Characteristics of the MicroRNA Expression Profile of Exosomes Released by Vero Cells Infected with Porcine Epidemic Diarrhea Virus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Virus
2.2. Exosome Isolation
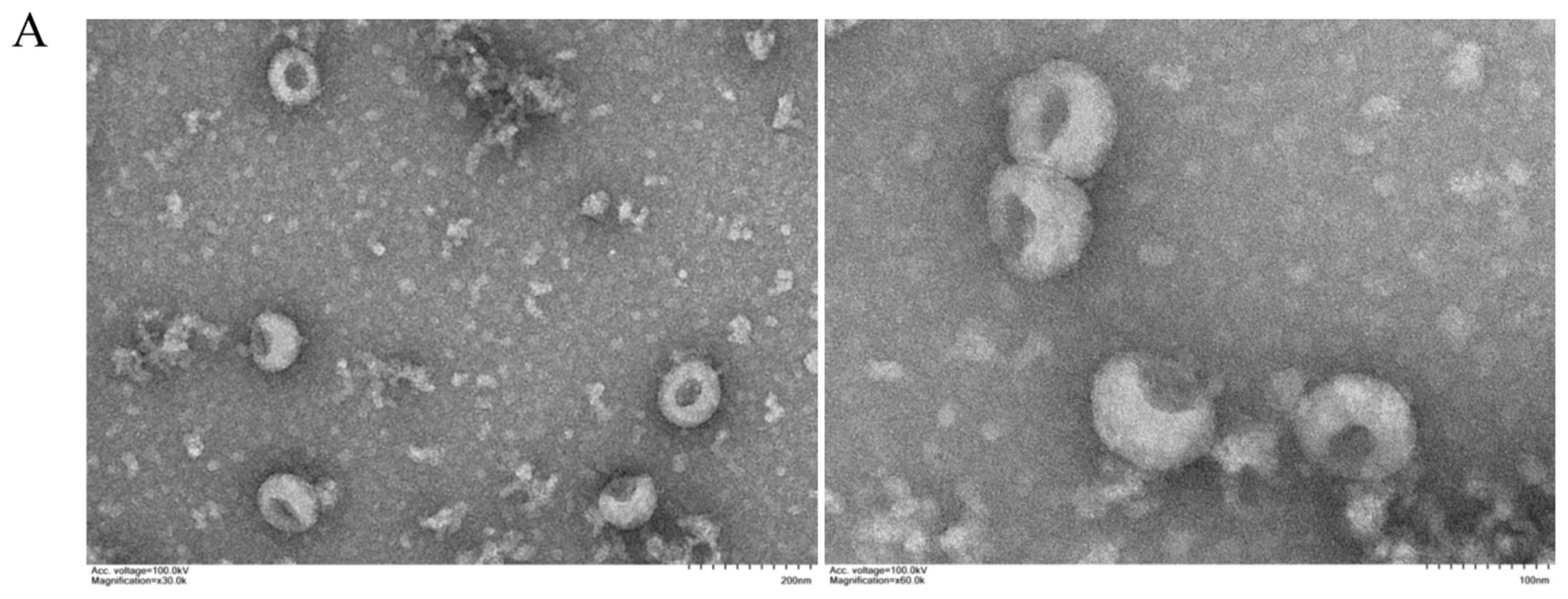
2.3. Transmission Electron Microscopy (TEM)
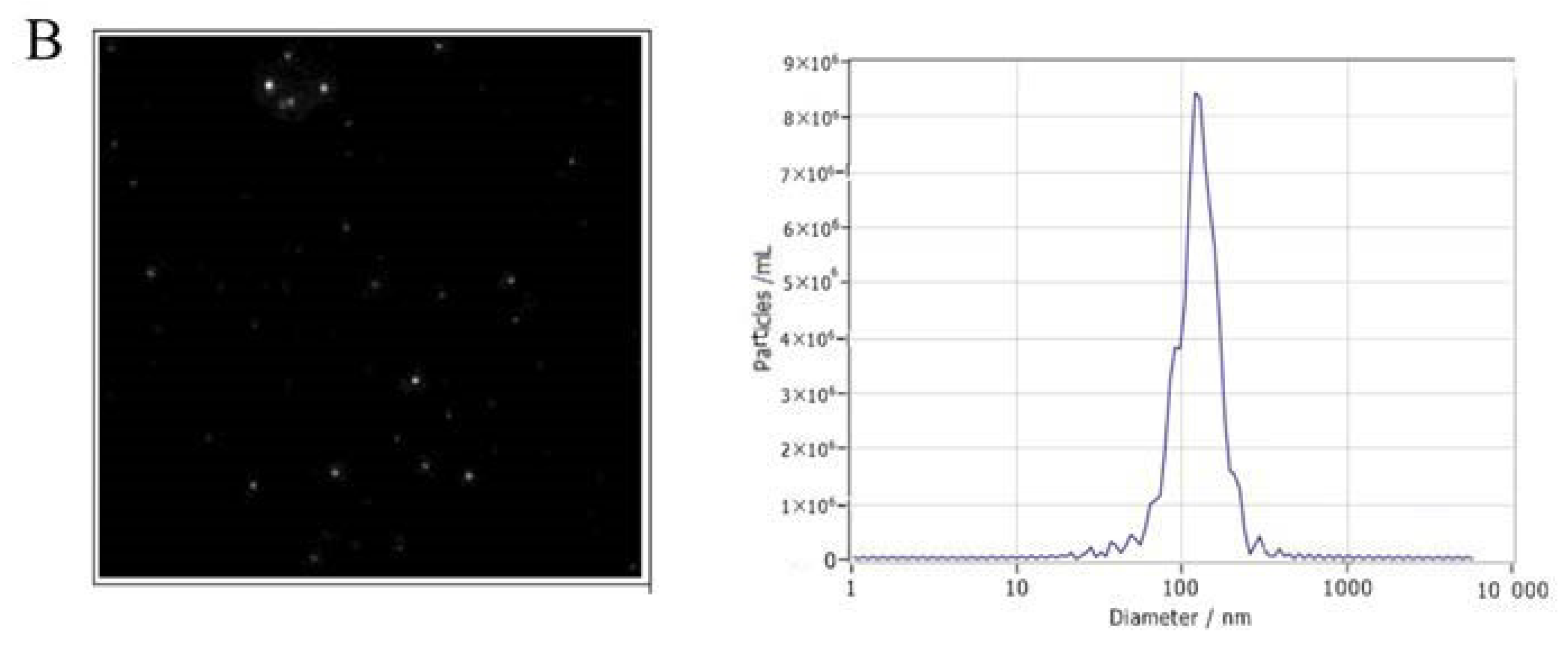
2.4. Nanoparticle Tracking Analysis (NTA)
2.5. MiRNA Microarray Assay and Bioinformatics Analysis of Target Genes
2.6. Analysis of the miRNAs by qRT-PCR
2.7. Statistical Analysis
3. Results
3.1. Characterization of Exosomes
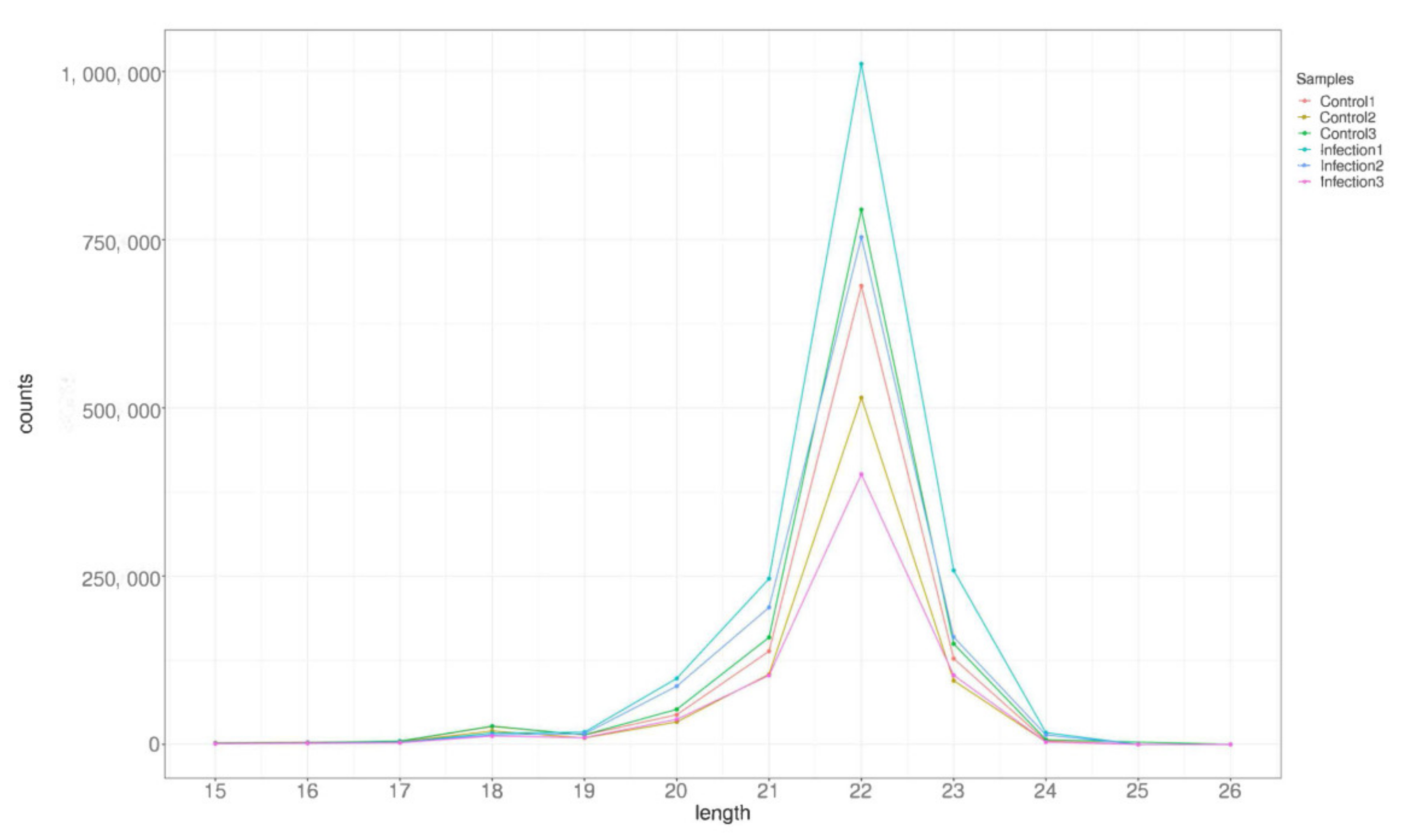
3.2. Analysis of Small RNA Sequencing Library Data
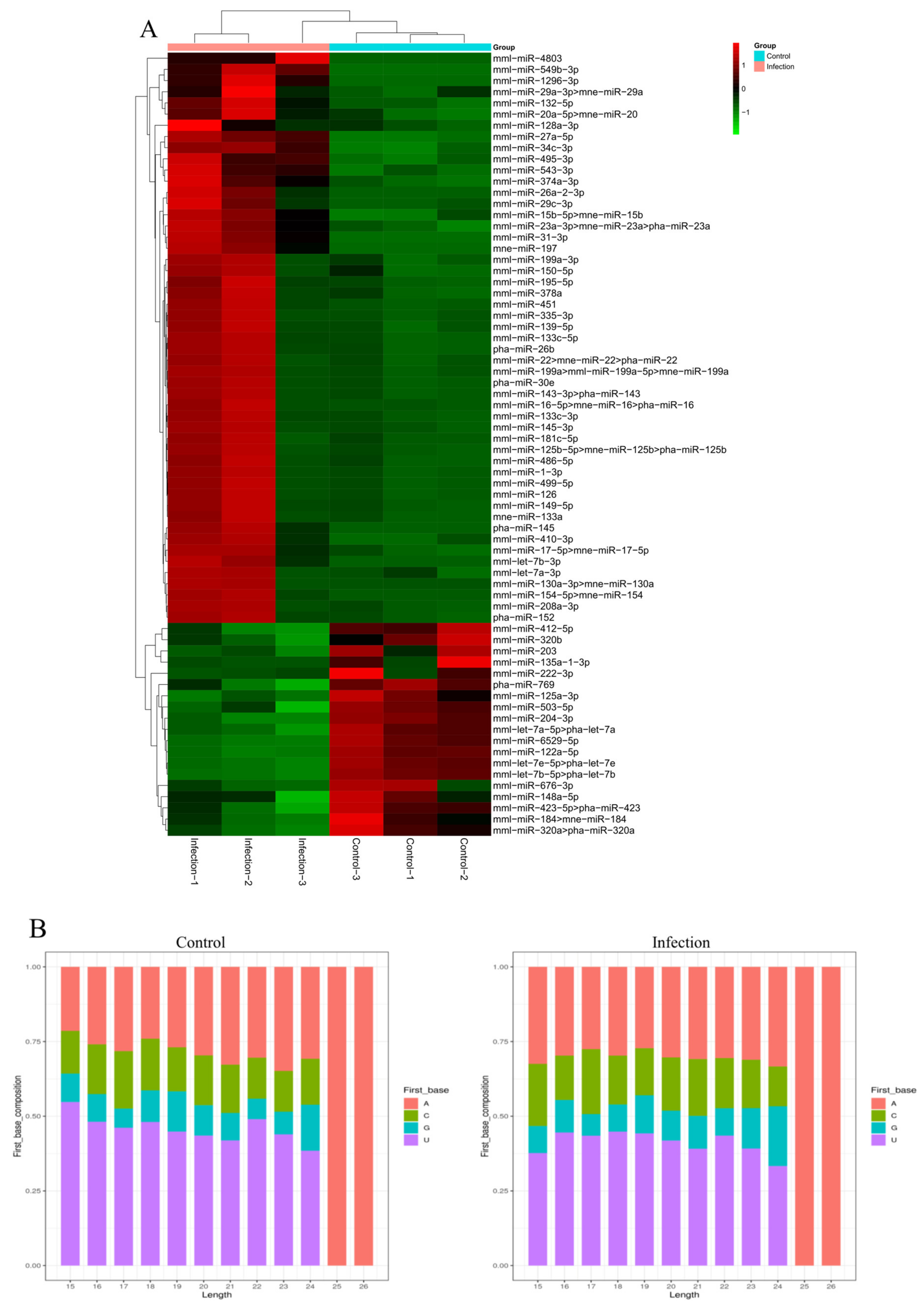
3.3. Identification of Known MiRNAs in Exosomes
3.4. Identification of Novel MiRNAs in Exosomes
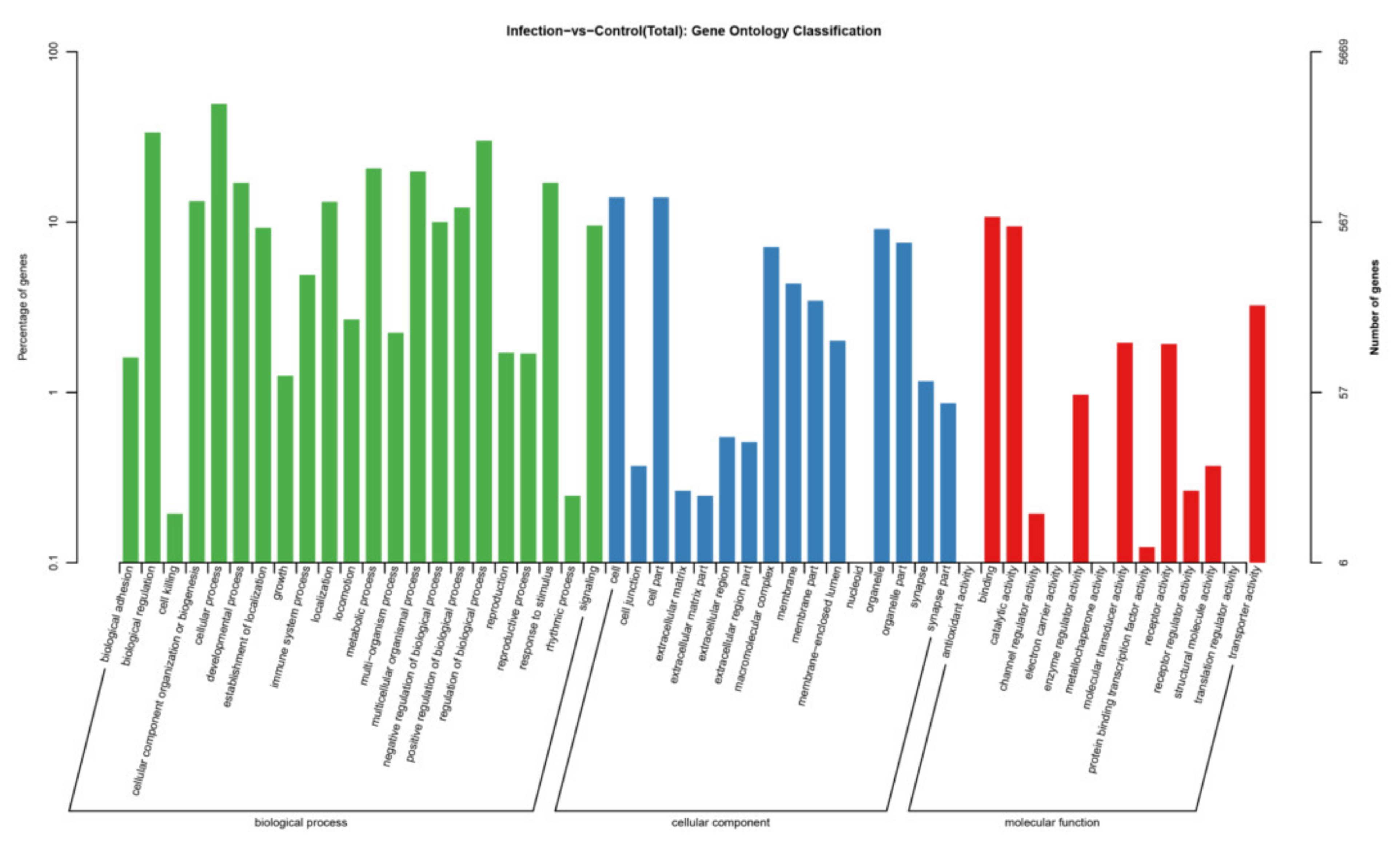
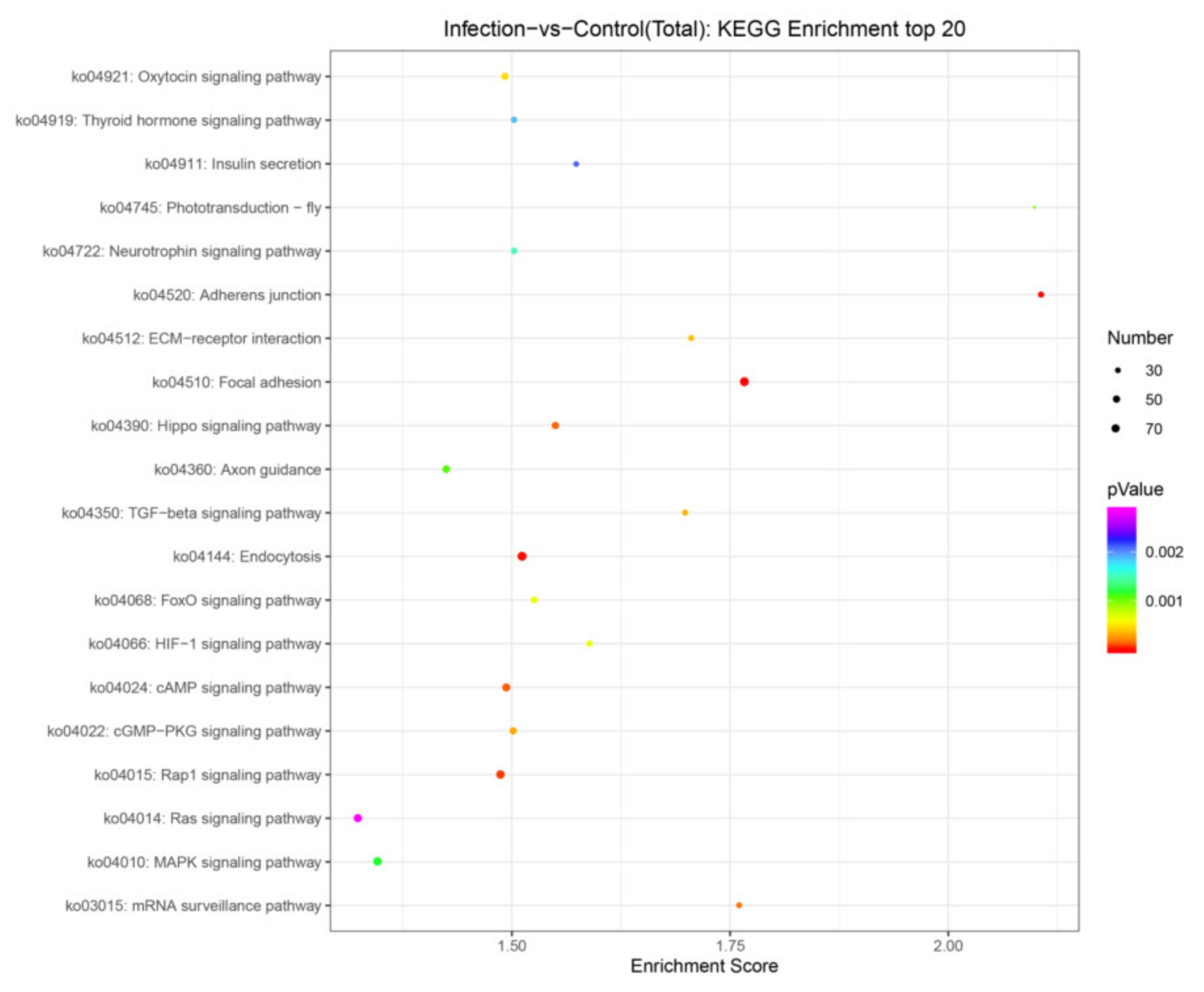
3.5. Target Gene Prediction and Pathway Enrichment Analysis of DEmiRNAs
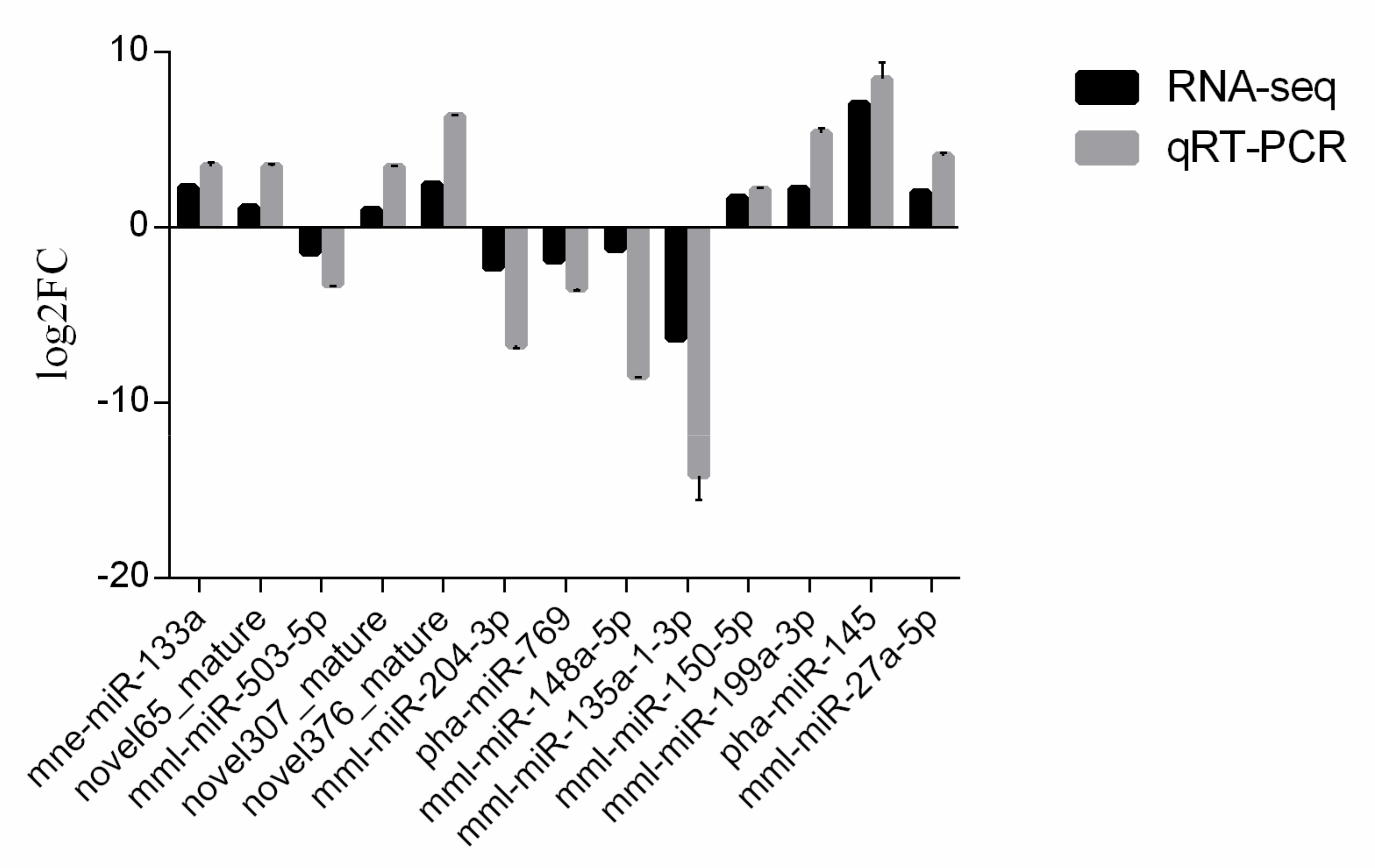
3.6. Validation of MiRNAs by qRT-PCR
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Salamat, S.E.A.; Collantes, T.M.A.; Lumbera, W.M.L.; Tablizo, F.A.; Mutia, C.T.M.; Ong, J.D.P.; Bandoy, D. Sequence analysis of new variants of porcine epidemic diarrhea virus in Luzon, Philippines, in 2017. Arch. Virol. 2021, 166, 1859–1867. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wang, C.; Shu, J.; Feng, H.; He, Y.; Chen, J.; Shu, J. Porcine Epidemic Diarrhea Virus Induces Vero Cell Apoptosis via the p53-PUMA Signaling Pathway. Viruses 2021, 13, 1218. [Google Scholar] [CrossRef] [PubMed]
- Wen, F.; Yang, J.; Li, A.; Gong, Z.; Yang, L.; Cheng, Q.; Wang, C.; Zhao, M.; Yuan, S.; Chen, Y.; et al. Genetic characterization and phylogenetic analysis of porcine epidemic diarrhea virus in Guangdong, China, between 2018 and 2019. PLoS ONE 2021, 16, e0253622. [Google Scholar] [CrossRef] [PubMed]
