Differential Pathogenesis of SARS-CoV-2 Variants of Concern in Human ACE2-Expressing Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. In Vivo Mouse Challenge Experiments
2.2. Infectious Virus Titration by Plaque Assay
2.3. RNA Extraction and Quantitative RT-PCR
2.4. Statistical Analysis
3. Results
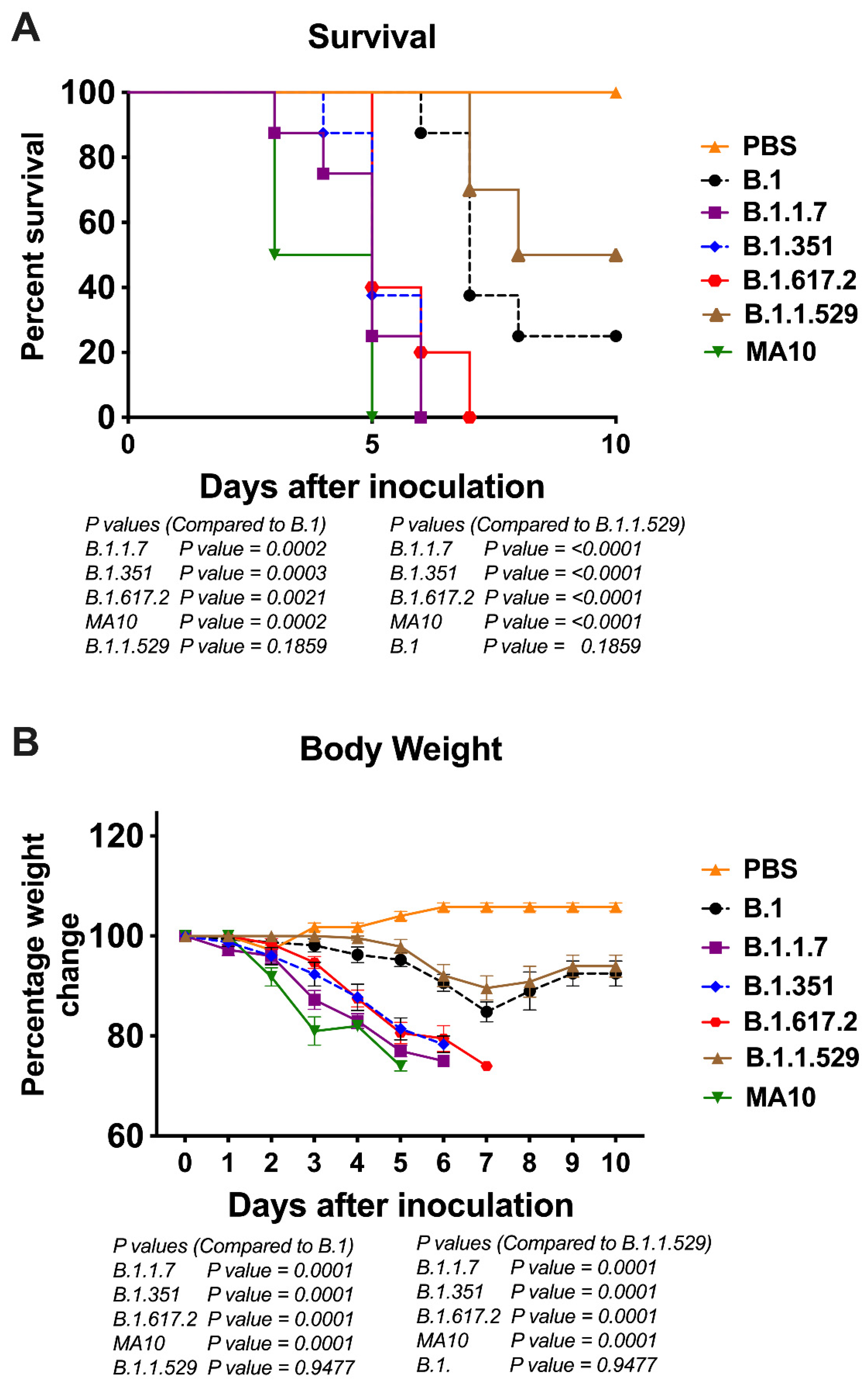
3.1. Clinical Disease Progression of K18-hACE2 Mice Infected with SARS-CoV-2 VoC
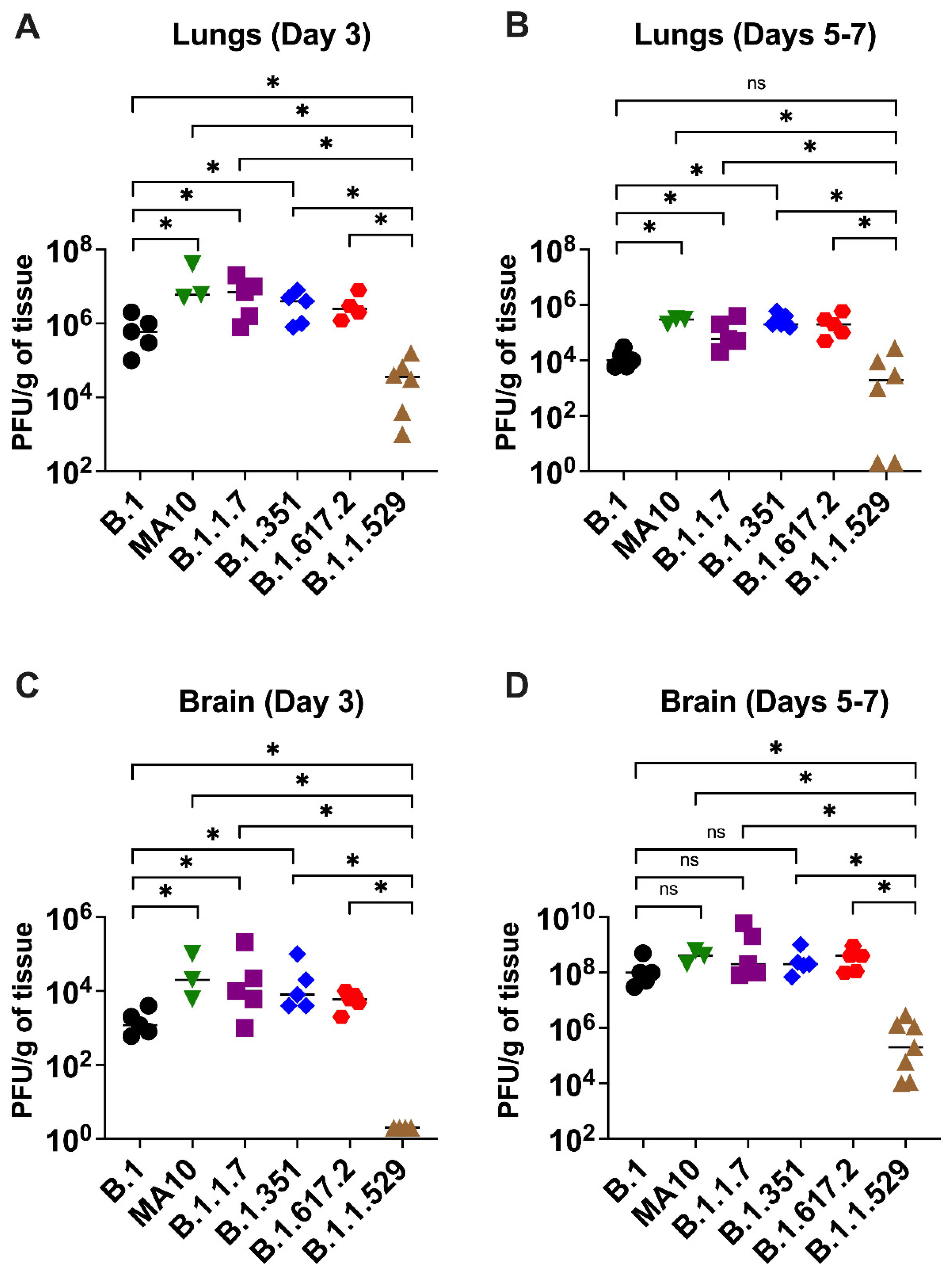
3.2. Viral Load in K18-hACE2 Mice Infected with SARS-CoV-2 VoC
3.3. Inflammation in the Lungs following Infection with SARS-CoV-2 VoC
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rothan, H.A.; Byrareddy, S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020, 109, 102433. [Google Scholar] [CrossRef]
- Rothan, H.A.; Acharya, A.; Reid, S.P.; Kumar, M.; Byrareddy, S.N. Molecular Aspects of COVID-19 Differential Pathogenesis. Pathogens 2020, 9, 538. [Google Scholar] [CrossRef]
- Khan, A.; Zia, T.; Suleman, M.; Khan, T.; Ali, S.S.; Abbasi, A.A.; Mohammad, A.; Wei, D.Q. Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: An insight from structural data. J. Cell Physiol. 2021, 236, 7045–7057. [Google Scholar] [CrossRef]
- Walensky, R.P.; Walke, H.T.; Fauci, A.S. SARS-CoV-2 Variants of Concern in the United States-Challenges and Opportunities. JAMA 2021, 325, 1037–1038. [Google Scholar] [CrossRef]
- Kumar, M.; Iyer, S.S. ASSURED-SQVM diagnostics for COVID-19: Addressing the why, when, where, who, what and how of testing. Expert Rev. Mol. Diagn. 2021, 21, 349–362. [Google Scholar] [CrossRef]
- Stolp, B.; Stern, M.; Ambiel, I.; Hofmann, K.; Morath, K.; Gallucci, L.; Cortese, M.; Bartenschlager, R.; Ruggieri, A.; Graw, F.; et al. SARS-CoV-2 variants of concern display enhanced intrinsic pathogenic properties and expanded organ tropism in mouse models. Cell Rep. 2022, 38, 110387. [Google Scholar] [CrossRef]
- Shuai, H.; Chan, J.F.; Yuen, T.T.; Yoon, C.; Hu, J.C.; Wen, L.; Hu, B.; Yang, D.; Wang, Y.; Hou, Y.; et al. Emerging SARS-CoV-2 variants expand species tropism to murines. EBioMedicine 2021, 73, 103643. [Google Scholar] [CrossRef]
