Cross Strain Protection against Cytomegalovirus Reduces DISC Vaccine Efficacy against CMV in the Guinea Pig Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Virus, Cells, Synthetic Genes and Oligonucleotides
2.2. Animal Studies
2.3. GPCMV Glycoprotein ELISAs
2.4. GPCMV Neutralization Assays
2.5. Real Time PCR
2.6. Statistical Analysis
3. Results
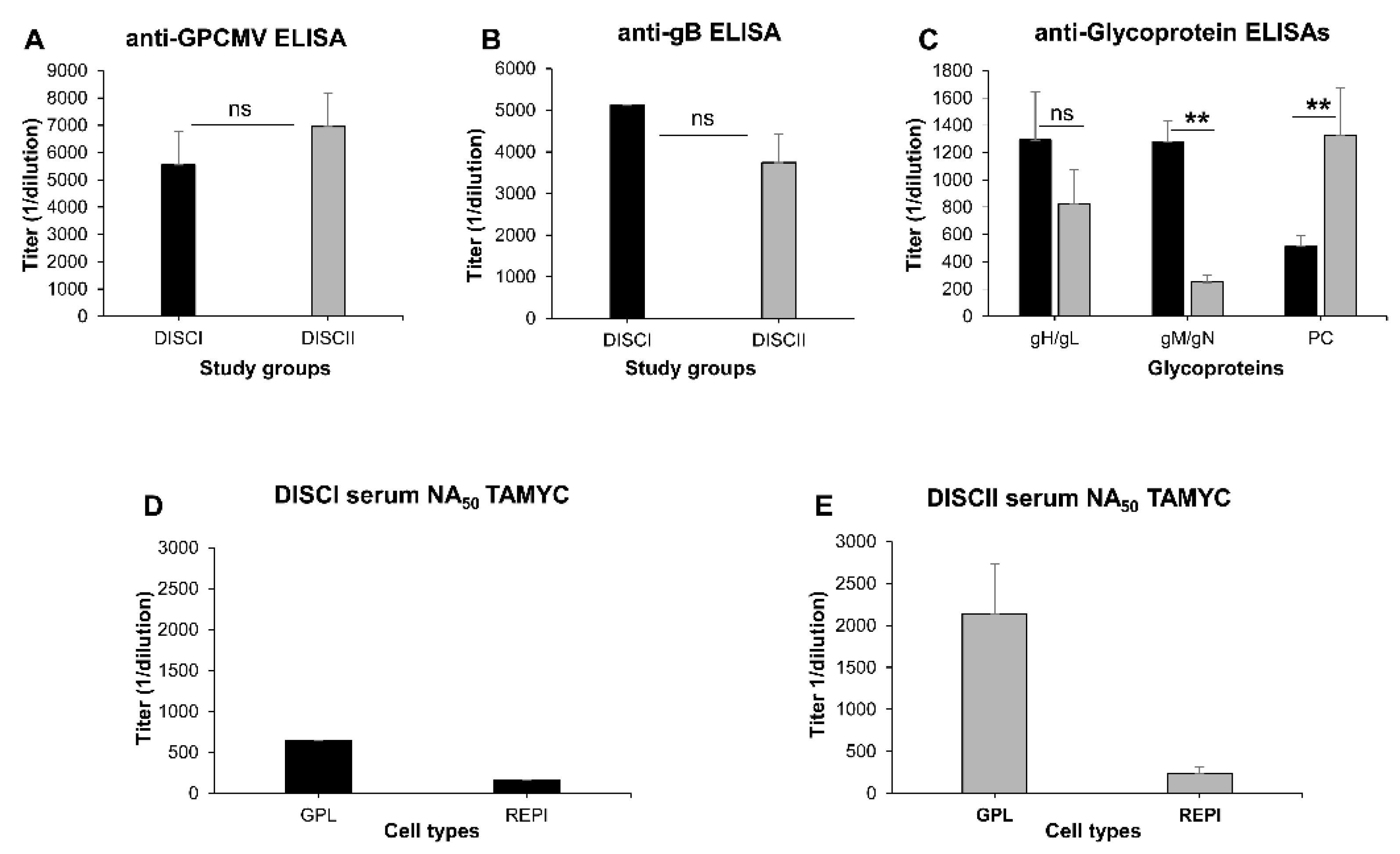
3.1. Comparative Ability of GPCMV DISC Vaccine (PC+ or PC−) to Neutralize TAMYC Strain
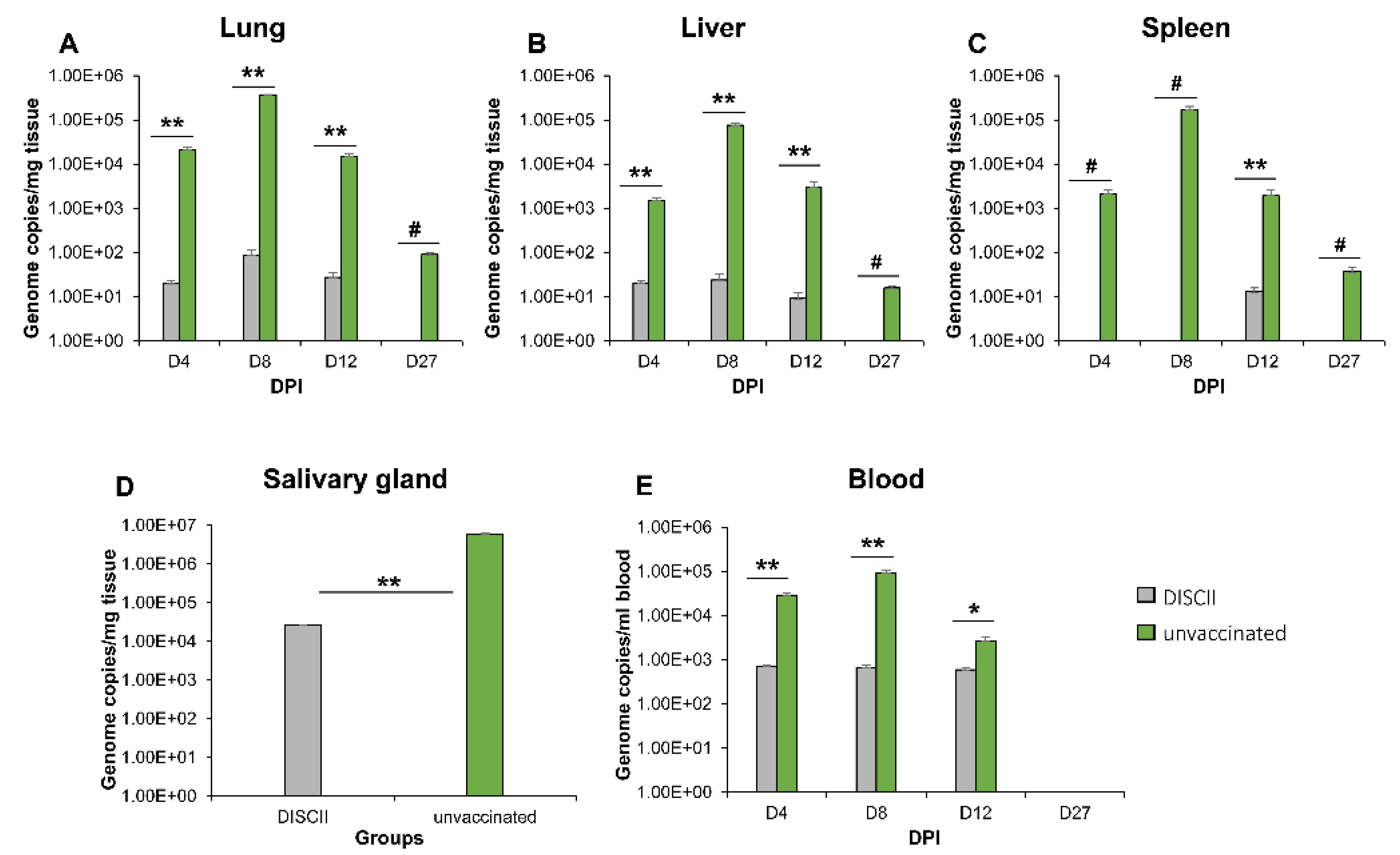
3.2. DISC Vaccine Cross Protection against GPCMV (TAMYC Strain) Virus Challenge
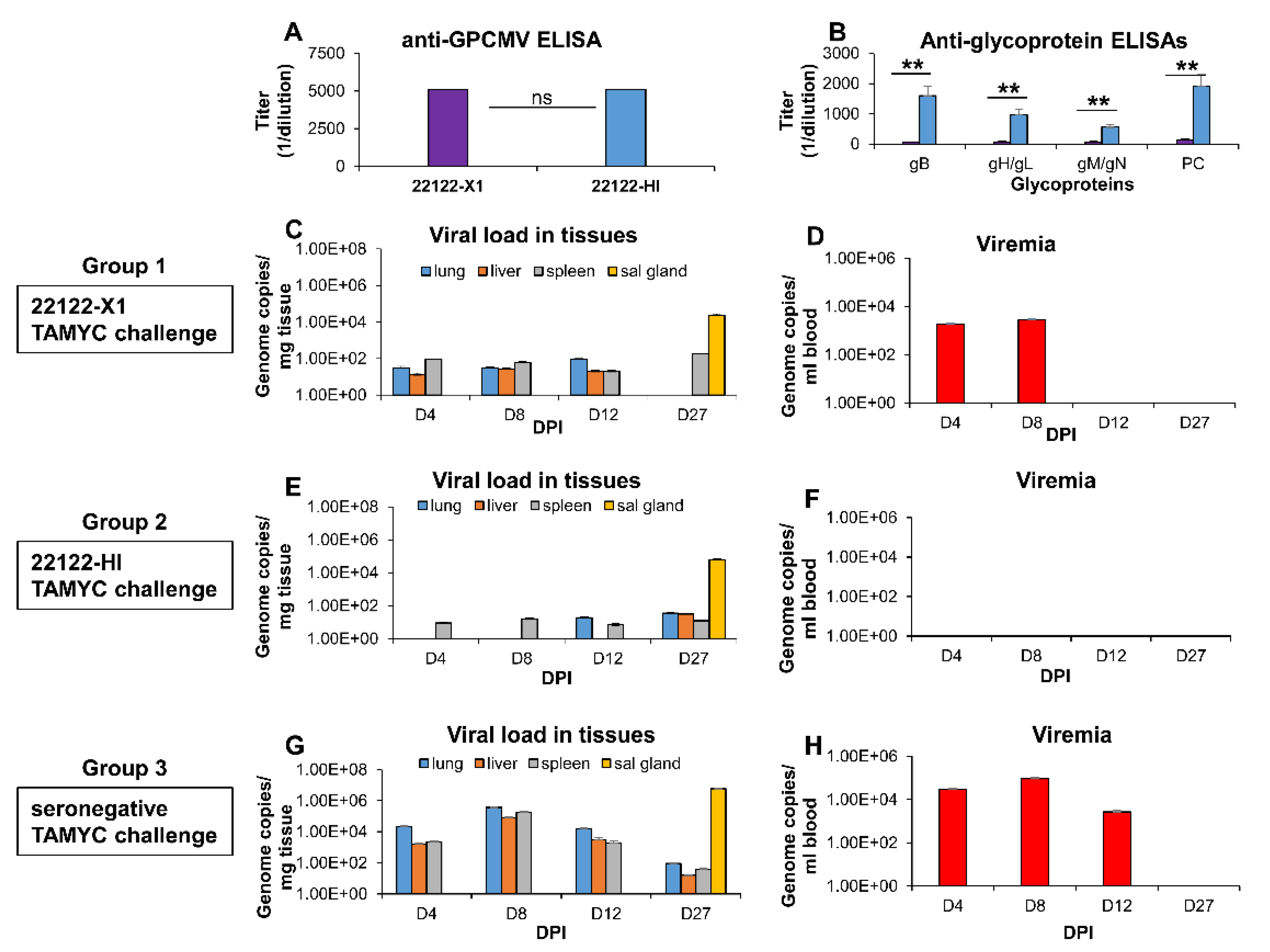
3.3. Limitation of Cross Strain Protection in Animals Convalescent for 22122 Strain
3.4. Comparative Antibody Neutralization of 22122 and TAMYC GPCMV Strains by Hyperimmune Convalescent Sera from Animals (22122 or TAMYC Strain)
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ross, S.A.; Boppana, S.B. Congenital cytomegalovirus infection: Outcome and diagnosis. Semin. Pediatr. Infect. Dis. 2005, 16, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, P.D.; Walter, S. Cytomegalovirus. Curr. Opin. Infect. Dis. 2005, 18, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Fowler, K.B.; Dahle, A.J.; Boppana, S.B.; Pass, R.F. Newborn hearing screening: Will children with hearing loss caused by congenital cytomegalovirus infection be missed? J. Pediatr. 1999, 135, 60–64. [Google Scholar] [CrossRef]
- Manicklal, S.; Emery, V.C.; Lazzarotto, T.; Boppana, S.B.; Gupta, R.K. The “silent” global burden of congenital cytomegalovirus. Clin. Microbiol. Rev. 2013, 26, 86–102. [Google Scholar] [CrossRef]
- Enders, G.; Daiminger, A.; Bader, U.; Exler, S.; Enders, M. Intrauterine transmission and clinical outcome of 248 pregnancies with primary cytomegalovirus infection in relation to gestational age. J. Clin. Virol. 2011, 52, 244–246. [Google Scholar] [CrossRef]
- Fowler, K.B.; Stagno, S.; Pass, R.F. Maternal immunity and prevention of congenital cytomegalovirus infection. JAMA 2003, 289, 1008–1011. [Google Scholar] [CrossRef]
- Sylwester, A.W.; Mitchell, B.L.; Edgar, J.B.; Taormina, C.; Pelte, C.; Ruchti, F.; Sleath, P.R.; Grabstein, K.H.; Hosken, N.A.; Kern, F.; et al. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J. Exp. Med. 2005, 202, 673–685. [Google Scholar] [CrossRef]
- Yue, Y.; Barry, P.A. Rhesus cytomegalovirus a nonhuman primate model for the study of human cytomegalovirus. Adv. Virus Res. 2008, 72, 207–226. [Google Scholar] [CrossRef]
- Griffith, B.P.; McCormick, S.R.; Fong, C.K.; Lavallee, J.T.; Lucia, H.L.; Goff, E. The placenta as a site of cytomegalovirus infection in guinea pigs. J. Virol. 1985, 55, 402–409. [Google Scholar] [CrossRef]
