Molecular Characterization and Identification of Potential Inhibitors for ‘E’ Protein of Dengue Virus
Abstract
1. Introduction
2. Materials and Methods
2.1. Dengue Serotyping by rRT-PCR
2.2. Dengue Envelope Gene Characterization
2.3. Homology Modelling
2.4. Protein Processing
2.4.1. Ligand Processing and Docking of Published Inhibitors
2.4.2. Virtual Screening and Docking of Analogs
2.4.3. Molecular Dynamics Simulations
3. Results
3.1. Dengue NS1, IgM, and IgG Enzyme Linked Immunosorbent Assay (ELISA)
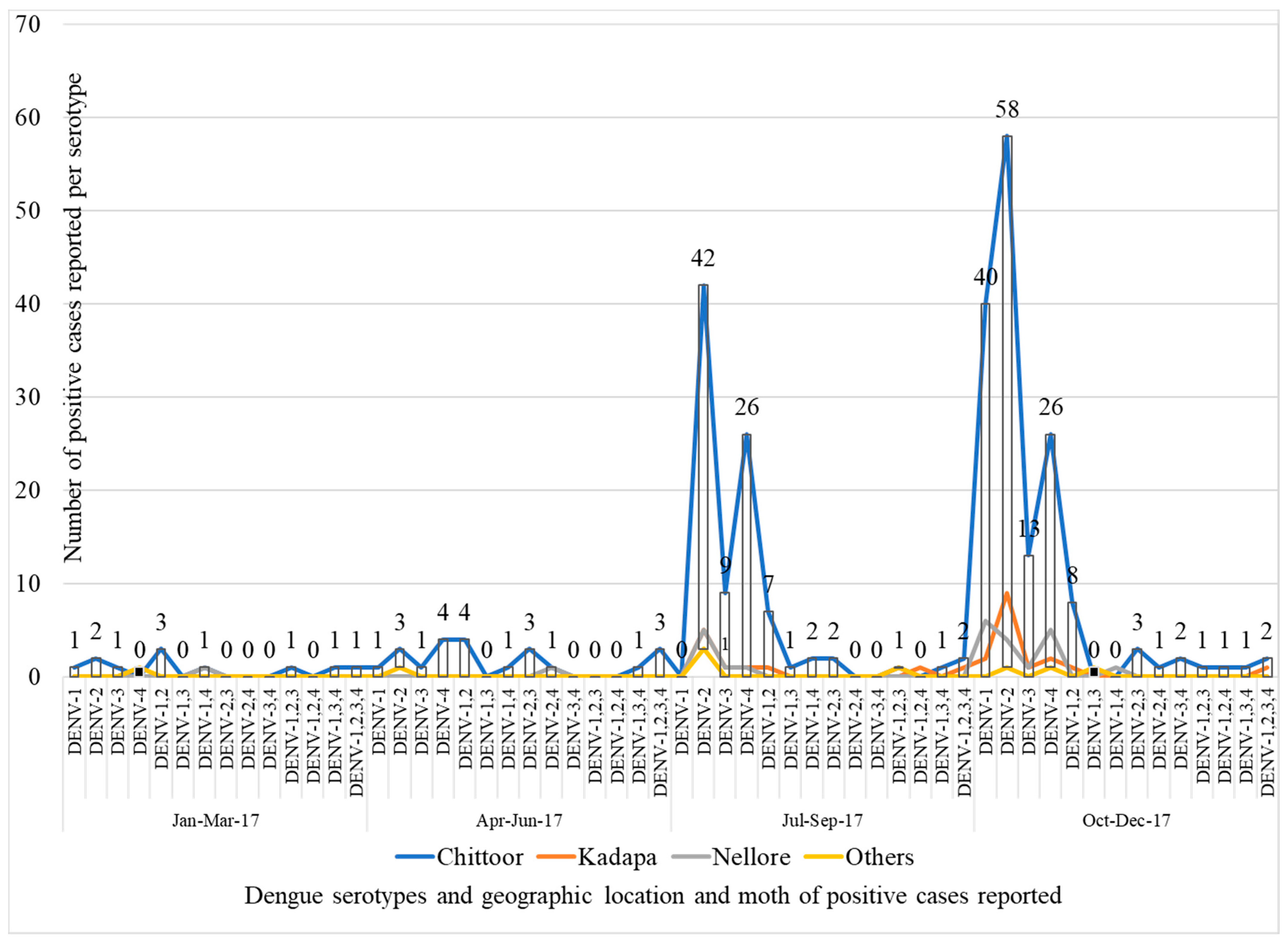
3.2. Dengue Serotyping by Real-Time Reverse Transcriptase Polymerase Chain Reaction (rRT-PCR)
3.3. Clinical Manifestations
3.3.1. Dengue Envelope Gene Characterization
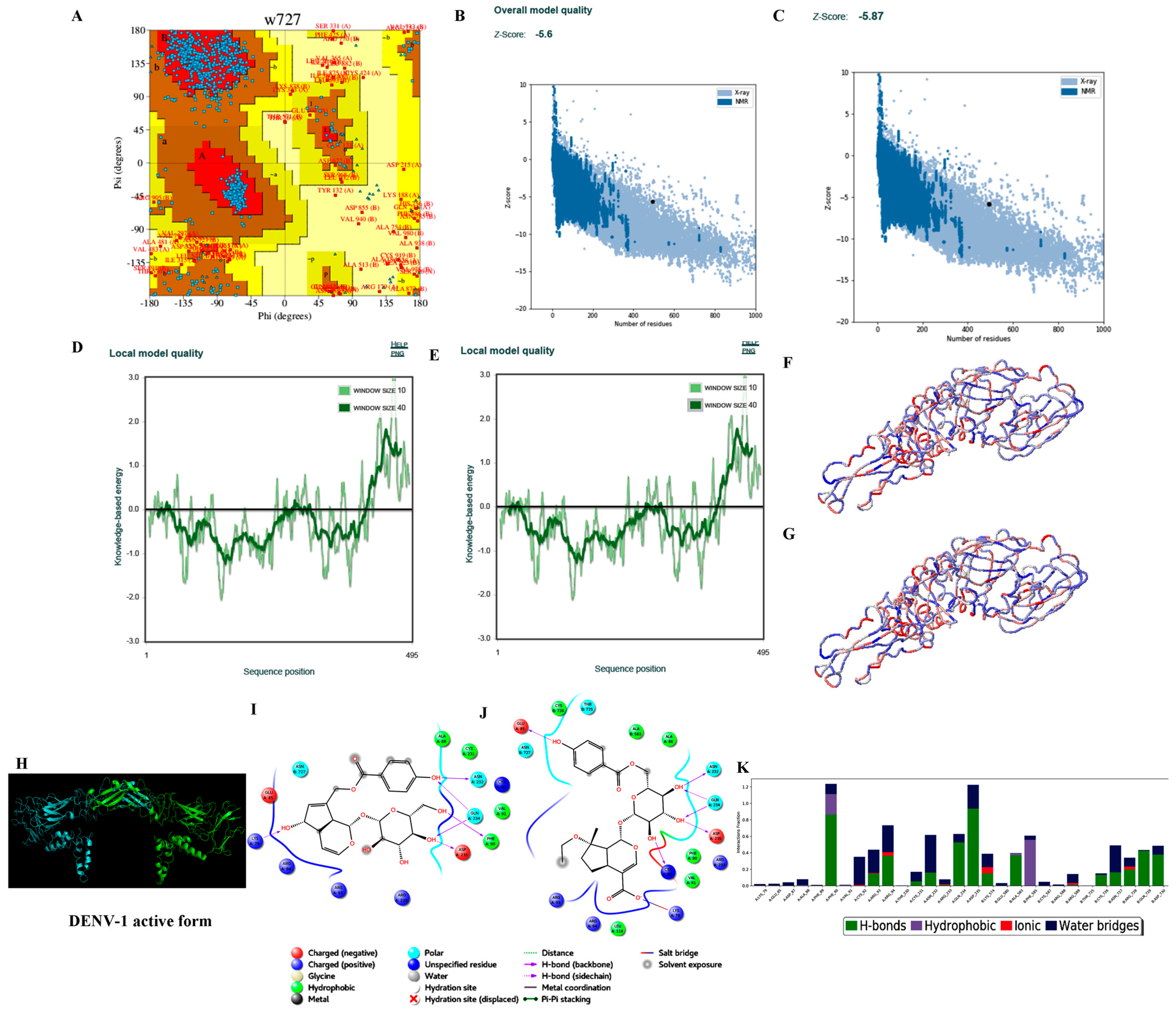
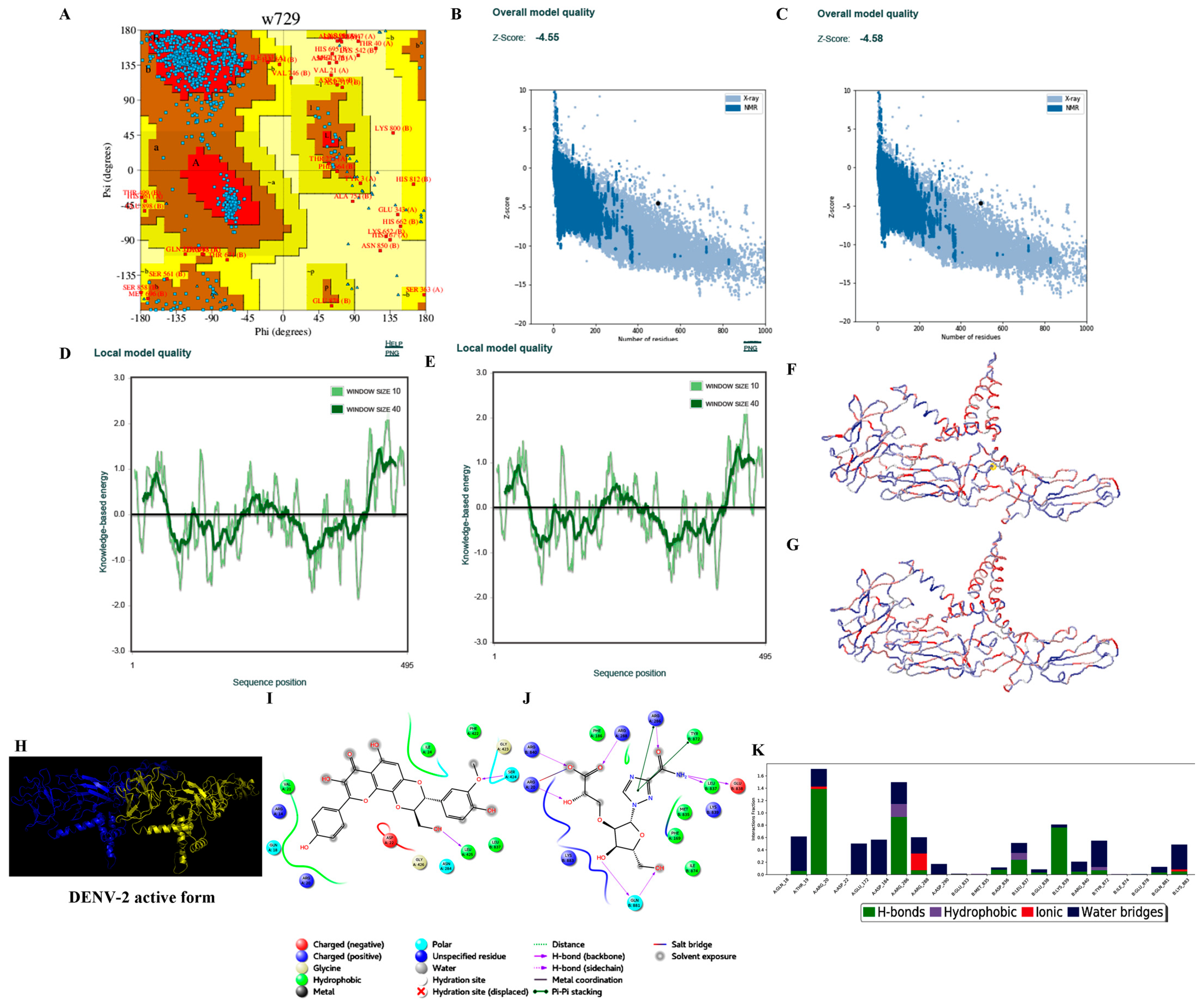
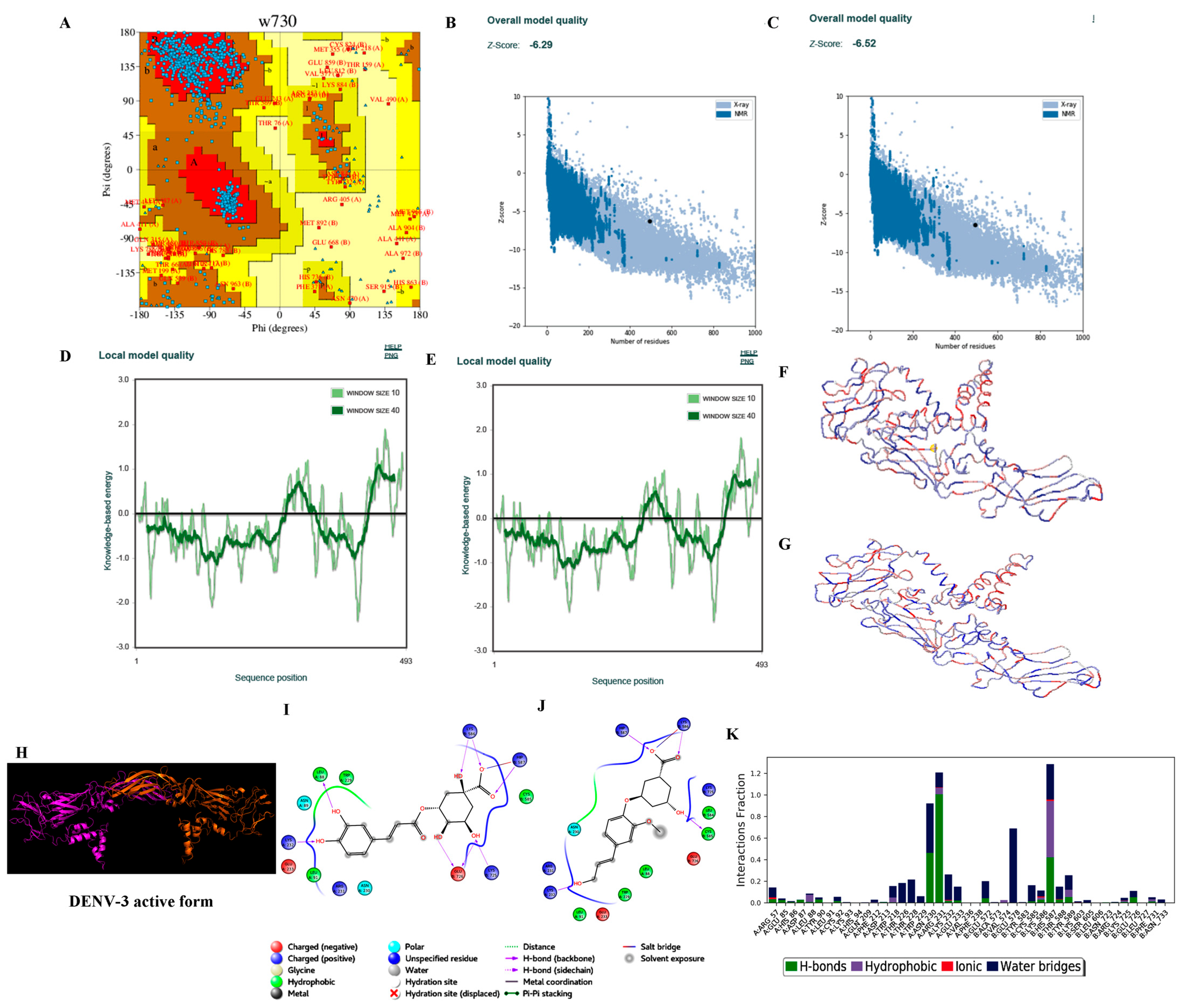
3.3.2. Homology Modelling
3.3.3. Protein Preparation
3.3.4. Ligand Processing and Docking of Published Inhibitors
3.3.5. Virtual Screening and Docking of Analogs
3.3.6. Molecular Dynamics Simulations
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Racherla, R.G.; Pamireddy, M.L.; Mohan, A.; Mudhigeti, N.; Mahalakshmi, P.A.; Nallapireddy, U.; Kalawat, U. Co-Circulation of Four Dengue Serotypes at South Eastern Andhra Pradesh, India: A Prospective Study. Indian J. Med. Microbiol. 2018, 36, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Gubler, D.J. Epidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st century. Trends Microbiol. 2002, 10, 100–103. [Google Scholar] [CrossRef]
- National Vector Borne Diseases Control Programme, New Delhi. Clinical Management of Dengue Fever (DF) and Dengue Haemorrhagic Fever (DHF). 2008. Available online: http://www.nvbdcp.gov.in/Doc/Clinical%20Guidelines.pdf (accessed on 15 November 2021).
