Genetic Diversity of Porcine Circovirus 2 in Wild Boar and Domestic Pigs in Ukraine
Abstract
1. Introduction
1.1. Epidemiology of Porcine Circovirus 2-Associated Diseases in Swine
1.2. PCV2 Genome and Genotypes
2. Materials and Methods
2.1. Sample Collection
2.2. Diagnostic PCR to Detect PCV2
2.3. Amplification and Sequencing of ORF2 (Cap) and PCV2 Genomes
2.4. Nanopore Sequencing and Bioinformatics
2.5. Sequence Curation and Availability
2.6. Phylogenetics Analysis
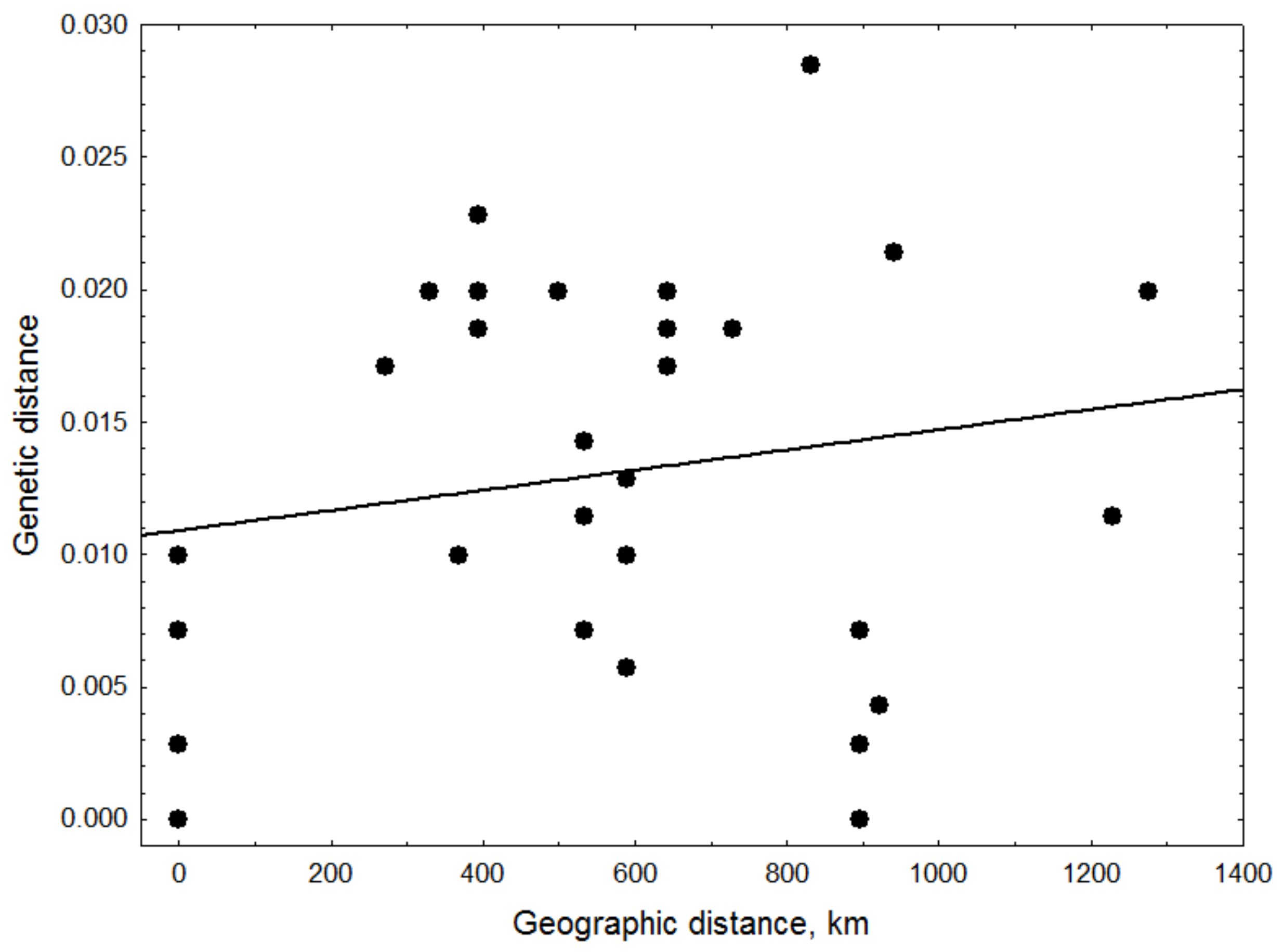
2.7. Mantel Test
3. Results
3.1. Detection of PCV2 in Wild Boar and Domestic Pigs in Ukraine
3.2. Subtyping of PCV2 by Sequencing
3.3. PCV2 Full-Length Genome from Ukraine
3.4. Co-Infections with Two PCV2 Genotypes