- Woolf, N.K.; Koehrn, F.J.; Harris, J.P.; Richman, D.D. Congenital cytomegalovirus labyrinthitis and sensorineural hearing loss in guinea pigs. J. Infect. Dis. 1989, 160, 929–937. [Google Scholar] [CrossRef]
- Hartley, J.W.; Rowe, W.P.; Huebner, R.J. Serial propagation of the guinea pig salivary gland virus in tissue culture. Proc. Soc. Exp. Biol. Med. 1957, 96, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.Y.; El-Hamdi, N.S.; McGregor, A. Convalescent Immunity to Guinea Pig Cytomegalovirus Induces Limited Cross Strain Protection against Re-Infection but High-Level Protection against Congenital Disease. Int. J. Mol. Sci. 2020, 21, 5997. [Google Scholar] [CrossRef] [PubMed]
- Schleiss, M.R.; Permar, S.R.; Plotkin, S.A. Progress toward Development of a Vaccine against Congenital Cytomegalovirus Infection. Clin. Vaccine Immunol. 2017, 24, e00268-17. [Google Scholar] [CrossRef] [PubMed]
- Tabata, T.; Petitt, M.; Fang-Hoover, J.; Freed, D.C.; Li, F.; An, Z.; Wang, D.; Fu, T.M.; Pereira, L. Neutralizing Monoclonal Antibodies Reduce Human Cytomegalovirus Infection and Spread in Developing Placentas. Vaccines 2019, 7, 135. [Google Scholar] [CrossRef] [PubMed]
- Sandonis, V.; Garcia-Rios, E.; McConnell, M.J.; Perez-Romero, P. Role of Neutralizing Antibodies in CMV Infection: Implications for New Therapeutic Approaches. Trends Microbiol. 2020, 28, 900–912. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.S.; Huffman, T.; Jenks, J.A.; Cisneros de la Rosa, E.; Xie, G.; Vandergrift, N.; Pass, R.F.; Pollara, J.; Permar, S.R. HCMV glycoprotein B subunit vaccine efficacy mediated by nonneutralizing antibody effector functions. Proc. Natl. Acad. Sci. USA 2018, 115, 6267–6272. [Google Scholar] [CrossRef]
- Pass, R.F.; Zhang, C.; Evans, A.; Simpson, T.; Andrews, W.; Huang, M.L.; Corey, L.; Hill, J.; Davis, E.; Flanigan, C.; et al. Vaccine prevention of maternal cytomegalovirus infection. N. Engl. J. Med. 2009, 360, 1191–1199. [Google Scholar] [CrossRef]
- Coleman, S.; Hornig, J.; Maddux, S.; Choi, K.Y.; McGregor, A. Viral Glycoprotein Complex Formation, Essential Function and Immunogenicity in the Guinea Pig Model for Cytomegalovirus. PLoS ONE 2015, 10, e0135567. [Google Scholar] [CrossRef][Green Version]
- Coleman, S.; Choi, K.Y.; Root, M.; McGregor, A. A Homolog Pentameric Complex Dictates Viral Epithelial Tropism, Pathogenicity and Congenital Infection Rate in Guinea Pig Cytomegalovirus. PLoS Pathog. 2016, 12, e1005755. [Google Scholar] [CrossRef]
- Auerbach, M.; Yan, D.; Fouts, A.; Xu, M.; Estevez, A.; Austin, C.D.; Bazan, F.; Feierbach, B. Characterization of the guinea pig CMV gH/gL/GP129/GP131/GP133 complex in infection and spread. Virology 2013, 441, 75–84. [Google Scholar] [CrossRef]