- Klungthong, C.; Gibbons, R.V.; Thaisomboonsuk, B.; Nisalak, A.; Kalayanarooj, S.; Thirawuth, V.; Nutkumhang, N.; Mammen, M.P., Jr.; Jarman, R.G. Dengue Virus Detection Using Whole Blood for Reverse Transcriptase PCR and Virus Isolation. J. Clin. Microbiol. 2007, 45, 2480–2485. [Google Scholar] [CrossRef] [PubMed]
- Shukla, M.K.; Barde, P.V.; Singh, N. Utility of CDC DENV 1–4 real-time RT-PCR assay for the diagnosis of dengue. Dengue 2018, 40, 15. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Fonseca, V.; Libin, P.J.K.; Theys, K.; Faria, N.R.; Nunes, M.R.T.; Restovic, M.I.; Freire, M.; Giovanetti, M.; Cuypers, L.; Nowe, A.; et al. A computational method for the identification of Dengue, Zika and Chikungunya virus species and genotypes. PLOS Neglected Trop. Dis. 2019, 13, e0007231. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinform. 2016, 54, 1–37. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [PubMed]
- Wallner, B.; Elofsson, A. Can correct protein models be identified? Protein Sci. 2003, 12, 1073–1086. [Google Scholar] [CrossRef] [PubMed]
- Release, S. 1: Maestro; Schrödinger, LLC: New York, NY, USA, 2020; 1p. [Google Scholar]
- Jacobson, M.P.; Pincus, D.L.; Rapp, C.S.; Day, T.J.F.; Honig, B.; Shaw, D.E.; Friesner, R.A. A hierarchical approach to all-atom protein loop prediction. Proteins Struct. Funct. Bioinform. 2004, 55, 351–367. [Google Scholar] [CrossRef]
- Greenwood, J.R.; Calkins, D.; Sullivan, A.P.; Shelley, J.C. Towards the comprehensive, rapid, and accurate prediction of the favorable tautomeric states of drug-like molecules in aqueous solution. J. Comput. Aided Mol. Des. 2010, 24, 591–604. [Google Scholar] [CrossRef]
- Harder, E.; Damm, W.; Maple, J.; Wu, C.; Reboul, M.; Xiang, J.Y.; Wang, L.; Lupyan, D.; Dahlgren, M.K.; Knight, J.L.; et al. OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins. J. Chem. Theory Comput. 2016, 12, 281–296. [Google Scholar] [CrossRef]