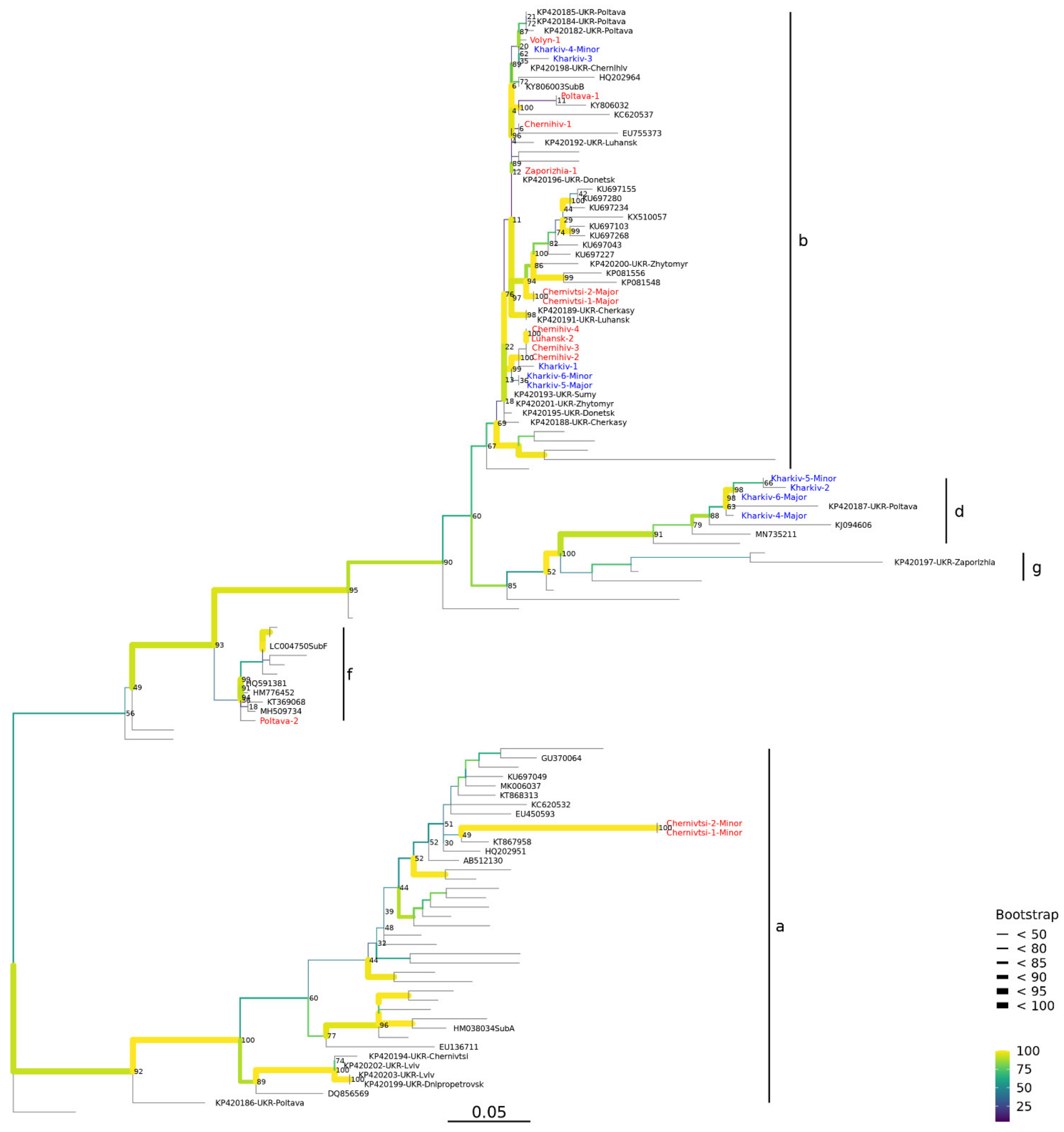
3.5. Phylogenetic Analysis of PCV2 in Ukraine
3.6. Co-Circulation of PCV2 Genotypes
3.7. Genetic Structure of PCV2 Genotype b
4. Discussion
4.1. PCV2 Genotypes Detected in Ukraine
4.2. Transmission and Epidemiology
4.3. A Wild Boar Transmission Chain
4.4. PCV2 Genotypes in Ukraine Reflect Global Diversity
4.5. Limitations
4.6. Nanopore’s Co-Infection Accuracy
4.7. Application of MinION Capacity Building for One Health
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- USDA. Livestock and Poultry: World Markets and Trade. Global Market Analysis. 2021. Available online: https://apps.fas.usda.gov/psdonline/circulars/livestock_poultry.pdf (accessed on 1 April 2022).