- Choi, K.Y.; El-Hamdi, N.; McGregor, A. Endothelial cell infection by guinea pig cytomegalovirus is a lytic or persistent infection dependent upon tissue origin but requires viral pentamer complex and pp65 tegument protein. J. Virol. 2021. [Google Scholar]
- Coleman, S.; Choi, K.Y.; McGregor, A. Cytomegalovirus UL128 homolog mutants that form a pentameric complex produce virus with impaired epithelial and trophoblast cell tropism and altered pathogenicity in the guinea pig. Virology 2017, 509, 205–221. [Google Scholar] [CrossRef]
- Choi, K.Y.; El-Hamdi, N.S.; McGregor, A. Requirements for guinea pig cytomegalovirus tropism and antibody neutralization on placental amniotic sac cells. J. Gen. Virol. 2020, 101, 426–439. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.Y.; Root, M.; McGregor, A. A Novel Non-Replication-Competent Cytomegalovirus Capsid Mutant Vaccine Strategy Is Effective in Reducing Congenital Infection. J. Virol. 2016, 90, 7902–7919. [Google Scholar] [CrossRef] [PubMed]
- Britt, W.J.; Harrison, C. Identification of an abundant disulfide-linked complex of glycoproteins in the envelope of guinea pig cytomegalovirus. Virology 1994, 201, 294–302. [Google Scholar] [CrossRef]
- Schleiss, M.R.; Jensen, N.J. Cloning and expression of the guinea pig cytomegalovirus glycoprotein B (gB) in a recombinant baculovirus: Utility for vaccine studies for the prevention of experimental infection. J. Virol. Methods 2003, 108, 59–65. [Google Scholar] [CrossRef]
- Choi, K.Y.; El-Hamdi, N.S.; McGregor, A. Neutralizing antibodies to gB based CMV vaccine requires full length antigen but reduced virus neutralization on non-fibroblast cells limits vaccine efficacy in the guinea pig model. Vaccine 2020, 38, 2340–2349. [Google Scholar] [CrossRef]
- Schleiss, M.R.; Bourne, N.; Stroup, G.; Bravo, F.J.; Jensen, N.J.; Bernstein, D.I. Protection against congenital cytomegalovirus infection and disease in guinea pigs, conferred by a purified recombinant glycoprotein B vaccine. J. Infect. Dis. 2004, 189, 1374–1381. [Google Scholar] [CrossRef]
- Hashimoto, K.; Yamada, S.; Katano, H.; Fukuchi, S.; Sato, Y.; Kato, M.; Yamaguchi, T.; Moriishi, K.; Inoue, N. Effects of immunization of pregnant guinea pigs with guinea pig cytomegalovirus glycoprotein B on viral spread in the placenta. Vaccine 2013, 31, 3199–3205. [Google Scholar] [CrossRef]
- Cardin, R.D.; Bravo, F.J.; Pullum, D.A.; Orlinger, K.; Watson, E.M.; Aspoeck, A.; Fuhrmann, G.; Guirakhoo, F.; Monath, T.; Bernstein, D.I. Replication-defective lymphocytic choriomeningitis virus vectors expressing guinea pig cytomegalovirus gB and pp65 homologs are protective against congenital guinea pig cytomegalovirus infection. Vaccine 2016, 34, 1993–1999. [Google Scholar] [CrossRef]