- Natarajan, P.; Priyadarshini, V.; Pradhan, D.; Manne, M.; Swargam, S.; Kanipakam, H.; Bhuma, V.; Amineni, U. E-pharmacophore-based virtual screening to identify GSK-3β inhibitors. J. Recept. Signal Transduct. 2015, 36, 445–458. [Google Scholar] [CrossRef]
- Hema, K.; Priyadarshini, I.V.; Swargam, S.; Pradeep, N.; Chiranjeevi, P.; Umamaheswari, A. 202 Subunit vaccine design against pathogens causing atherosclerosis. J. Biomol. Struct. Dyn. 2015, 33, 135–136. [Google Scholar] [CrossRef]
- Swargam, S.; Pradhan, D.; Pradeep, N.; Hema, K.; Krishna, V.S.; Umamaheswari, A. 201 Structure guided novel lead molecules against ERK proteins: Application of multiple docking and molecular dynamics studies. J. Biomol. Struct. Dyn. 2015, 33, 134–135. [Google Scholar] [CrossRef][Green Version]
- Pradhan, D.; Priyadarshini, V.; Aggrawal, S.; Pradeep, N.; Nayek, A.; Jain, A.K.; Umamaheswari, A. 181 Discovery of potential inhibitors of BMX non-receptor tyrosine kinase through e-pharmacophore based virtual screening. J. Biomol. Struct. Dyn. 2015, 33, 118–120. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein−Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [PubMed]
- Satuluri, S.H.; Katari, S.K.; Pasala, C.; Amineni, U. Novel and potent inhibitors for dihydropteroate synthase of Helicobacter pylori. J. Recept. Signal Transduct. 2020, 40, 246–256. [Google Scholar] [CrossRef]
- Bowers, K.J.; Chow, D.E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossvary, I.; Moraes, M.A.; Sacerdoti, F.D.; et al. Scalable algorithms for molecular dynamics simu-lations on commodity clusters. In Proceedings of the SC ’06: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, Tampa, FL, USA, 11–17 November 2006; IEEE: Piscataway, NJ, USA, 2006; p. 43. [Google Scholar]
- Dengue Virus Typing Tool, n.d. Available online: https://www.genomedetective.com/app/typingtool/dengue/ (accessed on 10 December 2021).