- Gaudreault, N.N.; Madden, D.W.; Wilson, W.C.; Trujillo, J.D.; Richt, J.A. African Swine Fever Virus: An Emerging DNA Arbovirus. Front. Vet. Sci. 2020, 7, 215. [Google Scholar] [CrossRef] [PubMed]
- Gebhardt, J.T.; Tokach, M.D.; Dritz, S.S.; DeRouchey, J.M.; Woodworth, J.C.; Goodband, R.D.; Henry, S.C. Postweaning mortality in commercial swine production II: Review of infectious contributing factors. Transl. Anim. Sci. 2020, 4, 485–506. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, T.; Zhang, X.; Liu, X.; Ren, L. Co-Infection of Swine with Porcine Circovirus Type 2 and Other Swine Viruses. Viruses 2019, 11, 185. [Google Scholar] [CrossRef] [PubMed]
- Karuppannan, A.K.; Opriessnig, T. Porcine Circovirus Type 2 (PCV2) Vaccines in the Context of Current Molecular Epidemiology. Viruses 2017, 9, 99. [Google Scholar] [CrossRef] [PubMed]
- Larochelle, R.; Bielanski, A.; Müller, P.; Magar, R.; Larochelle, R.; Bielanski, A.; Müller, P.; Magar, R. PCR Detection and Evidence of Shedding of Porcine Circovirus Type 2 in Boar Semen. J. Clin. Microbiol. 2000, 38, 4629–4632. [Google Scholar] [CrossRef] [PubMed]
- Segalés, J.; Calsamiglia, M.; Olvera, A.; Sibila, M.; Badiella, L.; Domingo, M. Quantification of porcine circovirus type 2 (PCV2) DNA in serum and tonsillar, nasal, tracheo-bronchial, urinary and faecal swabs of pigs with and without postweaning multisystemic wasting syndrome (PMWS). Vet. Microbiol. 2005, 111, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Patterson, A.R.; Opriessnig, T. Epidemiology and horizontal transmission of porcine circovirus type 2 (PCV2). Anim. Health Res. Rev. 2010, 11, 217–234. [Google Scholar] [CrossRef]
- Madson, D.M.; Opriessnig, T. Effect of porcine circovirus type 2 (PCV2) infection on reproduction: Disease, vertical transmission, diagnostics and vaccination. Anim. Health Res. Rev. 2011, 12, 47–65. [Google Scholar] [CrossRef]
- Rose, N.; Opriessnig, T.; Grasland, B.; Jestin, A. Epidemiology and transmission of porcine circovirus type 2 (PCV2). Virus Res. 2012, 164, 78–89. [Google Scholar] [CrossRef]
- Bolin, S.R.; Stoffregen, W.C.; Nayar, G.P.S.; Hamel, A.L. Postweaning Multisystemic Wasting Syndrome Induced after Experimental Inoculation of Cesarean-Derived, Colostrum-Deprived Piglets with Type 2 Porcine Circovirus. J. Veter Diagn. Investig. 2001, 13, 185–194. [Google Scholar] [CrossRef]
- Franzo, G.; Segalés, J. Porcine Circovirus 2 Genotypes, Immunity and Vaccines: Multiple Genotypes but One Single Serotype. Pathogens 2020, 9, 1049. [Google Scholar] [CrossRef]
- Franzo, G.; Tinello, S.; Grassi, L.; Tucciarone, C.M.; Legnardi, M.; Cecchinato, M.; Dotto, G.; Mondin, A.; Martini, M.; Pasotto, D.; et al. Free to Circulate: An Update on the Epidemiological Dynamics of Porcine Circovirus 2 (PCV-2) in Italy Reveals the Role of Local Spreading, Wild Populations, and Foreign Countries. Pathogens 2020, 9, 221. [Google Scholar] [CrossRef]
- Kekarainen, T.; Gonzalez, A.; Llorens, A.; Segalés, J. Genetic variability of porcine circovirus 2 in vaccinating and non-vaccinating commercial farms. J. Gen. Virol. 2014, 95, 1734–1742. [Google Scholar] [CrossRef]
