Abstract
Numerous large DNA viruses have evolved sophisticated countermeasures to hijack the premature programmed cell death of host cells post-infection, including the expression of proteins homologous in sequence, structure, or function to cellular Bcl-2 proteins. Kaposi sarcoma herpes virus (KSHV), a member of the gammaherpesvirinae, has been shown to encode for KsBcl-2, a potent inhibitor of Bcl-2 mediated apoptosis. KsBcl-2 acts by directly engaging host pro-apoptotic Bcl-2 proteins including Bak, Bax and Bok, the BH3-only proteins; Bim, Bid, Bik, Hrk, Noxa and Puma. Here we determined the crystal structures of KsBcl-2 bound to the BH3 motif of pro-apoptotic proteins Bid and Puma. The structures reveal that KsBcl-2 engages pro-apoptotic BH3 motif peptides using the canonical ligand binding groove. Thus, the presence of the readily identifiable conserved BH1 motif sequence “NWGR” of KsBcl-2, as well as highly conserved Arg residue (R86) forms an ionic interaction with the conserved Asp in the BH3 motif in a manner that mimics the canonical ionic interaction seen in host Bcl-2:BH3 motif complexes. These findings provide a structural basis for KSHV mediated inhibition of host cell apoptosis and reveal the flexibility of virus encoded Bcl-2 proteins to mimic key interactions from endogenous host signalling pathways.
1. Introduction
Programmed cell death forms part of a suite of immune response strategies utilized by multicellular organisms to eliminate invading pathogens [1]. In order to counter host cell death-based defence mechanisms viruses have co-evolved genes to either counter or enhance host programmed cell death to ensure their own proliferation and survival [2]. One such strategy for viral manipulation of the host-cell programmed cell death response has been the acquisition of pro-survival homologs of the B-cell lymphoma 2 (Bcl-2) family that mediate intrinsic or mitochondrial initiated apoptosis [3]. Bcl-2 proteins are critical arbiters of intrinsic apoptosis and are characterized by the presence of one or more conserved Bcl-2 homology (BH) sequence motifs and a unique Bcl-2 fold, though the viral Bcl-2 homologs frequently lack any sequence conservation with their mammalian counterparts, the protein fold is maintained [4,5].
The mammalian Bcl-2 family can be divided into pro-survival or pro-apoptotic members. Mammalian pro-survival members comprise Bcl-2, Bcl-xL, Mcl-1, Bcl-w, A1 and Bcl-B all of which harbor multiple BH motifs in addition to a C-terminal transmembrane region that targets them to the outer mitochondrial membrane [4,6]. The other members of the family are pro-apoptotic Bcl-2 proteins, and these are subdivided into two groups: those that feature multiple BH motifs such as Bak, Bax and Bok; and others that contain only the BH3 motif and are denoted as BH3-only proteins [7]. The mammalian BH3-only proteins include Bad, Bid, Bik, Bim, Bmf, Hrk, Noxa and Puma, and act either by directly activating Bak and Bax, or they counter the ability of pro-survival Bcl-2 proteins to hold Bak and Bax in check [8]. A number of herpes viruses have acquired pro-survival Bcl-2 homologs among their survival genes [9], such as Epstein–Barr virus (EBV) encoded BHRF-1 [10,11] and BALF-1 [12], Kaposi-sarcoma associated herpesvirus (KSHV) apoptosis regulator KsBcl-2 [13], Herpesvirus saimiri Bcl-2 homolog HVS-1 ORF16 [14], Turkey herpesvirus encoded HVT079 [15,16] and murine gammaherpesvirus 68 encoded M11 [17].
Interactions between pro-survival Bcl-2 proteins and the BH3 motif of pro-apoptotic proteins occur via a conserved hydrophobic ligand binding groove on the pro-survival Bcl-2 protein neutralising their pro-survival activity allowing pro-apoptotic Bcl-2 proteins to initiate apoptosis. Key contributors to the cell death are the pro-apoptotic Bax and Bak proteins. After activation, Bax and Bak multimerize on the mitochondrial outer membrane to form large pores [18] that results in the release of pro-apoptotic factors including cytochrome c. Cytochrome c together with the adapter protein APAF-1 forms the apoptosome [19], which provides a platform for initiator caspases and enables activation of downstream executioner caspases and subsequent dismantling of the cell. The presence of pro-survival viral Bcl-2 homologs inhibits the action of Bax and Bak to preserve the cell for viral replication [20].