- Carey, D.E.; Myers, R.M.; Reuben, R.; Rodrigues, F.M. Studies on dengue in Vellore, south India. Am. J. Trop. Med. Hyg. 1966, 15, 580–587. [Google Scholar] [CrossRef]
- Rao, M.R.K.; Padhy, R.N.; Das, M.K. Episodes of the epidemiological factors correlated with prevailing viral infections with dengue virus and molecular characterization of serotype-specific dengue virus circulation in eastern India. Infect. Genet. Evol. 2018, 58, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Vaddadi, K.; Gandikota, C.; Jain, P.K.; Prasad, V.S.V.; Venkataramana, M. Co-circulation and co-infections of all dengue virus sero-types in Hyderabad, India 2014. Epidemiol. Infect 2017, 145, 2563–2574. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.D.; Dandawate, C.N.; Banerjee, K.; Krishnamurthy, K. Virological and serological studies on an outbreak of dengue-like illness in Visakhapatnam, Andhra Pradesh. Indian J. Med. Res. 1965, 53, 777–789. [Google Scholar]
- Dash, P.K.; Sharma, S.; Srivastava, A.; Santhosh, S.R.; Parida, M.M.; Neeraja, M.; Subbalaxmi, M.V.S.; Lakshmi, V.; Rao, P.V.L. Emergence of dengue virus type 4 (genotype I) in India. Epidemiol. Infect. 2010, 139, 857–861. [Google Scholar] [CrossRef]
- Shastri, J.; Williamson, M.; Vaidya, N.; Agrawal, S.; Shrivastav, O. Nine year trends of dengue virus infection in Mumbai, Western India. J. Lab. Physicians 2017, 9, 296–302. [Google Scholar] [CrossRef]
- Bharaj, P.; Chahar, H.S.; Pandey, A.; Diddi, K.; Dar, L.; Guleria, R.; Kabra, S.K.; Broor, S. Concurrent infections by all four dengue virus serotypes during an outbreak of dengue in 2006 in Delhi, India. Virol. J. 2008, 5, 1. [Google Scholar] [CrossRef]
- Mishra, G.; Jain, A.; Prakash, O.; Prakash, S.; Kumar, R.; Garg, R.K.; Pandey, N.; Singh, M. Molecular characterization of dengue viruses circulating during 2009-2012 in Uttar Pradesh, India. J. Med. Virol. 2014, 87, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.; Taraphdar, D.; Chatterjee, S. Molecular Typing of Dengue Virus Circulating in Kolkata, India in 2010. J. Trop. Med. 2012, 2012, 960329. [Google Scholar] [CrossRef] [PubMed]
- Afreen, N.; Deeba, F.; Naqvi, I.; Shareef, M.; Ahmed, A.; Broor, S.; Parveen, S. Molecular Investigation of 2013 Dengue Fever Outbreak from Delhi, India. PLoS Curr. 2014. [Google Scholar] [CrossRef] [PubMed]
- Reddy, M.N.; Dungdung, R.; Valliyott, L.; Pilankatta, R. Occurrence of concurrent infections with multiple serotypes of dengue viruses during 2013–2015 in northern Kerala, India. PeerJ 2017, 5, e2970. [Google Scholar] [CrossRef]
- Garg, S.; Chakravarti, A.; Singh, R.; Masthi, N.R.; Goyal, R.C.; Jammy, G.R.; Ganguly, E.; Sharma, N.; Singh, M.M.; Ferreira, G.; et al. Dengue serotype-specific seroprevalence among 5- to 10-year-old children in India: A community-based cross-sectional study. Int. J. Infect. Dis. 2017, 54, 25–30. [Google Scholar] [CrossRef]
- Cecilia, D.; Kakade, M.B.; Bhagat, A.B.; Vallentyne, J.; Singh, A.; Patil, J.A.; Todkar, S.M.; Varghese, S.B.; Shah, P.S. Detection of dengue-4 virus in Pune, Western India after an absence of 30 years-its association with two severe cases. Virol. J. 2011, 8, 1–4. [Google Scholar] [CrossRef]
- Chakravarti, A.; Arora, R.; Luxemburger, C. Fifty years of dengue in India. Trans. R. Soc. Trop. Med. Hyg. 2012, 106, 273–282. [Google Scholar] [CrossRef]
- VinodKumar, C.; Kalapannavar, N.; Basavarajappa, K.; Sanjay, D.; Gowli, C.; Nadig, N.G.; Prasad, B. Episode of coexisting infections with multiple dengue virus serotypes in central Karnataka, India. J. Infect. Public Health 2013, 6, 302–306. [Google Scholar] [CrossRef]
- Manakkadan, A.; Joseph, I.; Prasanna, R.R.; Kunju, R.I.; Kailas, L.; Sreekumar, E. Lineage shift in Indian strains of Dengue virus serotype-3 (Genotype III), evidenced by detection of lineage IV strains in clinical cases from Kerala. Virol. J. 2013, 10, 37. [Google Scholar] [CrossRef]
- Neeraja, M.; Lakshmi, V.; Dash, P.K.; Parida, M.M.; Rao, P.V.L. The clinical, serological and molecular diagnosis of emerging dengue infection at a tertiary care institute in southern, India. J. Clin. Diagn. Res. JCDR 2013, 7, 457. [Google Scholar] [CrossRef]
- Anoop, M.; Issac, A.; Mathew, T.; Philip, S.; Kareem, N.A.; Unnikrishnan, R.; Sreekumar, E. Genetic characterization of dengue virus serotypes causing concurrent infection in an outbreak in Ernakulam, Kerala, South India. Indian J. Exp. Biol. 2010, 48, 849–857. [Google Scholar] [PubMed]