- Wang, Y.; Noll, L.; Lu, N.; Porter, E.; Stoy, C.; Zheng, W.; Liu, X.; Peddireddi, L.; Niederwerder, M.; Bai, J. Genetic diversity and prevalence of porcine circovirus type 3 (PCV3) and type 2 (PCV2) in the Midwest of the USA during 2016–2018. Transbound. Emerg. Dis. 2019, 67, 1284–1294. [Google Scholar] [CrossRef]
- Jang, G.; Yoo, H.; Kim, Y.; Yang, K.; Lee, C. Genetic and phylogenetic analysis of porcine circovirus type 2 on Jeju Island, South Korea, 2019–2020: Evidence of a novel intergenotypic recombinant. Arch. Virol. 2021, 166, 1093–1102. [Google Scholar] [CrossRef]
- Zheng, G.; Lu, Q.; Wang, F.; Xing, G.; Feng, H.; Jin, Q.; Guo, Z.; Teng, M.; Hao, H.; Li, D.; et al. Phylogenetic analysis of porcine circovirus type 2 (PCV2) between 2015 and 2018 in Henan Province, China. BMC Veter Res. 2020, 16, 6. [Google Scholar] [CrossRef]
- Breitbart, M.; Delwart, E.; Rosario, K.; Segalés, J.; Varsani, A. ICTV Report Consortium ICTV Virus Taxonomy Profile: Circoviridae. J. Gen. Virol. 2017, 98, 1997–1998. [Google Scholar] [CrossRef]
- He, J.; Cao, J.; Zhou, N.; Jin, Y.; Wu, J.; Zhou, J. Identification and Functional Analysis of the Novel ORF4 Protein Encoded by Porcine Circovirus Type 2. J. Virol. 2013, 87, 1420–1429. [Google Scholar] [CrossRef]
- Liu, J.; Chen, I.; Kwang, J. Characterization of a Previously Unidentified Viral Protein in Porcine Circovirus Type 2-Infected Cells and Its Role in Virus-Induced Apoptosis. J. Virol. 2005, 79, 8262–8274. [Google Scholar] [CrossRef]
- Li, D.; Wang, J.; Xu, S.; Cai, S.; Ao, C.; Fang, L.; Xiao, S.; Chen, H.; Jiang, Y. Identification and functional analysis of the novel ORF6 protein of porcine circovirus type 2 in vitro. Veter Res. Commun. 2017, 42, 1–10. [Google Scholar] [CrossRef]
- Franzo, G.; Segalés, J. Porcine circovirus 2 (PCV-2) genotype update and proposal of a new genotyping methodology. PLoS ONE 2018, 13, e0208585. [Google Scholar] [CrossRef] [PubMed]
- Rudova, N.G.; Bolotin, V.I.; Solodiankin, O.S.; Gerilovych, P. Porcine circovirus type II screening in feral swine population in Ukraine. J. Veter-Med. Biotechnol. Biosaf. 2019, 5, 10–12. [Google Scholar] [CrossRef][Green Version]
- Kovalenko, G.; Molozhanova, A.; Halka, I.; Nychyk, S. Antibody Prevalence to Influenza Type A in Wild Boar of Northern Ukraine. Vector-Borne Zoonotic Dis. 2017, 17, 836–839. [Google Scholar] [CrossRef] [PubMed]
- Sytiuk, M.P.; Bezymennyy, M.V.; Halka, I.V.; Uhovskyy, V.V.; Muzykina, L.M.; Lavalley, M.; Nychyk, S.A.; Nedosekov, V.V.; Howard, M.W.; Bortz, E. Seroprevalence of Enzootic Teschen Disease in the Wild Boar Population in Ukraine. Vector-Borne Zoonotic Dis. 2022, 22, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Kovalenko, G.; Ducluzeau, A.-L.; Ishchenko, L.; Sushko, M.; Sapachova, M.; Rudova, N.; Solodiankin, O.; Gerilovych, A.; Dagdag, R.; Redlinger, M.; et al. Complete Genome Sequence of a Virulent African Swine Fever Virus from a Domestic Pig in Ukraine. Microbiol. Resour. Announc. 2019, 8, e00883-19. [Google Scholar] [CrossRef]