- Swanson, E.C.; Gillis, P.; Hernandez-Alvarado, N.; Fernandez-Alarcon, C.; Schmit, M.; Zabeli, J.C.; Wussow, F.; Diamond, D.J.; Schleiss, M.R. Comparison of monovalent glycoprotein B with bivalent gB/pp65 (GP83) vaccine for congenital cytomegalovirus infection in a guinea pig model: Inclusion of GP83 reduces gB antibody response but both vaccine approaches provide equivalent protection against pup mortality. Vaccine 2015, 33, 4013–4018. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Martin, N.; Marcandalli, J.; Huang, C.S.; Arthur, C.P.; Perotti, M.; Foglierini, M.; Ho, H.; Dosey, A.M.; Shriver, S.; Payandeh, J.; et al. An Unbiased Screen for Human Cytomegalovirus Identifies Neuropilin-2 as a Central Viral Receptor. Cell 2018, 174, 1158–1171.e1119. [Google Scholar] [CrossRef] [PubMed]
- Xiaofei, E.; Meraner, P.; Lu, P.; Perreira, J.M.; Aker, A.M.; McDougall, W.M.; Zhuge, R.; Chan, G.C.; Gerstein, R.M.; Caposio, P.; et al. OR14I1 is a receptor for the human cytomegalovirus pentameric complex and defines viral epithelial cell tropism. Proc. Natl. Acad. Sci. USA 2019, 116, 7043–7052. [Google Scholar] [CrossRef]
- Vanarsdall, A.L.; Pritchard, S.R.; Wisner, T.W.; Liu, J.; Jardetzky, T.S.; Johnson, D.C. CD147 Promotes Entry of Pentamer-Expressing Human Cytomegalovirus into Epithelial and Endothelial Cells. mBio 2018, 9, e00781-18. [Google Scholar] [CrossRef]
- Stein, K.R.; Gardner, T.J.; Hernandez, R.E.; Kraus, T.A.; Duty, J.A.; Ubarretxena-Belandia, I.; Moran, T.M.; Tortorella, D. CD46 facilitates entry and dissemination of human cytomegalovirus. Nat. Commun. 2019, 10, 2699. [Google Scholar] [CrossRef]
- Feire, A.L.; Koss, H.; Compton, T. Cellular integrins function as entry receptors for human cytomegalovirus via a highly conserved disintegrin-like domain. Proc. Natl. Acad. Sci. USA 2004, 101, 15470–15475. [Google Scholar] [CrossRef]
- Wu, Y.; Prager, A.; Boos, S.; Resch, M.; Brizic, I.; Mach, M.; Wildner, S.; Scrivano, L.; Adler, B. Human cytomegalovirus glycoprotein complex gH/gL/gO uses PDGFR-alpha as a key for entry. PLoS Pathog. 2017, 13, e1006281. [Google Scholar] [CrossRef]
- Choi, K.Y.; El-Hamdi, N.S.; McGregor, A. Inclusion of the Viral Pentamer Complex in a Vaccine Design Greatly Improves Protection against Congenital Cytomegalovirus in the Guinea Pig Model. J. Virol. 2019, 93, e01442-19. [Google Scholar] [CrossRef]
- El-Hamdi, N.S.; Choi, K.Y.; McGregor, A. Guinea pig cytomegalovirus trimer complex gH/gL/gO uses PDGFRA as universal receptor for cell fusion and entry. Virology 2020, 548, 236–249. [Google Scholar] [CrossRef]
- Freed, D.C.; Tang, Q.; Tang, A.; Li, F.; He, X.; Huang, Z.; Meng, W.; Xia, L.; Finnefrock, A.C.; Durr, E.; et al. Pentameric complex of viral glycoprotein H is the primary target for potent neutralization by a human cytomegalovirus vaccine. Proc. Natl. Acad. Sci. USA 2013, 110, E4997–E5005. [Google Scholar] [CrossRef]