KSHV or human herpesvirus-8 (HHV8) is an oncogenic virus of the herpesviridae family belonging to the subfamily gammaherpesvirinae and genus rhadinovirus. Other members of rhadinovirus include the closely related herpesvirus saimiri as well as murine gammaherpesvirus 68 and Macaca mulatta rhadinovirus. While oncogenic EBV belongs to the same subfamily of gammaherpesvirinae, it is not a member of the genus rhadinovirus. KSHV primarily infects host B-lymphocytes and endothelial cells causing a lifelong latent infection in humans. KSHV is a large linear double stranded DNA virus whose genome size is about ~165 kbp [21,22] that encodes a number of immunomodulatory genes [22,23] including a homolog of the cellular Bcl-2 protein, KsBcl-2 [13,24]. KsBcl-2 lacks any significant shared sequence identity with mammalian Bcl-2 proteins [13] though it does contain a recognizable “NWGR” sequence motif in the BH1 region, which is crucial for pro-survival function of majority of Bcl-2 family proteins. The highly conserved arginine of the “NWGR” sequence makes contact with the BH3 motif of pro-apoptotic Bcl-2 proteins. However, this interaction does not lead to heterodimerization with pro-apoptotic Bcl-2 proteins Bak and Bax at the mitochondrial outer membrane [13]. Furthermore, the ksbcl-2 [orf16] gene is expressed in the viral lytic replication cycle [13] and inhibits both cellular apoptosis and autophagy when transiently expressed in cells as well as in in-vitro studies [25,26], similar to that previously observed for murine gammaherpesvirus 68 protein M11 [17,25,26]. In addition, KsBcl-2 is an essential element in KSHV replication, and the virus cannot complete its lytic replication cycle, proliferate and reactivate when mutated [27,28,29]. Moreover, KsBcl-2 is important for virion assembly via tegument protein (ORF55) through its N-terminal amino acid residues 11–20 [30]. Here, we report the structural and biochemical characterization of KSHV encoded KsBcl-2, and its potential role in modulating apoptosis in humans. KsBcl-2 shares 17% sequence identity with its EBV homolog BHRF-1. KsBcl-2 is able to bind to peptides spanning the BH3 motif of human pro-apoptotic Bcl-2 proteins including those of Bax, Bak, Bok, Bim, Bid, Bik, Hrk, Noxa and Puma with a wide range of affinities from nanomolar to sub-micromolar range. We also show KsBcl-2 adopts a globular conserved monomeric Bcl-2-fold structure similar to other characterized herpes virus encoded Bcl-2 homologs and binds BH3 motif peptides using the canonical Bcl-2 ligand binding groove. Combined, these findings establish KsBcl-2 as a potent inhibitor of host pro-apoptotic proteins.
2. Materials and Methods
Synthetic cDNA codon optimized for Escherichia coli encoding for KsBcl-2 mutant V117A from the Kaposi’s sarcoma-associated herpesvirus lacking the C-terminal 29 residues (spanning residues 1–146) was cloned into the bacterial expression vector pGEX-6P3 (Genscript) and transformed into E. coli Codon plus (RIL). Cells were grown in 2YT medium containing 1 μg mL−1 ampicillin at 37 °C in a shaker incubator until an OD600 of 0.6 was reached, and KsBcl-2 expression was induced by adding isopropyl β-D-1-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM for 20 h at 18 °C. Cells were harvested by ultracentrifugation at 5000 rpm (JLA 9.1000 rotor, Beckman Coulter Avanti J-E) for 20 min prior to resuspension in 100 mL lysis buffer A (50 mM Tris pH 8.0, 300 mM NaCl and 5 mM DTT (dithiothreitol)). Harvested cells were lysed via sonication (Model 705 Sonic Dismembrator, Fisher Scientific, Hampton, NH, USA) with 6 s pulses up to 1.5 min with 1 min pulse off time between each pulse at an amplitude of 60. The resultant lysate was transferred to SS34 tubes for centrifugation at 18,000 rpm (JA-25.50 rotor, Beckman Coulter Avanti J-E) for 30 min. The supernatant was decanted and loaded onto 2 mL of Glutathione Sepharose 4B resin in a gravity flow column (GE Healthcare) equilibrated with buffer A. Following sample application, the column was washed with 100 mL of buffer A followed by HRV 3C protease cleavage overnight at 4 °C in the presence of 25 mL of buffer A (Addition of excess buffer is essential to stop the after-cleavage aggregation of protein). The liberated target protein was eluted using 30 mL of buffer A and concentrated to a 1.0 mg mL−1 using a centrifugal concentrator with a 3 kDa molecular weight cut-off (Amicon® Ultra 15). Concentrated KsBcl-2 protein was subjected to multiple size-exclusion chromatography runs using a Superdex S75 16/60 column mounted on an ÄKTAExpress system (GE Healthcare) equilibrated in 25 mM Tris-HCl pH 8.5, 200 mM NaCl, 5 mM TCEP (Tris(2-carboxyethyl)phosphine hydrochloride), where it eluted as a single peak in the volume corresponding to a monomeric species. The final sample purity was estimated to be higher than 95% based on SDS–PAGE analysis.
2.1. Measurement of Dissociation Constants
All affinity measurements were performed in triplicate. Protein concentrations were measured using UV spectrophotometer (Nanodrop, Thermo Scientific) at a wavelength of 280 nm. Peptide concentrations were calculated based on dry peptide weight after synthesis. BH3 domain peptides used were commercially synthesized using liquid phase peptide synthesis (Genscript) and purified to a final purity of 95%, and comprised the following sequences [31]: hsBim (UniProtID: O43521-3 51-DMRPEIWIAQELRRIGDEFNAYYARR-76), hsBak (UniProtID: Q16611-1 67-PSSTMGQVGRQLAIIGDDINRRYDSE-92), hsBax (UniProtID: Q07812-1 50-VPQDASTKKLSECLKRIGDELDSNMELQ-77), hsPuma (UniProtID: Q9BXH1-1 130-EEQWAREIGAQLRRMADDLNAQYERR-155), hsNoxa (UniProtID: Q13794-1 18-PAELEVECATQLRRFGDKLNFRQKLL-43), hsBad (UniProtID: Q92934-1 103-NLWAAQRYGRELRRMSDEFVDSFKKG-128), hsBid (UniProtID: P55957-179-SESQEDIIRNIARHLAQVGDSMDRSIPPGLVNGL-104), hsBik (UniProtID: Q13323-1 51-MEGSDALALRLACIGDEMDVSLRAP-75), hsHrk (UniProtID: O00198-1 26-RSSAAQLTAARLKAIGDELHQRTMWR-51), hsBmf (UniProtID: Q96LC9-1 125-QHQAEVQIARKLQCIADQFHRLHVQQ-151), hsBok: 59-VPGRLAEVCAVLLRLGDELE MIRPSV-84 (UniProtID Q9UMX3). Solution competition assays measuring dissociation constants using a T200 Biacore optical biosensor using BH3 domain peptides and 10 nM KsBcl-2 were performed as previously described [32] and are summarized in Table 1. Assays were performed at room temperature with 0.15 M NaCl, 3 mM EDTA, 0.005% Surfactant P20, 5% (v/v) DMSO, 0.01 M Hepes pH 7.4 as the running buffer. Bim BH3 peptide was immobilized on CM5 sensorchips using amine-coupling chemistry. Recombinant KsBcl-2 (10 nM) was pre-incubated on ice with varying concentrations of competitor BH3 peptides then injected at the flow rate of 10 uL/min to determine IC50 values. All BH3 domain peptides were prepared in running buffer.