- Muñoz-Gómez, V.; Solodiankin, O.; Rudova, N.; Gerilovych, A.; Nychyk, S.; Hudz, N.; Ukhovska, T.; Sytiuk, M.; Polischuk, V.; Mustra, D.; et al. Supporting control programs on African swine fever in Ukraine through a knowledge, attitudes, and practices survey targeting backyard farmers. Veter Med. Sci. 2021, 7, 1786–1799. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Park, G.-N.; Choe, S.; Cha, R.M.; Kim, S.-Y.; Hyun, B.-H.; Park, B.-K.; An, D.-J. Genetic Diversity of Porcine Circovirus Isolated from Korean Wild Boars. Pathogens 2020, 9, 457. [Google Scholar] [CrossRef]
- Kleymann, A.; Soto, E.; Illanes, O.; Malik, Y.S.; Fuentealba, C.; Ghosh, S. High rates of detection and complete genomic analysis of porcine circovirus 2 (PCV2) in the Lesser Antilles island of St. Kitts: Identification of PCV2b-PCV2d recombinants. Transbound. Emerg. Dis. 2020, 67, 2282–2289. [Google Scholar] [CrossRef]
- Dudar, L.V.; Budzanivska, I.G.; Polishchuk, V.P. Genetic characterization of porcine circovirus type 2 (PCV2) from wild boars detected in different regions of Ukraine. Biopolym. Cell 2018, 34, 41–48. [Google Scholar] [CrossRef]
- Xiao, C.-T.; Harmon, K.M.; Halbur, P.G.; Opriessnig, T. PCV2d-2 is the predominant type of PCV2 DNA in pig samples collected in the U.S. during 2014–2016. Veter-Microbiol. 2016, 197, 72–77. [Google Scholar] [CrossRef]
- Arefiev, V.; Kovalenko, G.; Frant, M.; Chumachenko, T.; Polyvianna, Y.; Pivnenko, S.; Bolotin, V.; Mayboroda, O.; Solodiankin, O.; Tarasov, O.; et al. Complete Genome Sequence of Salmonella enterica subspenterica Serovar Kottbus Strain Kharkiv, Isolated from a Commercial Pork Production Facility in Ukraine. Microbiol. Resour. Announc. 2020, 9, e01171-20. [Google Scholar] [CrossRef]
- Yang, S.; Yin, S.; Shang, Y.; Liu, B.; Yuan, L.; Zafar Khan, M.U.; Liu, X.; Cai, J. Phylogenetic and genetic variation analyses of porcine circovirus type 2 isolated from China. Transbound. Emerg. Dis. 2018, 65, e383–e392. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Kazutaka, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Chernomor, O.; Von Haeseler, A.; Minh, B.Q. Terrace Aware Data Structure for Phylogenomic Inference from Supermatrices. Syst. Biol. 2016, 65, 997–1008. [Google Scholar] [CrossRef]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar]
- Diniz-Filho, J.A.; Soares, T.N.; Lima, J.S.; Dobrovolski, R.; Lemes Landeiro, V.; Pires de Campos Telles, M.; Rangel, T.F.; Bini, L.M. Mantel test in population genetics. Genet. Mol. Biol. 2013, 36, 475–485. [Google Scholar] [CrossRef]
- Firth, C.; Charleston, M.A.; Duffy, S.; Shapiro, B.; Holmes, E.C. Insights into the evolutionary history of an emerging livestock pathogen: Porcine circovirus 2. J. Virol. 2009, 83, 12813–12821. [Google Scholar] [CrossRef]
- Saporiti, V.; Huerta, E.; Correa-Fiz, F.; Liesner, B.G.; Duran, O.; Segalés, J.; Sibila, M. Detection and genotyping of Porcine circovirus 2 (PCV-2) and detection of Porcine circovirus 3 (PCV-3) in sera from fattening pigs of different European countries. Transbound. Emerg. Dis. 2020, 67, 2521–2531. [Google Scholar] [CrossRef] [PubMed]