- Ha, S.; Li, F.; Troutman, M.C.; Freed, D.C.; Tang, A.; Loughney, J.W.; Wang, D.; Wang, I.M.; Vlasak, J.; Nickle, D.C.; et al. Neutralization of Diverse Human Cytomegalovirus Strains Conferred by Antibodies Targeting Viral gH/gL/pUL128-131 Pentameric Complex. J. Virol. 2017, 91, e02033-16. [Google Scholar] [CrossRef] [PubMed]
- Chiuppesi, F.; Wussow, F.; Johnson, E.; Bian, C.; Zhuo, M.; Rajakumar, A.; Barry, P.A.; Britt, W.J.; Chakraborty, R.; Diamond, D.J. Vaccine-Derived Neutralizing Antibodies to the Human Cytomegalovirus gH/gL Pentamer Potently Block Primary Cytotrophoblast Infection. J. Virol. 2015, 89, 11884–11898. [Google Scholar] [CrossRef] [PubMed]
- Wills, M.R.; Mason, G.M.; Sissons, J.G.P. Adaptive Cellular Immunity to Human Cytomegalovirus. In Cytomegaloviruses: From Molecular Pathogenesis to Intervention; Reddehase, M.J., Ed.; Caister Academic Press: Norfolk, UK, 2013; Volume II, pp. 142–172. [Google Scholar]
- McGregor, A.; Liu, F.; Schleiss, M.R. Molecular, biological, and in vivo characterization of the guinea pig cytomegalovirus (CMV) homologs of the human CMV matrix proteins pp71 (UL82) and pp65 (UL83). J. Virol. 2004, 78, 9872–9889. [Google Scholar] [CrossRef]
- Schleiss, M.; Lacayo, J.; Belkaid, Y.; McGregor, A.; Stroup, G.; Rayner, J.; Alterson, K.; Chulay, J.; Smith, J. Preconceptual administration of an alphavirus replicon UL83 (pp65 homolog) vaccine induces humoral and cellular immunity and improves pregnancy outcome in the guinea pig model of congenital cytomegalovirus infection. J. Infect. Dis. 2007, 195, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.Y.; El-Hamdi, N.; Hornig, J.; McGregor, A. Guinea pig cytomegalovirus protective T cell antigen GP83 is a functional pp65 homolog for innate immune evasion and pentamer dependent virus tropism. J. Virol. 2021, 95, e00324-21. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Freed, D.C.; He, X.; Li, F.; Tang, A.; Cox, K.S.; Dubey, S.A.; Cole, S.; Medi, M.B.; Liu, Y.; et al. A replication-defective human cytomegalovirus vaccine for prevention of congenital infection. Sci. Transl. Med. 2016, 8, 362ra145. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.Y.; El-Hamdi, N.S.; McGregor, A. A trimeric capable gB CMV vaccine provides limited protection against a highly cell associated and epithelial tropic strain of cytomegalovirus in guinea pigs. J. Gen. Virol. 2021, 102, 001579. [Google Scholar] [CrossRef]
- Yue, Y.; Zhou, S.S.; Barry, P.A. Antibody responses to rhesus cytomegalovirus glycoprotein B in naturally infected rhesus macaques. J. Gen. Virol. 2003, 84, 3371–3379. [Google Scholar] [CrossRef]
- Ryckman, B.J.; Chase, M.C.; Johnson, D.C. HCMV gH/gL/UL128-131 interferes with virus entry into epithelial cells: Evidence for cell type-specific receptors. Proc. Natl. Acad. Sci. USA 2008, 105, 14118–14123. [Google Scholar] [CrossRef]