2.2. Crystallization and Data Collection
A complex of KsBcl-2 with Bid BH3 was reconstituted as previously described [33] by adding Bid BH3-motif peptide at a 1:1.25 molar ratio to KsBcl-2. The reconstituted complex was concentrated to 4.5 mg mL−1 using a centrifugal concentrator with a 3 kDa molecular weight cut-off (Amicon® Ultra 0.5). Crystals of KsBcl-2: Bid BH3 were obtained at a protein concentration of 4.5 mg mL−1 using the sitting-drop method at 20 °C in 0.1 M Sodium acetate pH 4.6, 8% w/v PEG 4000. The crystals were KsBcl-2: Bid BH3complex and the hexagonal shaped crystals belonging to space group P64 with a = 89.42 Å, b = 89.42 Å, c = 50.98 Å, α = 90.00 Å, β = 90.00 Å, γ = 120.00 Å in the hexagonal crystal system. Diffraction data were collected on the MX2 beamline at the Australian Synchrotron using an EIGER 16 M detector at a wavelength of 0.9537 Å and an oscillation range of 0.1° per frame [34]. Diffraction data were integrated using DIALS [35] and scaled using AIMLESS [36]. Crystals of KsBcl-2 with Bid BH3 contained one chain of KsBcl-2 and one chain of Bid BH3 in the asymmetric unit, with a calculated solvent content of 46.12%. The structure was phased by molecular replacement using an alphafold [37] generated model for KsBcl-2 as a search model. The final KsBcl-2:Bid BH3 complex was manually built using Coot [38] and refined using PHENIX [39] with final Rwork/Rfree of 20.1/22.9%, with 98.1% of residues in Ramachandran favoured region of the plot and no outliers.
KsBcl-2: Puma BH3 crystals were grown as described above for the KsBcl-2: Bid BH3 complex. Crystals were obtained in 0.2 M sodium bromide, 0.1 M Bis-tris propane pH 6.5, 20% PEG 3350. The crystals were flash cooled at −173 °C in mother liquor supplemented with 20% ethylene glycol as a cryo-protectant. The KsBcl-2: Puma BH3 complex formed single thin long needle shaped (20 um, 2 um, 1 um) crystals belong to space group C2 with a = 59.22 Å, b = 48.52 Å, c = 57.38 Å, α = 90.00 Å, β = 97.06 Å, γ = 90.00 Å in the monoclinic crystal system. Diffraction data collection, integration and scaling were performed as described above. The molecular replacement was carried out using PHASER with the previously solved structure of KsBcl-2:Bid BH3 as a search model. KsBcl-2: Puma BH3 crystals contain one molecule of KsBcl-2 and one Puma BH3 peptide, with 39.3% solvent content and final TFZ and LLG values of 13.6 and 276 respectively. The final model of KsBcl-2: Puma BH3 was built manually over several cycles using Coot and refined using PHENIX with final Rwork/Rfree of 23.8/26.6% and 98.3% of residues in Ramachandran favoured region of the plot and no outliers. Details of the data-collection and refinement statistics are summarized in Table 2. All images were generated using PyMOL. All software was accessed via SBGrid [40]. Raw images were deposited with the SBGrid Data Bank [41].
2.3. Sequence Search, Alignment, and Interface Analysis
Sequence alignments were performed using MUSCLE [42] with the default settings, and sequence identities were calculated based on the total number of conserved residues in KsBcl-2 against the full sequence. Protein sequences were sourced from Uniprot and have the following accession codes: KSHV KsBcl-2 (UniProt accession number Q76RI8), human Mcl-1 (UniProt Q07820) EBV BHRF1 (UniProt P03182), Zebrafish pro-survival Bcl-2 protein NRZ1 (UniProt Q8UWD5). Protein interfaces were analysed using PISA [43].
2.4. Dali 3D Structure Analysis
To identify structural homologs of KsBcl-2, we performed a full search using the Dali webserver (http://ekhidna2.biocenter.helsinki.fi/dali/) [44].