- Arunkumar, G.; Rao, C.; Kaur, H.; Gupta, N.; Sabeena, S.P.; Ambica, R.; Jain, A.; Yadav, A.; Dwibedi, B.; Malhotra, B.; et al. Geographical distribution of primary & secondary dengue cases in India—2017: A cross-sectional multicentric study. Indian J. Med. Res. 2019, 149, 548–553. [Google Scholar] [CrossRef]
- Shrivastava, S.; Tiraki, D.; Diwan, A.; Lalwani, S.K.; Modak, M.; Mishra, A.C.; Arankalle, V.A. Co-circulation of all the four dengue virus serotypes and detection of a novel clade of DENV-4 (genotype I) virus in Pune, India during 2016 season. PLoS ONE 2018, 13, e0192672. [Google Scholar] [CrossRef] [PubMed]
- Mutheneni, S.R.; Morse, A.P.; Caminade, C.; Upadhyayula, S.M. Dengue burden in India: Recent trends and importance of climatic parameters. Emerg. Microbes. Infect. 2017, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Cecilia, D. Current status of dengue and chikungunya in India. WHO South-East Asia J. Public Health 2014, 3, 22–26. [Google Scholar] [CrossRef]
- Imai, N.; Ferguson, N.M. Targeting vaccinations for the licensed dengue vaccine: Considerations for serosurvey design. PLoS ONE 2018, 13, e0199450. [Google Scholar] [CrossRef]
- Powers, C.N.; Setzer, W.N. An in silico investigation of phytochemicals as antiviral agents against dengue fever. Comb Chem High Throughput Screen 2016, 19, 516–536. [Google Scholar] [CrossRef]
- Anasir, M.I.; Ramanathan, B.; Poh, C.L. Structure-Based Design of Antivirals against Envelope Glycoprotein of Dengue Virus. Viruses 2020, 12, 367. [Google Scholar] [CrossRef]
- Radhakrishnan, N.; Lam, K.W.; Norhaizan, M.E. Molecular docking analysis of Carica papaya Linn constituents as antiviral agent. Int. Food Res. J. 2017, 24, 1819–1825. [Google Scholar]

| Subtype | Primer | Sequence (5′-3′) | Position | Target Size (bp) | Annealing Temp. |
|---|---|---|---|---|---|
| First Half (FH) primers | |||||
| DENV-1 | Forward | TCTAGCACATGCCATAGGAACA | 853–874 | 917 | 55.9 °C |
| Reverse | AAATTGTTGTCGTTCCAGACGTT | 1769–1749 | |||
| DENV-2 | Forward | GGCATACACCATAGGAACGACA | 858–879 | 923 | 55.9 °C |
| Reverse | GTCCTGTGAAGAGTAAGTTTCCTGA | 1780–1756 | |||
| DENV-3 | Forward | ACTAGCCCTATTTCTCGCCCA | 8411–861 | 939 | 60.1 °C |
| Reverse | TTTAAGTGCCCCGCGAAAATG | 1779–1759 | |||
| DENV-4 | Forward | CGCTCTTGGCAGGATTTATGG | 841–861 | 927 | 55.9 °C |
| Reverse | GATTTCCATCACCGGAGTCCA | 1767–1747 | |||
| Second Half (SH) primers | |||||
| DENV-1 | Forward | GGGGGCTTCAACATCCCAAG | 1600–1619 | 910 | 55.9 °C |
| Reverse | CTCTGTCCAGGTGTGGACTTC | 2509–2489 | |||
| DENV-2 | Forward | CCGGAGCGGACACACAAG | 1601–1618 | 907 | 60.1 °C |
| Reverse | GTCCATGTGTGCACGTTGTCT | 2507–2487 | |||
| DENV-3 | Forward | ACAGAAACACCAACCTGGAACA | 1604–1625 | 918 | 55.9 °C |
| Reverse | TGCTTGGAATTTGTATTGCTCTGT | 2521–2498 | |||
| DENV-4 | Forward | AGCAGGAGCAGACACATCAGA | 1601–1621 | 921 | 55.9 °C |
| Reverse | TTGTACTGTTCTGTCCAAGTGTGC | 2521–2498 | |||
| Serotype (n) | Case Type (n) | Age ≤ 18 Years | Age > 18 Years | Male | Female | Urban | Rural | p-Value |
|---|---|---|---|---|---|---|---|---|
| 1 (50) | primary (10) | 7 | 3 | 4 | 6 | 4 | 6 | 0.0002 * |
| secondary (40) | 19 | 21 | 22 | 18 | 14 | 26 | ||
| 2 (140) | primary (74) | 28 | 46 | 31 | 43 | 34 | 40 | 0.574 |
| secondary (66) | 36 | 30 | 38 | 28 | 23 | 43 | ||
| 3 (30) | primary (14) | 10 | 4 | 9 | 5 | 7 | 7 | 0.621 |
| secondary (16) | 8 | 8 | 11 | 5 | 2 | 14 | ||
| 4 (69) | primary (10) | 4 | 6 | 8 | 2 | 3 | 7 | 0.0001 * |
| secondary (59) | 36 | 23 | 31 | 28 | 23 | 36 | ||
| 1 and 2 (24) | primary (13) | 3 | 10 | 7 | 6 | 3 | 10 | 0.607 |
| secondary (11) | 10 | 1 | 4 | 7 | 1 | 10 | ||
| 1, 2, and 3 (4) | primary (4) | 4 | 0 | 0 | 4 | 2 | 2 | 0.006 * |
| secondary (0) | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 1, 2, 3, and 4 (10) | primary (10) | 10 | 0 | 5 | 5 | 2 | 8 | 0.002 * |
| secondary (0) | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 1, 2, and 4 (2) | primary (2) | 2 | 0 | 2 | 0 | 1 | 1 | 0.006 * |
| secondary (0) | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 1 and 3 (2) | primary (2) | 1 | 1 | 1 | 1 | 0 | 2 | 0.009 * |
| secondary (0) | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 1, 3, and 4 (4) | primary (4) | 3 | 1 | 3 | 1 | 3 | 1 | 0.0004 * |
| secondary (0) | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 1 and 4(6) | primary (6) | 4 | 2 | 1 | 5 | 0 | 6 | 0.003 * |
| secondary (0) | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 2 and 3(8) | primary (8) | 7 | 1 | 5 | 3 | 3 | 5 | 0.0003 * |
| secondary (0) | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 2 and 4(3) | primary (3) | 1 | 2 | 1 | 2 | 0 | 3 | 0.002 * |