- McGregor, A.; Choi, K.Y.; Schleiss, M.R. Guinea pig cytomegalovirus GP84 is a functional homolog of the human cytomegalovirus (HCMV) UL84 gene that can complement for the loss of UL84 in a chimeric HCMV. Virology 2011, 410, 76–87. [Google Scholar] [CrossRef]
- Zhou, M.; Yu, Q.; Wechsler, A.; Ryckman, B.J. Comparative analysis of gO isoforms reveals that strains of human cytomegalovirus differ in the ratio of gH/gL/gO and gH/gL/UL128-131 in the virion envelope. J. Virol. 2013, 87, 9680–9690. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, M.; Stanton, R.; Kamil, J.; Ryckman, B.J. Expression Levels of Glycoprotein O (gO) Vary between Strains of Human Cytomegalovirus, Influencing the Assembly of gH/gL Complexes and Virion Infectivity. J. Virol. 2018, 92, e00606-18. [Google Scholar] [CrossRef] [PubMed]
- Day, L.Z.; Stegmann, C.; Schultz, E.P.; Lanchy, J.M.; Yu, Q.; Ryckman, B.J. Polymorphisms in Human Cytomegalovirus Glycoprotein O (gO) Exert Epistatic Influences on Cell-Free and Cell-to-Cell Spread and Antibody Neutralization on gH Epitopes. J. Virol. 2020, 94, e02051-19. [Google Scholar] [CrossRef] [PubMed]
- Lowen, A.C.; Mubareka, S.; Tumpey, T.M.; Garcia-Sastre, A.; Palese, P. The guinea pig as a transmission model for human influenza viruses. Proc. Natl. Acad. Sci. USA 2006, 103, 9988–9992. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.M.; McWhorter, A.R.; Masters, L.L.; Shellam, G.R.; Redwood, A.J. Laboratory strains of murine cytomegalovirus are genetically similar to but phenotypically distinct from wild strains of virus. J. Virol. 2008, 82, 6689–6696. [Google Scholar] [CrossRef]
- Wagner, F.M.; Brizic, I.; Prager, A.; Trsan, T.; Arapovic, M.; Lemmermann, N.A.; Podlech, J.; Reddehase, M.J.; Lemnitzer, F.; Bosse, J.B.; et al. The viral chemokine MCK-2 of murine cytomegalovirus promotes infection as part of a gH/gL/MCK-2 complex. PLoS Pathog. 2013, 9, e1003493. [Google Scholar] [CrossRef] [PubMed]
- Roark, H.K.; Jenks, J.A.; Permar, S.R.; Schleiss, M.R. Animal Models of Congenital Cytomegalovirus Transmission: Implications for Vaccine Development. J. Infect. Dis. 2020, 221, S60–S73. [Google Scholar] [CrossRef] [PubMed]
- Taher, H.; Mahyari, E.; Kreklywich, C.; Uebelhoer, L.S.; McArdle, M.R.; Mostrom, M.J.; Bhusari, A.; Nekorchuk, M.; E, X.; Whitmer, T.; et al. In vitro and in vivo characterization of a recombinant rhesus cytomegalovirus containing a complete genome. PLoS Pathog. 2020, 16, e1008666. [Google Scholar] [CrossRef]
- Nelson, C.S.; Cruz, D.V.; Tran, D.; Bialas, K.M.; Stamper, L.; Wu, H.; Gilbert, M.; Blair, R.; Alvarez, X.; Itell, H.; et al. Preexisting antibodies can protect against congenital cytomegalovirus infection in monkeys. JCI Insight 2017, 2, e94002. [Google Scholar] [CrossRef]