3. Results
To determine whether KSHV KsBcl-2 is able to interfere with Bcl-2 mediated apoptosis by engaging host proapoptotic Bcl-2, we expressed and purified recombinant KsBcl-2 lacking the C-terminal 29 residues comprising the transmembrane motif. We then examined the ability of recombinant KsBcl-2 to bind BH3 motif peptides from all human proapoptotic Bcl-2 proteins using surface plasmon resonance (SPR). SPR revealed a broad interaction spectrum for KsBcl-2 with nanomolar affinities for BH3 motifs from human Bax, Bak, Bok, Bim, Bid, Bik, Noxa, Puma and Hrk with affinities for the BH3-motifs from Bad and Bmf are higher than the 2 mM limit of detection (Table 1). A comparison of the relative affinities of BH3-peptides for KsBcl-2 and the closely related EBV BHRF1 [45], cellular homologue Mcl-1 [46] and fish homologue NRZ [47] is given in Table 3. KsBcl-2 has a much broader spectrum of BH3 binding than EBV BHRF-1 and a similar BH3-binding spectrum as Mcl-1 and NRZ. To establish the structural basis for BH3 ligand engagement by KsBcl-2 we then determined the crystal structure of KsBcl-2 complexes with two of the identified high affinity interactors, Bid and Puma BH3 (Figure 1). The KsBcl-2:Bid complex was refined to 1.41 Å, whereas the KsBcl-2:Puma complex was refined to 2.11 Å. Clear and continuous electron density was observed for KsBcl-2 residues 1–146 and Bid residues 79–104 in the KsBcl-2: Bid BH3 complex, as well as KsBcl-2 residues 1–146 and Puma residues 130–155 in the KsBcl-2: Puma BH3 complex (Figure 2). The crystal structure of a KsBcl-2: Bid BH3 complex revealed that as previously seen for the related BHRF-1 and cellular Mcl-1, KsBcl-2 adopts a typical Bcl-2 fold with globular monomeric topology with eight alpha-helices and alpha-helices 2–5 form the canonical hydrophobic ligand binding groove.

Table 1.
Binding affinities determined by Surface Plasmon Resonance of recombinant KsBcl-2 homolog KsBcl-2 with pro-apoptotic BH3 motif.
Table 1.
Binding affinities determined by Surface Plasmon Resonance of recombinant KsBcl-2 homolog KsBcl-2 with pro-apoptotic BH3 motif.
| Peptide | IC50 (nM) |
|---|---|
| Bak | 8 ± 0 |
| Bax | 16 ± 0 |
| Bok | 198 ± 13 |
| Bad | NB |
| Bid | 20 ± 9 |
| Bik | 64 ± 4 |
| Bim | 4 ± 1 |
| Bmf | 115 ± 10 |
| Hrk | 115 ± 10 |
| Puma | 26 ± 9 |
All BH3-peptides were 26-mers, except for a Bax BH3 28-mer and a Bid BH3 34-mer) peptides from human. All IC50 values (half maximal inhibitory constant in nM) are the means of three replicates with standard error. NB denotes no binding.

Table 2.
X-ray diffraction data collection and refinement statistics.
Table 2.
X-ray diffraction data collection and refinement statistics.
| KsBcl-2: Bid BH3 (PDB ID: 7QTW) | KsBcl-2: Puma BH3 (PDB ID: 7QTX) | |
|---|---|---|
| Data Collection | ||
| Space group | P64 | C2 |
| Cell dimensions | ||
| a, b, c (Å) | 89.48, 89.48, 51.06 | 59.23, 48.53, 57.39 |
| α, β, γ (°) | 90, 90, 120 | 90, 97.07, 90 |
| Wavelength (Å) | 0.9537 | 0.9537 |
| Resolution (Å) | 29.29–1.41 (1.46–1.41) * | 32.46–2.11 (8.97–2.11) * |
| Rsym or Rmerge | 0.12 (1.32) * | 0.13 (0.81) * |
| I/σI | 8.7 (0.2) * | 4.2 (0.8) * |
| Completeness (%) | 100.0 (99.6) * | 98.1 (96.1) * |
| CC1/2 | 0.99 (0.30) * | 0.98 (0.38) * |
| Redundancy | 19.3 (11.5) * | 2.4 (2.4) * |
| Refinement | ||
| Resolution (Å) | 29.29–1.41 (1.46–1.41) * | 32.46–2.11 (8.97–2.11) * |
| No. reflections | 44,973 | 9148 |
| Rwork/Rfree | 0.180/0.215 | 0.217/0.257 |
| Clashscore | 1.82 | 0.76 |
| No. atoms | ||
| Protein | 1357 | 1310 |
| Ligand/ion | 40 | 15 |
| Water | 148 | 95 |
| B-factors | ||
| Protein | 36.31 | 39.03 |
| Ligand/ion | 61.94 | 79.18 |
| Water | 49.45 | 42.92 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.013 | 0.002 |
| Bond angles (°) | 1.13 | 0.45 |
* Values in parentheses are for the highest resolution shell.
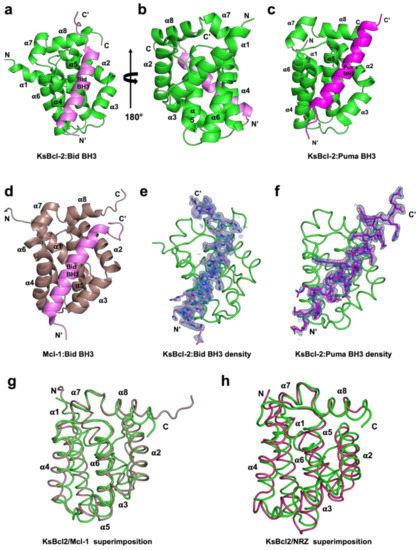
Figure 1.