| secondary (0) | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 3 and 4 (2) | primary (2) | 1 | 1 | 1 | 1 | 1 | 1 | 3.759 |
| secondary (0) | 0 | 0 | 0 | 0 | 0 | 0 |
| (a) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Serotypes | Case Type (n) | Feb% | Chi% | Mya% | Head% | Retro% | Arth% | Mal% | Rig% | Hem% | Rash% | p-Value |
| 1 (n = 50) | 1° dengue (10) | 100 | 80 | 90 | 80 | - | 10 | 20 | 10 | 30 | - | |
| 2° dengue (40) | 100 | 85 | 73 | 48 | 13 | 25 | 23 | 23 | 50 | 5 | 0.006 * | |
| ≤18 years (26) | 100 | 81 | 77 | 50 | 4 | 23 | 19 | 15 | 46 | 4 | ||
| >18 years (24) | 100 | 88 | 75 | 58 | 17 | 21 | 25 | 25 | 46 | 4 | 0.001 * | |
| all cases (50) | 100 | 84 | 76 | 54 | 10 | 22 | 22 | 20 | 46 | 4 | ||
| 2 (n = 140) | 1° dengue (74) | 100 | 80 | 88 | 58 | 12 | 30 | 20 | 16 | 43 | 7 | |
| 2° dengue (66) | 100 | 82 | 77 | 50 | 15 | 18 | 12 | 24 | 38 | 11 | 0.6 | |
| ≤18 years (64) | 100 | 84 | 81 | 42 | 8 | 14 | 14 | 17 | 36 | 14 | ||
| >18 years (76) | 100 | 78 | 84 | 64 | 18 | 33 | 18 | 22 | 45 | 4 | 0.39 | |
| all cases (140) | 100 | 81 | 83 | 54 | 14 | 24 | 16 | 20 | 41 | 9 | ||
| 3 (n = 30) | 1° dengue (14) | 100 | 93 | 71 | 86 | 7 | 43 | 29 | 21 | 21 | 7 | |
| 2° dengue (16) | 100 | 81 | 88 | 63 | 6 | 38 | - | 19 | 19 | 6 | 0.999 | |
| ≤18 years (18) | 100 | 78 | 72 | 67 | 6 | 28 | 17 | 17 | 17 | 6 | ||
| >18 years (12) | 100 | 100 | 92 | 83 | 8 | 58 | 8 | 25 | 25 | 8 | 0.6 | |
| all cases (30) | 100 | 87 | 80 | 73 | 7 | 40 | 13 | 20 | 20 | 7 | ||
| 4 (n = 69) | 1° dengue (10) | 100 | 80 | 90 | 80 | 20 | 40 | 20 | 20 | 50 | 10 | |
| 2° dengue (59) | 100 | 81 | 75 | 46 | 12 | 25 | 19 | 14 | 49 | 5 | 0.005 * | |
| ≤18 years (40) | 100 | 83 | 70 | 38 | 8 | 10 | 15 | 10 | 48 | 8 | ||
| >18 years (29) | 100 | 79 | 86 | 69 | 21 | 52 | 24 | 21 | 52 | 3 | 0.8 | |
| all cases (69) | 100 | 81 | 77 | 51 | 13 | 28 | 19 | 14 | 49 | 6 | ||
| 1/2 (n = 24) | 1° dengue (13) | 100 | 69 | 100 | 69 | - | 15 | 15 | 15 | 38 | 8 | |
| 2° dengue (11) | 100 | 100 | 55 | 18 | 18 | 18 | 18 | 18 | 73 | - | 0.6 | |
| ≤18 years (13) | 100 | 92 | 69 | 31 | 15 | 8 | 8 | 15 | 62 | - | ||
| >18 years (11) | 100 | 73 | 91 | 64 | - | 27 | 27 | 18 | 45 | 9 | 0.9 | |
| all cases (24) | 100 | 83 | 79 | 46 | 8 | 17 | 17 | 17 | 54 | 4 | ||
| 1/2/3 (n = 4) | 1° dengue (4) | 100 | 100 | 75 | 25 | - | - | - | - | - | 25 | |
| 2° dengue (nil) | - | - | - | - | - | - | - | - | - | - | 0.020 * | |
| ≤18 years (4) | 100 | 100 | 75 | 25 | - | - | - | - | - | 25 | ||
| >18 years (nil) | - | - | - | - | - | - | - | - | - | - | 0.020 * | |
| all cases (4) | 100 | 100 | 75 | 25 | - | - | - | - | - | 25 | ||
| 1/2/3/4 (n = 10) | 1° dengue (10) | 100 | 80 | 80 | 30 | - | - | - | - | 40 | - | |
| 2° dengue (nil) | - | - | - | - | - | - | - | - | - | - | 00017 * | |
| ≤18 years (10) | 100 | 80 | 80 | 30 | - | - | - | - | 40 | - | ||
| >18 years (nil) | - | - | - | - | - | - | - | - | - | - | 0.017 * | |
| all cases (10) | 100 | 80 | 80 | 30 | - | - | - | - | 40 | - | ||
| (b) | ||||||||||||
| Serotypes | Case Type (n) | Feb% | Chi% | Mya% | Head% | Retro% | Arth% | Mal% | Rig% | Hem% | Rash% | p-Value |
| 1/2/4 (n = 2) | 1° DENGUE (2) | 100 | 100 | 50 | - | - | - | - | - | 50 | - | |
| 2° DENGUE (nil) | - | - | - | - | - | - | - | - | - | - | 0.037 * | |
| ≤18 years (2) | 100 | 100 | 50 | - | - | - | - | - | 50 | - | ||
| >18 years (nil) | - | - | - | - | - | - | - | - | - | - | 0.037 * | |
| All cases (2) | 100 | 100 | 50 | - | - | - | - | - | 50 | - | ||
| 1/3 (n = 2) | 1° DENGUE (2) | 2 | 100 | 100 | - | - | - | 50 | - | 50 | - | |
| 2° DENGUE (nil) | - | - | - | - | - | - | - | - | - | - | 0.013 * | |
| ≤18 years (1) | 1 | 100 | 100 | - | - | - | - | - | - | - | ||
| >18 years (1) | 1 | 100 | 100 | - | - | - | 100 | - | 100 | - | 0.388 | |
| All cases (2) | 2 | 100 | 100 | - | - | - | 50 | - | 50 | - | ||
| 1/3/4 (n = 4) | 1° DENGUE (4) | 4 | 100 | 100 | 25 | - | - | - | - | - | - | |
| 2° DENGUE (nil) | - | - | - | - | - | - | - | - | - | - | 0.043 * | |
| ≤18 years (3) | 3 | 100 | 100 | - | - | - | - | - | - | - | ||
| >18 years (1) | 1 | 100 | 100 | 100 | - | - | - | - | - | - | 0.317 | |
| All cases (4) | 4 | 100 | 100 | 25 | - | - | - | - | - | - | ||
| ¼ (n = 6) | 1° DENGUE (6) | 6 | 67 | 83 | 50 | 17 | 33 | 33 | 33 | 83 | 17 | |
| 2° DENGUE (nil) | - | - | - | - | - | - | - | - | - | - | 0.001 * | |
| ≤18 years (4) | 4 | 75 | 75 | 25 | - | 25 | 25 | 25 | 75 | 25 | ||
| >18 years (2) | 2 | 50 | 100 | 100 | 50 | 50 | 50 | 50 | 100 | - | 0.299 | |
| All cases (6) | 6 | 67 | 83 | 50 | 17 | 33 | 33 | 33 | 83 | 17 | ||