- Revello, M.G.; Lazzarotto, T.; Guerra, B.; Spinillo, A.; Ferrazzi, E.; Kustermann, A.; Guaschino, S.; Vergani, P.; Todros, T.; Frusca, T.; et al. A randomized trial of hyperimmune globulin to prevent congenital cytomegalovirus. N. Engl. J. Med. 2014, 370, 1316–1326. [Google Scholar] [CrossRef]
- Hughes, B.L.; Clifton, R.G.; Rouse, D.J.; Saade, G.R.; Dinsmoor, M.J.; Reddy, U.M.; Pass, R.; Allard, D.; Mallett, G.; Fette, L.M.; et al. A Trial of Hyperimmune Globulin to Prevent Congenital Cytomegalovirus Infection. N. Engl. J. Med. 2021, 385, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Itell, H.L.; Kaur, A.; Deere, J.D.; Barry, P.A.; Permar, S.R. Rhesus monkeys for a nonhuman primate model of cytomegalovirus infections. Curr. Opin. Virol. 2017, 25, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Valencia, S.; Gill, R.B.; Dowdell, K.C.; Wang, Y.; Hornung, R.; Bowman, J.J.; Lacayo, J.C.; Cohen, J.I. Comparison of vaccination with rhesus CMV (RhCMV) soluble gB with a RhCMV replication-defective virus deleted for MHC class I immune evasion genes in a RhCMV challenge model. Vaccine 2019, 37, 333–342. [Google Scholar] [CrossRef]
- Hornig, J.; Choi, K.Y.; McGregor, A. The essential role of guinea pig cytomegalovirus (GPCMV) IE1 and IE2 homologs in viral replication and IE1-mediated ND10 targeting. Virology 2017, 504, 122–140. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.Y.; McGregor, A. A Fully Protective Congenital CMV Vaccine Requires Neutralizing Antibodies to Viral Pentamer and gB Glycoprotein Complexes but a pp65 T-Cell Response Is Not Necessary. Viruses 2021, 13, 1467. [Google Scholar] [CrossRef] [PubMed]
- Abel, K.; Strelow, L.; Yue, Y.; Eberhardt, M.K.; Schmidt, K.A.; Barry, P.A. A heterologous DNA prime/protein boost immunization strategy for rhesus cytomegalovirus. Vaccine 2008, 26, 6013–6025. [Google Scholar] [CrossRef]
- Gerna, G.; Revello, M.G.; Baldanti, F.; Percivalle, E.; Lilleri, D. The pentameric complex of human Cytomegalovirus: Cell tropism, virus dissemination, immune response and vaccine development. J. Gen. Virol. 2017, 98, 2215–2234. [Google Scholar] [CrossRef]
- Schleiss, M.R.; Buus, R.; Choi, K.Y.; McGregor, A. An Attenuated CMV Vaccine with a Deletion in Tegument Protein GP83 (pp65 Homolog) Protects against Placental Infection and Improves Pregnancy Outcome in a Guinea Pig Challenge Model. Future Virol. 2013, 8, 1151–1160. [Google Scholar] [CrossRef]
- Liu, Y.; Freed, D.C.; Li, L.; Tang, A.; Li, F.; Murray, E.M.; Adler, S.P.; McVoy, M.A.; Rupp, R.E.; Barrett, D.; et al. A Replication-Defective Human Cytomegalovirus Vaccine Elicits Humoral Immune Responses Analogous to Those with Natural Infection. J. Virol. 2019, 93, e00747-19. [Google Scholar] [CrossRef]