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: Extracellular organelles important in intercellular communication. J. Proteom. 2010, 73, 1907–1920. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Silverman, J.M.; Reiner, N.E. Exosomes and other microvesicles in infection biology: Organelles with unanticipated phenotypes. Cell. Microbiol. 2011, 13, 1–9. [Google Scholar] [CrossRef]
- Tkach, M.; Théry, C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 2016, 164, 1226–1232. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Zhang, D.; Ding, W.; Wang, W.; Jin, N.; Ding, Z. NDV related exosomes enhance NDV replication through exporting NLRX1 mRNA. Vet. Microbiol. 2021, 260, 109167. [Google Scholar] [CrossRef]
- Chen, J.; Jin, L.; Yan, M.; Yang, Z.; Wang, H.; Geng, S.; Gong, Z.; Liu, G. Serum Exosomes from Newborn Piglets Restrict Porcine Epidemic Diarrhea Virus Infection. J. Proteome Res. 2019, 18, 1939–1947. [Google Scholar] [CrossRef]
- Feng, Z.; Hensley, L.; McKnight, K.L.; Hu, F.; Madden, V.; Ping, L.; Jeong, S.H.; Walker, C.; Lanford, R.E.; Lemon, S.M. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 2013, 496, 367–371. [Google Scholar] [CrossRef] [Green Version]
- Yao, Z.; Qiao, Y.; Li, X.; Chen, J.; Ding, J.; Bai, L.; Shen, F.; Shi, B.; Liu, J.; Peng, L.; et al. Exosomes Exploit the Virus Entry Machinery and Pathway To Transmit Alpha Interferon-Induced Antiviral Activity. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [Green Version]
- Jung, K.O.; Youn, H.; Lee, C.H.; Kang, K.W.; Chung, J.K. Visualization of exosome-mediated miR-210 transfer from hypoxic tumor cells. Oncotarget 2017, 8, 9899–9910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mihelich, B.L.; Dambal, S.; Lin, S.; Nonn, L. miR-182, of the miR-183 cluster family, is packaged in exosomes and is detected in human exosomes from serum, breast cells and prostate cells. Oncol. Lett. 2016, 12, 1197–1203. [Google Scholar] [CrossRef] [PubMed]