- Stone, S.; Rothan, H.A.; Natekar, J.P.; Kumari, P.; Sharma, S.; Pathak, H.; Arora, K.; Auroni, T.T.; Kumar, M. SARS-CoV-2 Variants of Concern Infect the Respiratory Tract and Induce Inflammatory Response in Wild-Type Laboratory Mice. Viruses 2021, 14, 27. [Google Scholar] [CrossRef]
- Ramanathan, M.; Ferguson, I.D.; Miao, W.; Khavari, P.A. SARS-CoV-2 B.1.1.7 and B.1.351 spike variants bind human ACE2 with increased affinity. Lancet Infect. Dis. 2021, 21, 1070. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Plante, K.S.; Plante, J.A.; Xie, X.; Zhang, X.; Ku, Z.; An, Z.; Scharton, D.; Schindewolf, C.; et al. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature 2022, 602, 294–299. [Google Scholar] [CrossRef]
- Niu, Z.; Zhang, Z.; Gao, X.; Du, P.; Lu, J.; Yan, B.; Wang, C.; Zheng, Y.; Huang, H.; Sun, Q. N501Y mutation imparts cross-species transmission of SARS-CoV-2 to mice by enhancing receptor binding. Signal Transduct. Target. Ther. 2021, 6, 284. [Google Scholar] [CrossRef]
- Frampton, D.; Rampling, T.; Cross, A.; Bailey, H.; Heaney, J.; Byott, M.; Scott, R.; Sconza, R.; Price, J.; Margaritis, M.; et al. Genomic characteristics and clinical effect of the emergent SARS-CoV-2 B.1.1.7 lineage in London, UK: A whole-genome sequencing and hospital-based cohort study. Lancet Infect. Dis. 2021, 21, 1246–1256. [Google Scholar] [CrossRef]
- Faulkner, N.; Ng, K.W.; Wu, M.Y.; Harvey, R.; Margaritis, M.; Paraskevopoulou, S.; Houlihan, C.; Hussain, S.; Greco, M.; Bolland, W.; et al. Reduced antibody cross-reactivity following infection with B.1.1.7 than with parental SARS-CoV-2 strains. ELife 2021, 10, e69317. [Google Scholar] [CrossRef]
- Bayarri-Olmos, R.; Johnsen, L.B.; Idorn, M.; Reinert, L.S.; Rosbjerg, A.; Vang, S.; Hansen, C.B.; Helgstrand, C.; Bjelke, J.R.; Bak-Thomsen, T.; et al. The alpha/B.1.1.7 SARS-CoV-2 variant exhibits significantly higher affinity for ACE-2 and requires lower inoculation doses to cause disease in K18-hACE2 mice. ELife 2021, 10, e70002. [Google Scholar] [CrossRef]
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 2021, 592, 438–443. [Google Scholar] [CrossRef]