- Reiner, G.; Bronnert, B.; Hohloch, C.; Fresen, C.; Haack, I.; Willems, H.; Reinacher, M. Qualitative and quantitative distribution of PCV2 in wild boars and domestic pigs in Germany. Veter Microbiol. 2010, 145, 1–8. [Google Scholar] [CrossRef] [PubMed]
- González-Barrio, D.; Martín-Hernando, M.P.; Ruiz-Fons, F. Shedding patterns of endemic Eurasian wild boar (Sus scrofa) pathogens. Res. Veter-Sci. 2015, 102, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Cságola, A.; Kecskeméti, S.; Kardos, G.; Kiss, I.; Tuboly, T. Genetic characterization of type 2 porcine circoviruses detected in Hungarian wild boars. Arch. Virol. 2005, 151, 495–507. [Google Scholar] [CrossRef]
- Knell, S.; Willems, H.; Hertrampf, B.; Reiner, G. Comparative genetic characterization of Porcine Circovirus type 2 samples from German wild boar populations. Veter-Microbiol. 2005, 109, 169–177. [Google Scholar] [CrossRef]
- Cadar, D.; Cságola, A.; Spinu, M.; Dán, Á.; Ursu, K.; Lőrincz, M.; Tuboly, T. Prevalence of porcine circoviruses in Transylvanian wild boars, detected by real-time PCR—short communication. Acta Veter Hung. 2010, 58, 475–481. [Google Scholar] [CrossRef]
- Fabisiak, M.; Szczotka, A.; Podgórska, K.; Stadejek, T. Prevalence of infection and genetic diversity of porcine circovirus type 2 (pcv2) in wild boar (sus scrofa) in poland. J. Wildl. Dis. 2012, 48, 612–618. [Google Scholar] [CrossRef][Green Version]
- Toplak, I.; Grom, J.; Hostnik, P.; Barlič-Maganja, D. Phylogenetic analysis of type 2 porcine circoviruses identified in wild boar in Slovenia. Veter Rec. 2004, 155, 178–180. [Google Scholar] [CrossRef]
- Amoroso, M.G.; Serra, F.; Esposito, C.; D’Alessio, N.; Ferrara, G.; Cioffi, B.; Anzalone, A.; Pagnini, U.; De Carlo, E.; Fusco, G.; et al. Prevalence of Infection with Porcine Circovirus Types 2 and 3 in the Wild Boar Population in the Campania Region (Southern Italy). Animals 2021, 11, 3215. [Google Scholar] [CrossRef]
- Frant, M.; Gal, A.; Bocian, Ł; Ziętek-Barszcz, A.; Niemczuk, K.; Woźniakowski, G. African Swine Fever Virus (ASFV) in Poland in 2019—Wild Boars: Searching Pattern. Agriculture 2021, 11, 45. [Google Scholar] [CrossRef]
- Correa-Fiz, F.; Franzo, G.; Llorens, A.; Segalés, J.; Kekarainen, T. Porcine circovirus 2 (PCV-2) genetic variability under natural infection scenario reveals a complex network of viral quasispecies. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Forth, J.H.; Forth, L.F.; Blome, S.; Höper, D.; Beer, M. African swine fever whole-genome sequencing—Quantity wanted but quality needed. PLOS Pathog. 2020, 16, e1008779. [Google Scholar] [CrossRef]
- Olvera, A.; Cortey, M.; Segalés, J. Molecular evolution of porcine circovirus type 2 genomes: Phylogeny and clonality. Virology 2007, 357, 175–185. [Google Scholar] [CrossRef]

| Region in Ukraine (Oblast) | Blood Sera Collected | Positive for PCV2 (PCR) | PCV2 Prevalence (%) | Number Selected for Sequencing |
|---|---|---|---|---|
| Poltava | 15 | 3 | 20% | 2 |
| Sumy | 8 | 5 | 63% | - |