- Squadrito, M.L.; Baer, C.; Burdet, F.; Maderna, C.; Gilfillan, G.D.; Lyle, R.; Ibberson, M.; De Palma, M. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014, 8, 1432–1446. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Liu, W.; Xiao, J.; Cao, B. The role of exosomes and “exosomal shuttle microRNA” in tumorigenesis and drug resistance. Cancer Lett. 2015, 356, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Tan, L.; Sun, Y.; Qiu, X.; Liao, Y.; Song, C.; Liu, W.; Nair, V.; Ding, C. Exosomes Carry microRNAs into Neighboring Cells to Promote Diffusive Infection of Newcastle Disease Virus. Viruses 2019, 11, 527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bellingham, S.A.; Coleman, B.M.; Hill, A.F. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res. 2012, 40, 10937–10949. [Google Scholar] [CrossRef] [Green Version]
- Roth, W.W.; Huang, M.B.; Addae Konadu, K.; Powell, M.D.; Bond, V.C. Micro RNA in Exosomes from HIV-Infected Macrophages. Int. J. Environ. Res. Public Health 2015, 13, 32. [Google Scholar] [CrossRef] [Green Version]
- Salido-Guadarrama, I.; Romero-Cordoba, S.; Peralta-Zaragoza, O.; Hidalgo-Miranda, A.; Rodríguez-Dorantes, M. MicroRNAs transported by exosomes in body fluids as mediators of intercellular communication in cancer. OncoTargets Ther. 2014, 7, 1327–1338. [Google Scholar] [CrossRef] [Green Version]
- Rebustini, I.T.; Vlahos, M.; Packer, T.; Kukuruzinska, M.A.; Maas, R.L. An integrated miRNA functional screening and target validation method for organ morphogenesis. Sci. Rep. 2016, 6, 23215. [Google Scholar] [CrossRef] [Green Version]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 30, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Liang, X.; Yan, Y.; Lu, Y.; Zhang, D.; Yao, W.; Wu, W.; Yan, Z. Identification of Exosomal miRNAs in Rats With Pulmonary Neutrophilic Inflammation Induced by Zinc Oxide Nanoparticles. Front. Physiol. 