KsBcl-2 binds BH3 motif peptides of pro-apoptotic Bcl-2 proteins using the canonical ligand binding groove. Crystal structures of KsBcl-2 bound to Bid and Puma BH3 motifs. (a) KsBcl-2 (green) in complex with the Bid BH3 motif (magenta). KsBcl-2 helices are labelled α1–α8. The view is of the hydrophobic binding groove of one protomer formed by helices α3–α5, and (b) is the viewed along the 2-fold symmetry axis (rotated 180 degrees). (c) KsBcl-2 (green) in complex with the Puma BH3 domain (magenta) (d) human Mcl-1 (raspberry) in complex with the Bid BH3 motif (magenta). (e) 2Fo-Fc polder electron density map of KsBcl-2: Bid where electron density for Bid BH3 motif peptide shown in blue interfaces contoured at 1.5·σ (f) 2Fo-Fc polder electron density map of KsBcl-2: Puma complex shown as similar to (d). (g) structural superimposition of backbone of KsBcl-2 (green) onto human Mcl-1 (raspberry). The view is into the canonical hydrophobic binding groove formed by α2–α5. (h) structural superimposition of backbone of KsBcl-2 (green) onto zebrafish pro-survival Bcl-2 protein NRZ (warm pink) Images were generated using the PYMOL Molecular Graphics System, Version 1.8 Schrodinger, LLC. 6.
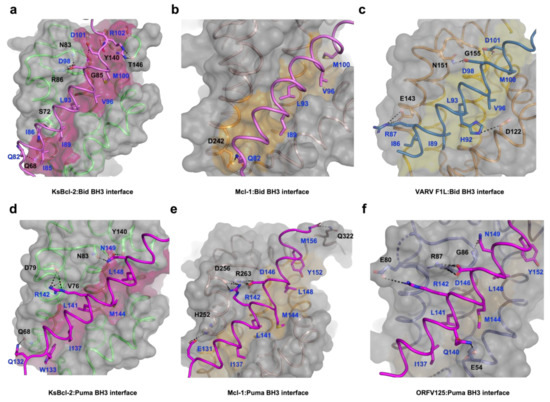
Figure 2.
Detailed view of the KsBcl-2:Bid BH3, KsBcl-2:Puma BH3, Mcl1:Bid BH3 and Mcl1:Puma BH3 interfaces. The KsBcl-2 surface, backbone and floor of the binding groove are shown in grey, green and hot pink, respectively. (a) KsBcl-2: Bid BH3 interface where Bid BH3 is shown in pink. The five key hydrophobic residues of Bid BH3 (I85, I89, L93, V96 and M100) protruding into the binding groove and the conserved salt-bridge formed by KsBcl-2 R86 and Bid BH3 D98 are labelled, as well as all other residues involved in additional ionic interactions and hydrogen bonds. Interactions are denoted as dashed black lines. (b) Molecular surface of human Mcl-1 (brown ribbon): Bid (pink) BH3 is shown as in (a). The four key hydrophobic residues of Bid BH3 (I89, L93, V96 and M100) are protruding into the binding groove, and the residues involved in hydrogen bonds are labelled. Interactions are denoted as black dotted lines. (c) VARV F1L (orange):Bid BH3 (pink) with the surface of VARV F1L is shown as in (a) and the floor of the binding grove is shown in yellow. The five key hydrophobic residues of Bid BH3 (I86, I89, L93, V96 and M100) are protruding into the binding groove and the salt bridge formed by Bid R87 and VARV F1L E143 is labelled, as well as residues involved in hydrogen bonding. (d) KsBcl-2: Puma BH3 with the surface of KsBcl-2 is shown as in (a), and Puma BH3 is shown in magenta. The five key hydrophobic residues of Puma BH3 (W133, I137, L141, M144 and L148) are protruding into the binding groove and the salt bridge formed by Puma R142 and KsBcl-2 D79 is labelled, as well as residues involved in hydrogen bonds. (e) Human Mcl-1 (brown):Puma BH3 is shown as in (b). The five key hydrophobic residues of Puma BH3 (I137, L141, M144, L148 and Y152) are protruding into the binding groove, and the conserved salt bridge formed by Puma D146 and Mcl-1 R263 is labelled, as well as residues involved in hydrogen bonds. (f) ORFV125 (purple):Puma BH3 is shown as in (b). The five key hydrophobic residues of Puma BH3 (I137, L141, M144, L148 and Y152) are protruding into the binding groove, and the conserved salt bridge formed by Puma D146 and ORFV125 R87 is labelled, as well as residues involved in hydrogen bonds. Interactions are denoted as black dotted lines.
A structural analysis using DALI [48] showed that complexes of cellular Mcl-1 (PDB ID 3MK8) [44] and zebrafish pro-survival protein NRZ-1 (PDB ID 6FBX) [47] are the closest structural homologs of KsBcl-2 with RMSD values of 1.8 Å and 2.7 Å over 133 Cα and 138 Cα atoms with sequence identities of 20% and 17%, respectively. In comparison to other closely related viral Bcl-2 proteins including EBV BHRF1 (PDB ID 2V6Q) [45] and gammaherpesvirus 68 encoded M11 (PDB ID 3BL2) [26] gave an RMSD value of 2.6 Å and 2.8 Å over 128 Cα and 116 Cα atoms with significantly low sequence identities of 13% and 12% respectively. Interestingly, superimposition with fowlpox virus encoded Bcl-2 homolog FPV039 (PDB ID 5TZQ) [49] yields an RMSD value of 2.3 Å over 124 Cα atoms with a sequence identity of 16%. As expected, the backbone structure of KsBcl-2 is highly similar to previously characterized homologous counterparts. Binding of Bid BH3 peptide to KsBcl-2 occurs via the canonical ligand binding groove that is formed by helices α2–α5 (Figure 1a). The four canonical BH3 motif defining residues from Bid, I89, L93 and V96 and M100, are bound in four hydrophobic pockets of the ligand binding groove (Figure 2a). A fifth hydrophobic pocket in KsBcl-2 is occupied by Bid I85. The hallmark ionic interaction between pro-survival Bcl-2 proteins and pro-apoptotic BH3 motif ligands between a conserved arginine (R86KsBcl-2) in the BH1 motif and aspartate (D98BID) of the Bid BH3-motif [4] is also present. This interaction is supplemented by multiple hydrogen bonding interactions between Q82BID and Q68KsBcl-2, I86BID and S72KsBcl-2, D98BID and N83KsBcl-2, D101BID and G85KsBcl-2, R102BID and Y140KsBcl-2 as well as between R102BID and T146KsBcl-2 (Figure 2a). Similarly, in the KsBcl-2: Puma BH3 complex Puma residues I137, L141, M144 and L148 protrude into the four hydrophobic pockets and an extra hydrophobic pocket is occupied by Puma residue W133 (Figure 2d). Additionally, an ionic interaction is formed by R142PUMA and D79KsBcl-2. However, the expected ionic interaction between pro-survival Bcl-2 proteins and pro-apoptotic BH3 motif ligands between a conserved arginine in the BH1 motif and aspartate of the BH3-motif [4] was surprisingly absent. Additional hydrogen bonds are found between Q132PUMA and Q68KsBcl-2, R142PUMA and V76KsBcl-2, N149PUMA and N83KsBcl-2 as well as between N149BID and Y140KsBcl-2 (Figure 2d).