- Halfmann, P.J.; Iida, S.; Iwatsuki-Horimoto, K.; Maemura, T.; Kiso, M.; Scheaffer, S.M.; Darling, T.L.; Joshi, A.; Loeber, S.; Singh, G.; et al. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature 2022, 603, 687–692. [Google Scholar] [CrossRef]
- VanBlargan, L.A.; Errico, J.M.; Halfmann, P.J.; Zost, S.J.; Crowe, J.E., Jr.; Purcell, L.A.; Kawaoka, Y.; Corti, D.; Fremont, D.H.; Diamond, M.S. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat. Med. 2022, 28, 490–495. [Google Scholar] [CrossRef]
- Shuai, H.; Chan, J.F.; Hu, B.; Chai, Y.; Yuen, T.T.; Yin, F.; Huang, X.; Yoon, C.; Hu, J.C.; Liu, H.; et al. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature 2022, 603, 693–699. [Google Scholar] [CrossRef]
- Zheng, J.; Wong, L.R.; Li, K.; Verma, A.K.; Ortiz, M.; Wohlford-Lenane, C.; Leidinger, M.R.; Knudson, C.M.; Meyerholz, D.K.; McCray, P.B., Jr.; et al. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature 2020, 589, 603–607. [Google Scholar] [CrossRef]
- Munoz-Fontela, C.; Dowling, W.E.; Funnell, S.G.P.; Gsell, P.S.; Riveros-Balta, A.X.; Albrecht, R.A.; Andersen, H.; Baric, R.S.; Carroll, M.W.; Cavaleri, M.; et al. Animal models for COVID-19. Nature 2020, 586, 509–515. [Google Scholar] [CrossRef]
- Kumari, P.; Rothan, H.A.; Natekar, J.P.; Stone, S.; Pathak, H.; Strate, P.G.; Arora, K.; Brinton, M.A.; Kumar, M. Neuroinvasion and Encephalitis Following Intranasal Inoculation of SARS-CoV-2 in K18-hACE2 Mice. Viruses 2021, 13, 132. [Google Scholar] [CrossRef] [PubMed]
- Rothan, H.A.; Arora, K.; Natekar, J.P.; Strate, P.G.; Brinton, M.A.; Kumar, M. Z-DNA-Binding Protein 1 Is Critical for Controlling Virus Replication and Survival in West Nile Virus Encephalitis. Front. Microbiol. 2019, 10, 2089. [Google Scholar] [CrossRef] [PubMed]
- Natekar, J.P.; Rothan, H.A.; Arora, K.; Strate, P.G.; Kumar, M. Cellular microRNA-155 Regulates Virus-Induced Inflammatory Response and Protects against Lethal West Nile Virus Infection. Viruses 2019, 12, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rothan, H.A.; Stone, S.; Natekar, J.; Kumari, P.; Arora, K.; Kumar, M. The FDA-approved gold drug auranofin inhibits novel coronavirus (SARS-CoV-2) replication and attenuates inflammation in human cells. Virology 2020, 547, 7–11. [Google Scholar] [CrossRef]