2018, 9, 217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Saini, H.K.; van Dongen, S.; Enright, A.J. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008, 36, D154–D158. [Google Scholar] [CrossRef] [Green Version]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef]
- Ye, J.; Zhang, Y.; Cui, H.; Liu, J.; Wu, Y.; Cheng, Y.; Xu, H.; Huang, X.; Li, S.; Zhou, A.; et al. WEGO 2.0: A web tool for analyzing and plotting GO annotations, 2018 update. Nucleic Acids Res. 2018, 46, W71–W75. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Shen, X.; Yin, L.; Pan, X.; Zhao, R.; Zhang, D. Porcine epidemic diarrhea virus infection blocks cell cycle and induces apoptosis in pig intestinal epithelial cells. Microb. Pathog. 2020, 147, 104378. [Google Scholar] [CrossRef]
- Zhang, X.; Li, C.; Zhang, B.; Li, Z.; Zeng, W.; Luo, R.; Cao, J.; Cheng, G.; Fan, S.; He, Q. Differential expression and correlation analysis of miRNA-mRNA profiles in swine testicular cells infected with porcine epidemic diarrhea virus. Sci. Rep. 2021, 11, 1868. [Google Scholar] [CrossRef]
- Zheng, H.; Xu, L.; Liu, Y.; Li, C.; Zhang, L.; Wang, T.; Zhao, D.; Xu, X.; Zhang, Y. MicroRNA-221-5p Inhibits Porcine Epidemic Diarrhea Virus Replication by Targeting Genomic Viral RNA and Activating the NF-κB Pathway. Int. J. Mol. Sci. 2018, 19, 3381. [Google Scholar] [CrossRef] [Green Version]
- Ge, F.F.; Yang, D.Q.; Li, X.; Ju, H.B.; Shen, H.X.; Liu, J.; Zhao, H.J.; Wang, J. Novel Method for Isolation of Porcine Epidemic Diarrhea Virus with the Use of Suspension Vero Cells and Immunogenicity Analysis. J. Clin. Microbiol. 2021, 59, 2. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.W.; Sun, X.F.; Tong, H.L.; Wang, Y.H.; Li, S.F.; Yan, Y.Q.; Li, G.P. Effect of differentiation on microRNA expression in bovine skeletal muscle satellite cells by deep sequencing. Cell. Mol. Biol. Lett. 2016, 21, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, H.; Cheng, A.; Zhang, X.; Pan, Y.; Wang, M.; Huang, J.; Zhu, D.; Chen, S.; Liu, M.; Zhao, X.; et al. DEF Cell-Derived Exosomal miR-148a-5p Promotes DTMUV Replication by Negative Regulating TLR3 Expression. Viruses 2020, 12, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Othumpangat, S.; Lindsley, W.G.; Beezhold, D.H.; Kashon, M.L.; Burrell, C.N.; Mubareka, S.; Noti, J.D. Differential Expression of Serum Exosome microRNAs and Cytokines in Influenza A and B Patients Collected in the 2016 and 2017 Influenza Seasons. Pathogens 2021, 10, 149. [Google Scholar] [CrossRef]
- Qian, X.; Xu, C.; Fang, S.; Zhao, P.; Wang, Y.; Liu, H.; Yuan, W.; Qi, Z. Exosomal MicroRNAs Derived From Umbilical Mesenchymal Stem Cells Inhibit Hepatitis C Virus Infection. Stem Cells Transl. Med. 2016, 5, 1190–1203. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Z.; Petree, J.R.; Lee, F.E.; Fan, X.; Salaita, K.; Guidot, D.M.; Sadikot, R.T. Macrophages exposed to HIV viral protein disrupt lung epithelial cell integrity and mitochondrial bioenergetics via exosomal microRNA shuttling. Cell Death Dis. 2019, 10, 580. [Google Scholar] [CrossRef]
- Wang, J.; Teng, Y.; Zhao, G.; Li, F.; Hou, A.; Sun, B.; Kong, W.; Gao, F.; Cai, L.; Jiang, C. Exosome-Mediated Delivery of Inducible miR-423-5p Enhances Resistance of MRC-5 Cells to Rabies Virus Infection. Int. J. Mol. Sci. 2019, 20, 1537. [Google Scholar] [CrossRef] [Green Version]
- Yuan, F.; Yun, Y.; Fan, H.; Li, Y.; Lu, L.; Liu, J.; Feng, W.; Chen, S.Y. MicroRNA-135a Protects Against Ethanol-Induced Apoptosis in Neural Crest Cells and Craniofacial Defects in Zebrafish by Modulating the Siah1/p38/p53 Pathway. Front. Cell Dev. Biol. 2020, 8, 583959. [Google Scholar] [CrossRef]