| 2/3 (n = 8) | 1° DENGUE (8) | 8 | 88 | 50 | 38 | - | 13 | - | 25 | - | 13 | |
| 2° DENGUE (nil) | - | - | - | - | - | - | - | - | - | - | 0.011 * | |
| ≤18 years (7) | 7 | 86 | 43 | 29 | - | 14 | - | 29 | - | 14 | ||
| >18 years (1) | 1 | 100 | 100 | 100 | - | - | - | - | - | - | 0.038 * | |
| All cases (8) | 8 | 88 | 50 | 38 | - | 13 | - | 25 | - | 13 | ||
| 2/4 (n = 3) | 1° DENGUE (3) | 3 | 67 | 67 | 100 | - | 33 | 33 | - | 33 | - | |
| 2° DENGUE (nil) | - | - | - | - | - | - | - | - | - | - | 0.002 * | |
| ≤18 years (1) | 1 | - | - | 100 | - | - | 100 | - | - | - | ||
| >18 years (2) | 2 | 100 | 100 | 100 | - | 50 | - | - | 50 | - | 0.051 * | |
| All cases (3) | 3 | 67 | 67 | 100 | - | 33 | 33 | - | 33 | - | ||
| 3/4 (n = 2) | 1° DENGUE (2) | 2 | 50 | 100 | - | - | - | - | - | 100 | - | |
| 2° DENGUE (nil) | - | - | - | - | - | - | - | - | - | - | 0.031 * | |
| ≤18 years (1) | 1 | - | 100 | - | - | - | - | - | 100 | - | ||
| >18 years (1) | 1 | 100 | 100 | - | - | - | - | - | 100 | - | 0.66 | |
| All cases (2) | 2 | 50 | 100 | - | - | - | - | - | 100 | - | ||
| Subtype | Genotype | Total | (%) |
|---|---|---|---|
| 1 | Could not assign | 3 | (2.94) |
| DENV-1 Genotype I | 30 | (29.41) | |
| DENV-1 Genotype IV | 4 | (3.92) | |
| Related to but not part of DENV-1 Genotype I | 24 | (23.53) | |
| Related to but not part of DENV-1 Genotype I and IV | 15 | (14.71) | |
| Related to but not part of DENV-1 Genotype IV | 26 | (25.49) | |
| Total | 109 | ||
| 2 | Could not assign | 17 | (8.90) |
| DENV-2 Genotype V—Asian I | 25 | (13.09) | |
| Related to but not part of DENV-2 Genotype II—Cosmopolitan | 145 | (75.92) | |
| Related to but not part of DENV-2 Genotype V—Asian I | 1 | (0.52) | |
| Related to but not part of DENV-2 Genotype VI—Sylvatic | 3 | (1.57) | |
| Total | 191 | ||
| 3 | DENV-3 Genotype III | 60 | |
| 4 | DENV-4 Genotype I | 10 | (10.42) |
| DENV-4 Genotype II | 60 | (62.50) | |
| Related to but not part of DENV-4 Genotype II | 26 | (27.08) | |
| Total | 96 | ||
| Grand total | 449 |
| S. No. | Parameters | DENV-1 | DENV-2 | DENV-3 | DENV-4 |
|---|---|---|---|---|---|
| 1. | Template (PDB_ID) used to build 3D structure of consensus sequence derived from this study isolates. | 4C2I (A and C chains) | 1P58 (A and B chains) | 3J6S (A and C chains) | 4CBF (A and C chains) |
| 2. | Query coverage with template | 100% | 100% | 100% | 100% |
| 3. | Consensus sequence identity with template | 63.43% | 62.22% | 88.84% | 84.44% |
| 4. | Best model | 8th model | 27th model | 19th model | 38th model |
| 5. | DOPE score of the best model | −84,972.195 kcal/mol | −84,226.984 kcal/mol | −87,818.578 kcal/mol | −83,635.703 kcal/mol |
| 6. | Ramachandran plot-residues falling under allowed regions (excluding Gly and Pro residues) | 850/872 (97.48%) * 854/872 (97.93%) # | 852/866 (98.38%) * 849/866 (94.8%) # | 831/848 (97.99%) * 833/848 (98.23%) # | 832/856 (97.20%) * 833/856 (97.31%) # |
| 7. | ProSA (Z-score) | −5.60 (−5.87) | −4.58 (−4.55) | −6.29 (−6.52) | −6.10 (−6.19) |
| 8. | ProQ | 11.362 | 11.476 | 11.454 | 10.319 |
| S. No. | Parameters/Properties during 1000 Trajectories of 100 ns MDS | DENV-1_Agnuside | DENV-1_RGBLD1 |
|---|---|---|---|
| 1. | Total Energy (kcal/mol) | −409,942.737 | −410,034.482 |
| 2. | Potential Energy (kcal/mol) | −583,489.002 | −583,632.685 |
| 3. | Degrees of freedom | 561,805 | 561,974 |
| 4. | Number of particles | 259,876 | 259,959 |
| 5. | Protein-Ligand RMSD: Cα, backbone, sidechain, protein hetero atoms, ligand with regard to protein, ligand with regard ligand (Å) | 5.181, 5.180, 6.021, 5.523, 5.556, 0.956 | 4.954, 4.958, 5.802, 5.314, 6.456, 2.419 |
| 6. | Protein RMSF: Cα, backbone, sidechain, protein hetero atoms (Å) | 2.447, 2.454, 2.818, 2.627 | 2.622, 2.632, 3.000, 2.810 |
| 7. | Ligand RMSF: ligand with regard to protein, ligand with regard to ligand (Å) | 2.582, 0.531 | 3.514, 1.341 |
| 8. | Hydrogen bonds | 3608 | 4900 |
| 9. | Hydrophobic interactions | 1844 | 410 |
| 10. | Ionic interactions | - | 196 |
| 11. | Metallic interactions | 11 | 24 |
| 12. | Pi–cation interactions | - | 4 |
| 13. | Pi–pi stacking interactions | 57 | 441 |
| 14. | Water bridge interactions | 3702 | 3251 |
| 15. | Total number of Interactions | 9222 | 9226 |
| S. No. | Parameters/Properties during 1000 Trajectories of 100 ns MDS | DENV-2_Rhodiolin | DENV-2_RGBLD2 |
|---|---|---|---|
| 1. | Total Energy (kcal/mol) | −384,355.621 | −394,785.237 |
| 2. | Potential Energy (kcal/mol) | −529,888.872 | −542,847.189 |
| 3. | Degrees of freedom | 470,928 | 479,089 |