- Thomas, M.; Kropff, B.; Schneider, A.; Winkler, T.H.; Gorzer, I.; Sticht, H.; Britt, W.J.; Mach, M.; Reuter, N. A Novel Strain-Specific Neutralizing Epitope on Glycoprotein H of Human Cytomegalovirus. J. Virol. 2021, 95, e0065721. [Google Scholar] [CrossRef]
- Wussow, F.; Yue, Y.; Martinez, J.; Deere, J.; Longmate, J.; Herrmann, A.; Barry, P.; Diamond, D. A vaccine based on the rhesus cytomegalovirus UL128 complex induces broadly neutralizing antibodies in rhesus macaques. J. Virol. 2013, 87, 1322–1332. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Ye, X.; Freed, D.C.; Li, L.; Ku, Z.; Xiong, W.; Gao, P.; Liu, X.; Montgomery, D.; Xu, W.; et al. Potent Bispecific Neutralizing Antibody Targeting Glycoprotein B and the gH/gL/pUL128/130/131 Complex of Human Cytomegalovirus. Antimicrob. Agents Chemother. 2021, 65, e02422-20. [Google Scholar] [CrossRef] [PubMed]
- Hansen, S.G.; Powers, C.J.; Richards, R.; Ventura, A.B.; Ford, J.C.; Siess, D.; Axthelm, M.K.; Nelson, J.A.; Jarvis, M.A.; Picker, L.J.; et al. Evasion of CD8+ T cells is critical for superinfection by cytomegalovirus. Science 2010, 328, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Abel, K.; Martinez, J.; Yue, Y.; Lacey, S.F.; Wang, Z.; Strelow, L.; Dasgupta, A.; Li, Z.; Schmidt, K.A.; Oxford, K.L.; et al. Vaccine-induced control of viral shedding following rhesus cytomegalovirus challenge in rhesus macaques. J. Virol. 2011, 85, 2878–2890. [Google Scholar] [CrossRef]
- Biolatti, M.; Dell’Oste, V.; Pautasso, S.; Gugliesi, F.; von Einem, J.; Krapp, C.; Jakobsen, M.R.; Borgogna, C.; Gariglio, M.; De Andrea, M.; et al. Human Cytomegalovirus Tegument Protein pp65 (pUL83) Dampens Type I Interferon Production by Inactivating the DNA Sensor cGAS without Affecting STING. J. Virol. 2018, 92, e01774-17. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, K.Y.; El-Hamdi, N.S.; McGregor, A. Cross Strain Protection against Cytomegalovirus Reduces DISC Vaccine Efficacy against CMV in the Guinea Pig Model. Viruses 2022, 14, 760. https://doi.org/10.3390/v14040760
Choi KY, El-Hamdi NS, McGregor A. Cross Strain Protection against Cytomegalovirus Reduces DISC Vaccine Efficacy against CMV in the Guinea Pig Model. Viruses. 2022; 14(4):760. https://doi.org/10.3390/v14040760
Chicago/Turabian StyleChoi, K. Yeon, Nadia S. El-Hamdi, and Alistair McGregor. 2022. "Cross Strain Protection against Cytomegalovirus Reduces DISC Vaccine Efficacy against CMV in the Guinea Pig Model" Viruses 14, no. 4: 760. https://doi.org/10.3390/v14040760
APA StyleChoi, K. Y., El-Hamdi, N. S., & McGregor, A. (2022). Cross Strain Protection against Cytomegalovirus Reduces DISC Vaccine Efficacy against CMV in the Guinea Pig Model. Viruses, 14(4), 760. https://doi.org/10.3390/v14040760