- Zhao, H.; Yang, J.; Wang, Q.; Cui, Z.; Li, D.; Niu, J.; Guo, Y.; Zhang, Q.; Zhang, S.; Zhao, Y.; et al. Exosomal miRNA-328-3p targets ZO-3 and inhibits porcine epidemic diarrhea virus proliferation. Arch. Virol. 2022, 167, 901–910. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Evripioti, A.A.; Ortega-Prieto, A.M.; Skelton, J.K.; Bazot, Q.; Dorner, M. Phosphodiesterase-induced cAMP degradation restricts hepatitis B virus infection. Philos. Trans. R. Soc. London. Ser. B Biol. Sci. 2019, 374, 20180292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Lu, W.; Zhang, Y.; Zou, F.; Jin, Z.; Zhao, T. The Hippo Pathway and Viral Infections. Front. Microbiol. 2019, 10, 3033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, L.L.; Li, J.R.; Li, H.; Tan, J.L.; Wang, M.X.; Liu, N.N.; Gao, R.M.; Yan, H.Y.; Wang, X.K.; Dong, B.; et al. TGF-β isoforms inhibit hepatitis C virus propagation in transforming growth factor beta/SMAD protein signalling pathway dependent and independent manners. J. Cell. Mol. Med. 2021, 25, 3498–3510. [Google Scholar] [CrossRef] [PubMed]
- Jahani, M.; Dokaneheifard, S.; Mansouri, K. Hypoxia: A key feature of COVID-19 launching activation of HIF-1 and cytokine storm. J. Inflamm. 2020, 17, 33. [Google Scholar] [CrossRef]
- Guo, F.; Yu, X.; Xu, A.; Xu, J.; Wang, Q.; Guo, Y.; Wu, X.; Tang, Y.; Ding, Z.; Zhang, Y.; et al. Japanese encephalitis virus induces apoptosis by inhibiting Foxo signaling pathway. Vet. Microbiol. 2018, 220, 73–82. [Google Scholar] [CrossRef]
- Motamedi, N.; Sewald, X.; Luo, Y.; Mothes, W.; DiMaio, D. SV40 Polyomavirus Activates the Ras-MAPK Signaling Pathway for Vacuolization, Cell Death, and Virus Release. Viruses 2020, 12, 1128. [Google Scholar] [CrossRef]
- Wang, H.D.; Trivedi, A.; Johnson, D.L. Hepatitis B virus X protein induces RNA polymerase III-dependent gene transcription and increases cellular TATA-binding protein by activating the Ras signaling pathway. Mol. Cell. Biol. 1997, 17, 6838–6846. [Google Scholar] [CrossRef] [Green Version]