| 4. | Number of particles | 219,287 | 223,321 |
| 5. | Protein-Ligand RMSD: Cα, backbone, sidechain, protein hetero atoms, ligand with regard to protein, ligand with regard to ligand (Å) | 7.855, 7.848, 8.671, 6.743, 2.043 | 7.591, 7.581, 8.484, 7.973, 4.701, 0.654 |
| 6. | Protein RMSF: Cα, backbone, sidechain, protein hetero atoms (Å) | 3.190, 3.215, 3.634, 3.423 | 2.916, 3.047, 3.433, 3.237 |
| 7. | Ligand RMSF: ligand with regard to protein, ligand with regard to ligand (Å) | 2.653, 0.784 | 1.699, 0.296 |
| 8. | Hydrogen bonds | 3209 | 3762 |
| 9. | Hydrophobic interactions | 462 | 165 |
| 10. | Ionic interactions | - | 342 |
| 11. | Metallic interactions | 23 | 2 |
| 12. | Pi–cation interactions | 95 | 213 |
| 13. | Pi–pi stacking interactions | 73 | 22 |
| 14. | Water bridge interactions | 4345 | 4100 |
| 15. | Total number of Interactions | 8207 | 8606 |
| S. No. | Parameters/Properties during 1000 Trajectories of 100 ns MDS | DENV-3_Chlorogenic Acid | DENV-3_RGBLD3 |
|---|---|---|---|
| 1. | Total Energy (kcal/mol) | −418,919.728 | −419,029.352 |
| 2. | Potential Energy (kcal/mol) | −589,515.379 | −589,610.425 |
| 3. | Degrees of freedom | 552,135 | 552,167 |
| 4. | Number of particles | 256,125 | 256,142 |
| 5. | Protein-Ligand RMSD: Cα, backbone, sidechain, protein hetero atoms, ligand with regard to protein, ligand with regard to ligand (Å) | 5.606, 5.607, 6.465, 5.975, 5.091, 2.382 | 4.584, 4.589, 5.439, 4.940, 8.760, 1.327 |
| 6. | Protein RMSF: Cα, backbone, sidechain, protein hetero atoms (Å) | 2.839, 2.848, 3.177, 3.008 | 2.843, 2.851, 3.194, 3.014 |
| 7. | Ligand RMSF: ligand with regard to protein, ligand with regard to ligand (Å) | 2.623, 0.978 | 5.472, 0.943 |
| 8. | Hydrogen bonds | 2625 | 2306 |
| 9. | Hydrophobic interactions | 6 | 225 |
| 10. | Ionic interactions | 20 | 46 |
| 11. | Metallic interactions | 2 | 11 |
| 12. | Pi–cation interactions | 40 | 353 |
| 13. | Pi–pi stacking interactions | 1 | 278 |
| 14. | Water bridge interactions | 3122 | 3553 |
| 15. | Total number of Interactions | 5816 | 6772 |
| S. No. | Parameters/Properties during 1000 Trajectories of 100 ns MDS | DENV-4_NITD448 | DENV-4_RGBLD4 |
|---|---|---|---|
| 1. | Total Energy (kcal/mol) | −401,751.525 | −402,105.698 |
| 2. | Potential Energy (kcal/mol) | −568,014.934 | −568,391.175 |
| 3. | Degrees of freedom | 538,245 | 538,175 |
| 4. | Number of particles | 249,220 | 249,195 |
| 5. | Protein-Ligand RMSD: Cα, backbone, sidechain, protein hetero atoms, ligand with regard to protein, ligand with regard to ligand (Å) | 5.775, 5.759, 6.665, 6.123, 8.256, 2.038 | 4.406, 4.403, 5.417, 4.853, 3.892, 1.834 |
| 6. | Protein RMSF: Cα, backbone, sidechain, protein hetero atoms (Å) | 2.800, 2.804, 3.194, 2.994 | 2.569, 2.580, 2.952, 2.761 |
| 7. | Ligand RMSF: ligand with regard to protein, ligand with regard to ligand (Å) | 8.993, 1.022 | 2.684, 1.346 |
| 8. | Hydrogen bonds | 2303 | 2293 |
| 9. | Hydrophobic interactions | 111 | 836 |
| 10. | Ionic interactions | 11 | 91 |
| 11. | Metallic interactions | 26 | 231 |
| 12. | Pi–cation interactions | 417 | 78 |
| 13. | Pi–pi stacking interactions | 38 | 415 |
| 14. | Water bridge interactions | 4269 | 5286 |
| 15. | Total number of Interactions | 7175 | 9230 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Racherla, R.G.; Katari, S.K.; Mohan, A.; Amineni, U.; Badur, M.; Chaudhury, A.; Nagaraja, M.; Kodavala, S.; Kante, M.; Kalawat, U. Molecular Characterization and Identification of Potential Inhibitors for ‘E’ Protein of Dengue Virus. Viruses 2022, 14, 940. https://doi.org/10.3390/v14050940
Racherla RG, Katari SK, Mohan A, Amineni U, Badur M, Chaudhury A, Nagaraja M, Kodavala S, Kante M, Kalawat U. Molecular Characterization and Identification of Potential Inhibitors for ‘E’ Protein of Dengue Virus. Viruses. 2022; 14(5):940. https://doi.org/10.3390/v14050940
Chicago/Turabian StyleRacherla, Rishi Gowtham, Sudheer Kumar Katari, Alladi Mohan, Umamaheswari Amineni, Manohar Badur, Abhijit Chaudhury, Mudhigeti Nagaraja, Sireesha Kodavala, Meenakshi Kante, and Usha Kalawat. 2022. "Molecular Characterization and Identification of Potential Inhibitors for ‘E’ Protein of Dengue Virus" Viruses 14, no. 5: 940. https://doi.org/10.3390/v14050940
APA StyleRacherla, R. G., Katari, S. K., Mohan, A., Amineni, U., Badur, M., Chaudhury, A., Nagaraja, M., Kodavala, S., Kante, M., & Kalawat, U. (2022). Molecular Characterization and Identification of Potential Inhibitors for ‘E’ Protein of Dengue Virus. Viruses, 14(5), 940. https://doi.org/10.3390/v14050940