- Leist, S.R.; Dinnon, K.H.; Schafer, A.; Tse, L.V.; Okuda, K.; Hou, Y.J.; West, A.; Edwards, C.E.; Sanders, W.; Fritch, E.J.; et al. A Mouse-Adapted SARS-CoV-2 Induces Acute Lung Injury and Mortality in Standard Laboratory Mice. Cell 2020, 183, 1070–1085.e12. [Google Scholar] [CrossRef]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.C.; Uhl, S.; Hoagland, D.; Moller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045.e9. [Google Scholar] [CrossRef]
- Mangalmurti, N.; Hunter, C.A. Cytokine Storms: Understanding COVID-19. Immunity 2020, 53, 19–25. [Google Scholar] [CrossRef]
- Gu, H.; Chen, Q.; Yang, G.; He, L.; Fan, H.; Deng, Y.Q.; Wang, Y.; Teng, Y.; Zhao, Z.; Cui, Y.; et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science 2020, 369, 1603–1607. [Google Scholar] [CrossRef]
- Nyberg, T.; Ferguson, N.M.; Nash, S.G.; Webster, H.H.; Flaxman, S.; Andrews, N.; Hinsley, W.; Bernal, J.L.; Kall, M.; Bhatt, S.; et al. Comparative analysis of the risks of hospitalisation and death associated with SARS-CoV-2 omicron (B.1.1.529) and delta (B.1.617.2) variants in England: A cohort study. Lancet 2022, 399, 1303–1312. [Google Scholar] [CrossRef]
- Mlcochova, P.; Kemp, S.A.; Dhar, M.S.; Papa, G.; Meng, B.; Ferreira, I.; Datir, R.; Collier, D.A.; Albecka, A.; Singh, S.; et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 2021, 599, 114–119. [Google Scholar] [CrossRef]
- Radvak, P.; Kwon, H.J.; Kosikova, M.; Ortega-Rodriguez, U.; Xiang, R.; Phue, J.N.; Shen, R.F.; Rozzelle, J.; Kapoor, N.; Rabara, T.; et al. SARS-CoV-2 B.1.1.7 (alpha) and B.1.351 (beta) variants induce pathogenic patterns in K18-hACE2 transgenic mice distinct from early strains. Nat. Commun. 2021, 12, 6559. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Mostafavi, H.; Ng, W.H.; Freitas, J.R.; King, N.J.C.; Zaid, A.; Taylor, A.; Mahalingam, S. The Delta SARS-CoV-2 Variant of Concern Induces Distinct Pathogenic Patterns of Respiratory Disease in K18-hACE2 Transgenic Mice Compared to the Ancestral Strain from Wuhan. mBio 2022, e0068322. [Google Scholar] [CrossRef] [PubMed]