| MiRNA Name | MiRNA Sequence (5′-3′) | RT Primer Sequence (5′-3′) | Forward PCR Primer Sequence (5′-3′) |
|---|---|---|---|
| mne-miR-133a | TTGGTCCCCTTCAACCAGCTGT | GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACACAGCT | CTCATTGGTCCCCTTCAACC |
| novel65_mature | GGTGGGGTCGGCGGGGGG | GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACCCCCCC | TCATTATAGGTGGGGTCGGC |
| mml-miR-503-5p | TAGCAGCGGGAACAGTTCTGCAG | GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACCTGCAG | ACTTAGCAGCGGGAACAGTT |
| novel307_mature | CGGCGGCGACGGTGGCGG | GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACCCGCCA | TATATTTACGGCGGCGACGG |
| novel376_mature | CAGGGGTGGAGCCTGCGGA | GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACTCCGCA | ATTACTTCAGGGGTGGAGCC |
| mml-miR-204-3p | GGCTGGGAAGGCAAAGGGACGT | GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACACGTCCC | AGTTAGGCTGGGAAGGCAAA |
| pha-miR-769 | TGAGACCTCTGGGTTCTGAGCT | GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACAGCTCA | TCAGTTGAGACCTCTGGGTTC |
| mml-miR-148a-5p | AAAGTTCTGAGACACTCCGACT | GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACAGTCGG | TGGCGAAAGTTCTGAGACACT |
| mml-miR-135a-1-3p | ATATAGGGATTGGAGCCGTGGC | GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACGCCACGG | CGCTCGATATAGGGATTGGAG |
| mml-miR-150-5p | TCTCCCAACCCTTGTACCAGTG | GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACCACTGG | TGCTGTCTCCCAACCCTTGT |
| mml-miR-199a-3p | ACAGTAGTCTGCACATTGGTTA | GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACTAACCAA | TCTCGCACAGTAGTCTGCACA |
| pha-miR-145 | GTCCAGTTTTCCCAGGAATCCCT | GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACAGGGATT | ACGTGTCCAGTTTTCCCAGG |
| mml-miR-27a-5p | AGGGCTTAGCTGCTTGTGAGCA | GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACTGCTCAC | GTGACAGGGCTTAGCTGCTT |
| MicroRNA U6 | AACGCTTCACGAATTTGCGT | CTCGCTTCGGCAGCACA |
| Category | Infected | Uninfected |
|---|---|---|