Table 3.
Summary of pro-apoptotic BH3 motif affinities for KsBcl-2 and its closely related homologs.
Table 3.
Summary of pro-apoptotic BH3 motif affinities for KsBcl-2 and its closely related homologs.
| Affinity (nM) | ||||||
|---|---|---|---|---|---|---|
| Peptide | KsBcl2 b (This Study) | BHRF-1 a [45] | M11 a [26] | Mcl-1 b [46] | NRZ a [47] | FPV039 a [49] |
| Bak | 8 | 150 | 76 | 10 | N/A | 76 |
| Bax | 16 | 1400 | 690 | 12 | 688 | 76 |
| Bok | 198 | N/A | N/A | N/A | N/A | N/A |
| Bad | NB | NB | N/A | >100,000 | 343 | 653 |
| Bid | 20 | 110 | 232 | 2100 | 409 | 2 |
| Bik | 64 | NB | N/A | 1700 | 12 | 30 |
| Bim | 4 | 18 | 131 | 2.4 | 41 | 10 |
| Bmf | NB | NB | 300 | 1100 | NB | 254 |
| Hrk | 115 | NB | 719 | 370 | N/A | 24 |
| Noxa | 220 | NB | 132 | 24 | 142 | 28 |
| Puma | 26 | 70 | 370 | 6.3 | 36 | 31 |
N/A-Not Applicable, NB-No Binding. a—Affinity measured in nM by Isothermal Titration Calorimetry. b—Affinity measured in nM by SPR.
4. Discussion
Bcl-2 homologs are widely used amongst large DNA viruses to ensure viral proliferation and/or survival [5,20,50]. Bcl-2 homologs have been described for members of the families of herpesviridae, poxviridae [51,52,53], asfaviridae [54] and iridoviridae [55]. Amongst the herpesviridae, the majority of gammaherpesvirinae genera have been shown to encode apoptosis inhibiting Bcl-2 homologs including EBV [45,56], KSHV, murine gammaherpesvirus 68 and herpesvirus saimiri [14]. Whilst many gammaherpesvirinae encode for sequence, structural or functional pro-survival Bcl-2 homologs, considerable sequence, structural and affinity profile diversity exists amongst these proteins [50]. There are widely differing interaction profiles with host proapoptotic Bcl-2 proteins and some differences in overall structure as well as detailed interactions at the atomic level, as might be expected from these highly sequence divergent Bcl-2 fold sequences [5]. Here, we report that the gammaherpesvirinae member KSHV Bcl-2 (KsBcl-2) is a potentially potent inhibitor of mammalian apoptosis. KsBcl-2 adopts a globular monomeric Bcl-2 fold (Figure 1a) and binds a number of BH3 motif peptides from human pro-apoptotic Bcl-2 proteins including those from Bax, Bak, Bok, Bim, Bid, Bik, Noxa, Puma and Hrk (Table 1) but we did not detect any affinity for Bad or Bmf. Among the cellular Bcl-2 proteins, Bcl-xL, Bcl-2 and Bcl-w bind a significant number of BH3-only proteins but show no affinity towards Noxa [4,32]. The previously characterized viral Bcl-2 proteins, tanapox virus TANV16L [53], Sheeppox virus SPP14 [57,58], fowlpox virus FPV039 [49], African swine fever virus A179L [54,59] and murine gammaherpesvirus M11 [26] were able to bind a substantial number of BH3-only proteins. KsBcl-2 displays the tightest binding to the key executioner Bcl-2 proteins Bax and Bak amongst vBcl-2 proteins examined to date with affinities that are comparable to human pro-survival Bcl-2 proteins such as Mcl-1 [46]. However we note that African swine fever virus A179L is nearly as potent a binder of Bak and Bax [54]. In contrast, other herpesviral vBcl-2 proteins, such as EBV BHRF1 (human herpes virus 4) [45] and murine herpesvirus 68 M11 proteins [26], harbor a more restricted overall pro-apoptotic Bcl-2 binding profile and bind Bak and Bax more weakly.
