longitudinal characterization of dysfunctional T cell-activation during human acute Ebola infection. Cell Death Dis. 2016, 7, e2164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruibal, P.; Oestereich, L.; Lüdtke, A.; Becker-Ziaja, B.; Wozniak, D.M.; Kerber, R.; Korva, M.; Cabeza-Cabrerizo, M.; Bore, J.A.; Koundouno, F.R.; et al. Unique human immune signature of Ebola virus disease in Guinea. Nature 2016, 533, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Cimini, E.; Viola, D.; Cabeza-Cabrerizo, M.; Romanelli, A.; Tumino, N.; Sacchi, A.; Bordoni, V.; Casetti, R.; Turchi, F.; Martini, F.; et al. Different features of Vδ2 T and NK cells in fatal and non-fatal human Ebola infections. PLoS Negl. Trop. Dis. 2017, 11, e0005645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cimini, E.; Agrati, C. γδ T Cells in Emerging Viral Infection: An Overview. Viruses 2022, 14, 1166. https://doi.org/10.3390/v14061166
Cimini E, Agrati C. γδ T Cells in Emerging Viral Infection: An Overview. Viruses. 2022; 14(6):1166. https://doi.org/10.3390/v14061166
Chicago/Turabian StyleCimini, Eleonora, and Chiara Agrati. 2022. "γδ T Cells in Emerging Viral Infection: An Overview" Viruses 14, no. 6: 1166. https://doi.org/10.3390/v14061166
APA StyleCimini, E., & Agrati, C. (2022). γδ T Cells in Emerging Viral Infection: An Overview. Viruses, 14(6), 1166. https://doi.org/10.3390/v14061166