| Raw reads | 30,572,744/26,165,923/27,571,400 | 27,070,230/21,158,763/26,987,232 |
| Clean reads | 24,269,195/21,385,578/22,875,953 | 20,920,004/16,414,523/20,535,191 |
| miRNAs’ reads | 1,673,115/1,253,338/675,418(Total) 2074/2003/1752(unique) | 1,047,301/788,983/1,212,878(Total) 1386/1208/1352(unique) |
| known miRNAs | 441/415/396 | 352/326/346 |
| novel miRNAs | 310/306/290 | 210/181/208 |
| rRNA reads | 189,890/164,385/185,472 | 115,653/98,041/103,065 |
| tRNA reads | 29,156/30,890/14,333 | 13,765/6017/8148 |
| snRNA reads | 27,653/31,092/33,635 | 36,342/26,198/34,540 |
| Cis-region reads | 55,409/67,613/84,276 | 86,221/61,990/85,314 |
| other_Rfam_RNA | 73,478/74,761/99,704 | 76,065/61,225/70,461 |
| unannotated | 9,493,236/8,307,505/8,358,603 | 9,414,281/7,302,434/9,459,270 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, L.; Shen, X.; Yin, D.; Wang, J.; Zhao, R.; Dai, Y.; Pan, X. Characteristics of the MicroRNA Expression Profile of Exosomes Released by Vero Cells Infected with Porcine Epidemic Diarrhea Virus. Viruses 2022, 14, 806. https://doi.org/10.3390/v14040806
Yin L, Shen X, Yin D, Wang J, Zhao R, Dai Y, Pan X. Characteristics of the MicroRNA Expression Profile of Exosomes Released by Vero Cells Infected with Porcine Epidemic Diarrhea Virus. Viruses. 2022; 14(4):806. https://doi.org/10.3390/v14040806
Chicago/Turabian StyleYin, Lei, Xuehuai Shen, Dongdong Yin, Jieru Wang, Ruihong Zhao, Yin Dai, and Xiaocheng Pan. 2022. "Characteristics of the MicroRNA Expression Profile of Exosomes Released by Vero Cells Infected with Porcine Epidemic Diarrhea Virus" Viruses 14, no. 4: 806. https://doi.org/10.3390/v14040806
APA StyleYin, L., Shen, X., Yin, D., Wang, J., Zhao, R., Dai, Y., & Pan, X. (2022). Characteristics of the MicroRNA Expression Profile of Exosomes Released by Vero Cells Infected with Porcine Epidemic Diarrhea Virus. Viruses, 14(4), 806. https://doi.org/10.3390/v14040806