- Meng, B.; Abdullahi, A.; Ferreira, I.; Goonawardane, N.; Saito, A.; Kimura, I.; Yamasoba, D.; Gerber, P.P.; Fatihi, S.; Rathore, S.; et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature 2022, 603, 706–714. [Google Scholar] [CrossRef] [PubMed]
- Hui, K.P.Y.; Ho, J.C.W.; Cheung, M.C.; Ng, K.C.; Ching, R.H.H.; Lai, K.L.; Kam, T.T.; Gu, H.; Sit, K.Y.; Hsin, M.K.Y.; et al. SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature 2022, 603, 715–720. [Google Scholar] [CrossRef] [PubMed]

| Gene (Accession No.) | Primer Sequence (5′–3′) |
|---|---|
| GAPDH (NM_008084) Forward Reverse IL-6 (NM_000600) | CAGTATGACTCCACTCAC GTAGACTCCACGACATAC |
| Forward | CCAGGAGCCCAGCTATGAAC |
| Reverse | CCCAGGGAGAAGGCAACTG |
| CCL2 (NM_011333) | |
| Forward | TCACCTGCTGCTACTCATTCACCA |
| Reverse | TACAGCTTCTTTGGGACACCTGCT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Natekar, J.P.; Pathak, H.; Stone, S.; Kumari, P.; Sharma, S.; Auroni, T.T.; Arora, K.; Rothan, H.A.; Kumar, M. Differential Pathogenesis of SARS-CoV-2 Variants of Concern in Human ACE2-Expressing Mice. Viruses 2022, 14, 1139. https://doi.org/10.3390/v14061139
Natekar JP, Pathak H, Stone S, Kumari P, Sharma S, Auroni TT, Arora K, Rothan HA, Kumar M. Differential Pathogenesis of SARS-CoV-2 Variants of Concern in Human ACE2-Expressing Mice. Viruses. 2022; 14(6):1139. https://doi.org/10.3390/v14061139
Chicago/Turabian StyleNatekar, Janhavi Prasad, Heather Pathak, Shannon Stone, Pratima Kumari, Shaligram Sharma, Tabassum Tasnim Auroni, Komal Arora, Hussin Alwan Rothan, and Mukesh Kumar. 2022. "Differential Pathogenesis of SARS-CoV-2 Variants of Concern in Human ACE2-Expressing Mice" Viruses 14, no. 6: 1139. https://doi.org/10.3390/v14061139
APA StyleNatekar, J. P., Pathak, H., Stone, S., Kumari, P., Sharma, S., Auroni, T. T., Arora, K., Rothan, H. A., & Kumar, M. (2022). Differential Pathogenesis of SARS-CoV-2 Variants of Concern in Human ACE2-Expressing Mice. Viruses, 14(6), 1139. https://doi.org/10.3390/v14061139