Interestingly, our structural homolog search using DALI [48] identified the closest structural homolog to KsBcl-2 as human Mcl-1 (PDB ID 3MK8) [44]. KsBcl-2 shares 21% sequence identity and structural superimposition of KsBcl-2 with Mcl-1 yielded an R.M.S.D value of 1.8 Å over 133 Cα atoms. The closest viral Bcl-2 homolog identified was fowlpox virus protein, FPV039 [49] with a sequence identity of 16% and structural superimposition yielding an R.M.S.D value of 2.3 Å over 130 Cα atoms. Sequence alignment of KsBcl-2 (Figure 3) with its closest structural homologs showed that KsBcl-2 shares sequence features of other multi-domain members of the Bcl-2 family and shares 24% sequence identity with NRZ, the zebrafish Bcl-2 protein [47], 17% identity with EBV BHRF1 [45]. Similarly, sequence alignment of KsBcl-2 with herpes virus encoded Bcl-2 homolog produces 19% identity for herpes virus saimiri encoded Bcl-2 homolog 16L (HVS 16L) [14], 10% identity for murine gammaherpes virus 68 Bcl-2 homolog M11 [17] and 27% identity for turkey herpes virus Bcl-2 homolog HVT079 [15,16]. However, the BH regions show a significantly lower level of conservation among these proteins [60] and these sequence variations that are located in the ligand binding groove create the basis in part for the selectivity differences observed for BH3-ligands compared to the cellular counterparts. Superimposition of KsBcl-2 with EBV BHRF1 produced an R.M.S.D of 2.8 Å over 83 residues whereas gammaherpesvirus 68 encoded M11 produced an R.M.S.D of 10.2 Å over 42 residues. EBV BHRF1 is very similar in structure to KsBcl-2, whereas M11 appears to be significantly different.
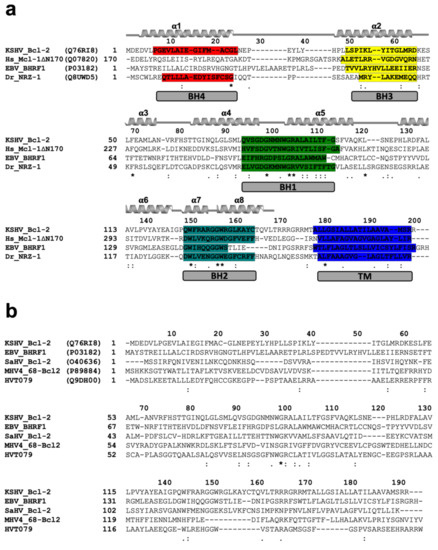
Figure 3.
Sequence alignments (a) Sequence alignment of KsBcl-2 with pro-survival Bcl-2 family members. The sequences of KSHV KsBcl-2 (UniProt accession number Q76RI8), human Mcl-1 (UniProt Q07820) EBV BHRF1 (UniProt P03182), Zebrafish pro-survival Bcl-2 protein NRZ1 (UniProt Q8UWD5) and were aligned using muscle [42]. Secondary structure elements are marked based on the crystal structure of KsBcl-2, and BH motifs are highlighted in colours: BH4, red; BH3, yellow; BH1, green; BH2, teal and trans-membrane region (TM) in blue. The regions of helix are marked and unstructured loops with a bar below the sequence, conserved residues are denoted by ‘*’, with highly conservative substitutions indicated by ‘:’ and conserved substitutions indicated by ‘.’. (b) Sequence alignment of pro-survival Bcl-2 proteins encoded by different herpes viruses. The sequences of KSHV KsBcl-2 (UniProt Q76RI8), EBV BHRF1 (UniProt P03182), Human Saimiri virus Bcl-2 protein 16L (HVS) (UniProt O40636), Murine gammaherpes virus 68 Bcl-2 protein, M11(UniProt: P89884) and Turkey herpes virus Bcl-2 protein HVT079 (UniProt: Q9DH00). Conserved residues, highly conservative residues and conserved residues are indicated as in (a).
Phasing of our crystal structures relied on a model generated using AlphaFold [37]. Superimposition of the experimentally determined structure compared with the AlphaFold model yielded an RMSD of 0.7 Å. The alpha fold model featured nearly identical side chain rotamers and secondary structure elements, with some minor variations in the alpha 3 helix. However, significant variation can be seen in the unstructured N-terminal end and the loop region between alpha 1–2 helices, which accounts for the overall RMSD difference observed.
A comparison of overall fold of the KsBcl-2: Bid BH3 complex reveals that it is very similar to that observed in other Bcl-2: BH3 complexes. Despite the overall similarity in fold, several key differences are observed in the crystal structures and protein: peptide interfaces of the KsBcl-2: Bid and Mcl-1: Bid (PDB ID: 2KBW) [61] complexes.
A comparison of structure and interactions of KsBcl-2:Puma (Figure 2d) complex reveals it is nearly identical to those observed in Mcl-1:Puma complex (Figure 2e) or ORFV125:Puma [62] complex (Figure 2f) whereas Mcl-1:Puma interaction is four-fold tighter than KsBcl-2:Puma and ORFV125: Puma interaction is approximately 60-fold weaker than KsBcl-2:Puma [63]. KsBcl-2 binding of either Puma BH3 or Bid BH3 utilizes an additional hydrophobic pocket from the base of the binding groove. For the KsBcl-2:Bid complex this additional hydrophobic pocket is utilized by N-terminal residue I85BID (Figure 2a and Figure 4a) and in KsBcl-2:Puma complex this additional hydrophobic pocket is utilized by W133PUMA (Figure 2b and Figure 4b). In comparison to previously reported viral Bcl-2: BH3 interactions, only three of the BH3 peptides (Bax, Bid and Puma (Figure 4c)) were shown to have this fifth pocket. A similar observation was noted previously for the variola virus VARV F1L:Bid complex (Figure 2c) [64]. However, this fifth hydrophobic pocket in the KsBcl-2 complexes is different to that observed for other complexes. For instance, both Mcl-1:Puma (Figure 4d) [65], ORFV125:Puma (Figure 4e) [62], SPPV14:Bax [58] and ORFV125:Bax [63] complexes, the C-terminal residue Y142PUMA and M74BAX utilized the fifth pocket respectively. Whereas, the fifth hydrophobic pocket is absent in both TANV16L:Puma or TANV16L:Bax complexes [53]. Furthermore, Puma is crucial for p53-independent and dependent apoptosis regulation against diversified stimuli [66], such as viral infection, dysregulated oncogene expression, radiation induced apoptosis, genotoxic stress and toxins. In contrast to other BH3-only proteins, Puma interacts with almost all viral Bcl-2 proteins [50] as well as five of the major cellular Bcl-2 proteins such as Bcl-xL, Bcl-2, Mcl-1, Bcl-w and A1 with high affinity interactions [67]. The combination of structural data and conserved interactions between Puma BH3 and pro-survival Bcl-2, BH3-mimetic drugs or selective peptide inhibitors against KsBcl-2 [68] was used to mimic these interactions [69].