| Zaporizhia | 13 | 12 | 92% | 1 |
| Chernihiv | 17 | 5 | 29% | 4 |
| Chernivtsi | 5 | 2 | 40% | 2 |
| Cherkasy | 10 | 1 | 10% | - |
| Kherson | 1 | 0 | 0% | - |
| Lviv | 13 | 3 | 23% | - |
| Volyn | 13 | 1 | 8% | 1 |
| Luhansk | 12 | 2 | 17% | 2 |
| Total | 107 | 34 | 31.8% | 12 |
| Sample | Host Type | Sequence Yield (Mbp) | Number of Reads | Mean Quality (Q-Score) | Major PCV2 Genotype |
|---|---|---|---|---|---|
| Chernihiv 1 | Wild Boar | 1349.3 | 170,304 | 13 | b |
| Chernihiv 2 | Wild Boar | 558.0 | 70,235 | 13.2 | b |
| Chernihiv 3 | Wild Boar | 568.1 | 71,070 | 13.2 | b |
| Chernihiv 4 | Wild Boar | 383.1 | 47,930 | 13.1 | b |
| Chernivtsi 1 | Wild Boar | 1149.3 | 144,338 | 13.1 | b |
| Chernivtsi 2 | Wild Boar | 1163.6 | 146,489 | 13.2 | b |
| Luhansk 1 | Wild Boar | NA | NA | NA | NA |
| Luhansk 2 | Wild Boar | 365.1 | 44,527 | 13.2 | b |
| Poltava 1 | Wild Boar | 696.9 | 87,848 | 13 | b |
| Poltava 2 | Wild Boar | 605.2 | 76,231 | 12.9 | f |
| Volyn | Wild Boar | 320.0 | 39,156 | 13.1 | b |
| Zaporizhzhia | Wild Boar | 121.1 | 14,941 | 13.1 | b |
| Kharkiv 1 | Domestic pig | 3599.6 | 453,425 | 14.5 | b |
| Kharkiv 2 | Domestic pig | 1240.9 | 156,283 | 14.6 | d |
| Kharkiv 3 | Domestic pig | 1828.5 | 229,888 | 14.6 | b |
| Kharkiv 4 | Domestic pig | 560.0 | 70,490 | 14.6 | d |
| Kharkiv 5 | Domestic pig | 5036.4 | 635,035 | 14.5 | b |
| Kharkiv 6 | Domestic pig | 213.9 | 26,903 | 14.6 | d |
| Sample | % Minor Genotype | No. Reads | Major Genotype | Minor Genotype |
|---|---|---|---|---|
| Chernivtsi 1 (wild boar) | 3.47 | 5191 | b | a |
| Chernivtsi 2 (wild boar) | 3.93 | 6000 | b | a |
| Karhkiv 4 (domestic pig) | 28.41 | 28356 | d | b |
| Karhkiv 5 (domestic pig) | 5.81 | 39560 | b | d |
| Karhkiv 6 (domestic pig) | 25.82 | 9398 | d | b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rudova, N.; Buttler, J.; Kovalenko, G.; Sushko, M.; Bolotin, V.; Muzykina, L.; Zinenko, O.; Stegniy, B.; Dunaiev, Y.; Sytiuk, M.; et al. Genetic Diversity of Porcine Circovirus 2 in Wild Boar and Domestic Pigs in Ukraine. Viruses 2022, 14, 924. https://doi.org/10.3390/v14050924
Rudova N, Buttler J, Kovalenko G, Sushko M, Bolotin V, Muzykina L, Zinenko O, Stegniy B, Dunaiev Y, Sytiuk M, et al. Genetic Diversity of Porcine Circovirus 2 in Wild Boar and Domestic Pigs in Ukraine. Viruses. 2022; 14(5):924. https://doi.org/10.3390/v14050924
Chicago/Turabian StyleRudova, Nataliia, Jeremy Buttler, Ganna Kovalenko, Mykola Sushko, Vitaliy Bolotin, Larysa Muzykina, Oleksandr Zinenko, Borys Stegniy, Yurii Dunaiev, Mykola Sytiuk, and et al. 2022. "Genetic Diversity of Porcine Circovirus 2 in Wild Boar and Domestic Pigs in Ukraine" Viruses 14, no. 5: 924. https://doi.org/10.3390/v14050924
APA StyleRudova, N., Buttler, J., Kovalenko, G., Sushko, M., Bolotin, V., Muzykina, L., Zinenko, O., Stegniy, B., Dunaiev, Y., Sytiuk, M., Gerilovych, A., Drown, D. M., Bortz, E., & Solodiankin, O. (2022). Genetic Diversity of Porcine Circovirus 2 in Wild Boar and Domestic Pigs in Ukraine. Viruses, 14(5), 924. https://doi.org/10.3390/v14050924