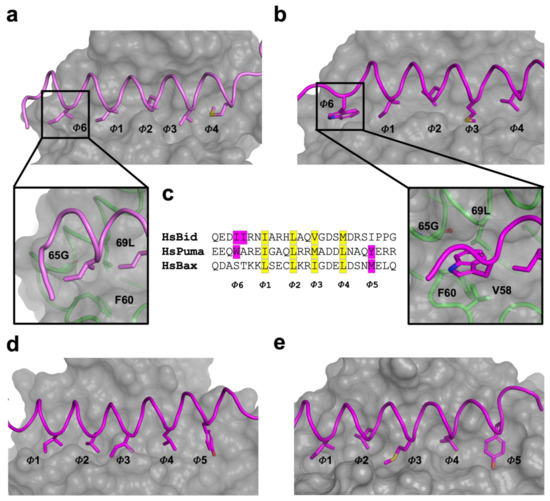
Figure 4.
Engagement of an additional hydrophobic pocket (fifth pocket) in viral Bcl-2 proteins with BH3motif peptides. (a) Hydrophobic binding groove of KsBcl-2:Bid BH3 complex. The hydrophobic pockets of the binding groove are labelled as Φ1–Φ6. The surface of the KsBcl-2 is shown in grey with side chains of the key hydrophobic residues located in the pocket Φ1–Φ6 shown as sticks. Bid BH3 backbone is shown as cartoon tube (pink). An enlarged view of the fifth hydrophobic pocket of KsBcl-2 is shown in the bottom panel with a cartoon tube of the KsBcl-2 backbone (green), with residues involved in forming the fifth hydrophobic pocket labelled. (b) Hydrophobic binding groove of KsBcl-2:Puma BH3 complex. Puma BH3 backbone is shown as cartoon tube (magenta). The surface, hydrophobic pockets of the binding groove and key residues are shown as in (a). (c) sequence of the key BH3 motif peptides (Bid, Puma and Bax) involved to engage fifth hydrophobic pocket with viral Bcl-2 proteins are shown. Conserved four hydrophobic residues engage with hydrophobic pockets are highlighted in yellow and labelled Φ1–Φ4. The additional hydrophobic residues engaging with the fifth hydrophobic pocket are highlighted in magenta and labelled as Φ5 (C-terminal pocket) and Φ6 (N-terminal pocket). (d) Hydrophobic binding groove of Mcl-1:Puma BH3 complex. Puma BH3 (magenta) backbone is shown as cartoon tube. (e) Hydrophobic binding groove of ORFV125:Puma BH3 complex. Puma BH3 (magenta) backbone is shown as cartoon tube. The surface, hydrophobic pockets of the binding groove and key residues are shown as in (a).
The BH1 motif of multi-domain Bcl-2 family proteins, along with pro-apoptotic Bcl-2 protein Bak, Bax and Bok plays a crucial structural role during cell death regulation and can be easily recognized by a short well conserved signature sequence motif “NWGR” in the BH1 motif at the N-terminal end the α5 helix [7] (Figure 4). The Gly and Arg residues of ‘NWGR” motif are important for protein–protein interactions within the Bcl-2 family proteins [65,70,71]. The Arg residue is located at the beginning of the α5 helix forms the canonical ionic/salt bridge interaction with the conserved Asp residue of BH3 motif of the pro-apoptotic Bcl-2 proteins. In the KsBcl-2: Bid complex, the corresponding R86KsBcl-2 residue in the NWGR motif interacts with D98 of the Bid BH3 peptide. However, this highly conserved interaction was not seen or is very weak in the KsBcl-2: Puma complex where the closest proximity for this interaction is 5.6 Å. In place of this interaction an alternative salt bridge is observed between D79 KsBcl-2 and R142Puma. An identical ionic interaction can be seen in Mcl-1: Puma complex [65] where D256Mcl-1 is conserved in both KsBcl-2 and Mcl-1. In contrast, human Mcl-1: Bid (PDB ID: 2KBW) [61] complex did not feature the canonical salt-bridge interaction, whereas the human Mcl-1: Puma complex formed this interaction between corresponding R263Mcl-1 residue in the NWGR motif interacts with D146 of the Puma BH3 peptide. Previous studies showed that a R263AMcl-1 mutation in Mcl-1 results significant loss of ability to interact with BH3 peptide [72]. Comparison of the binding mode of Bid BH3 or Puma BH3 with KsBcl-2 identified three structurally conserved residues: Q68, N83 and Y140 in the binding groove which make polar contacts with the BH3 motif of pro-apoptotic Bcl-2 proteins. This suggests that KsBcl-2 residues Q68, N83 and Y140 are important for stabilization of the BH3 motif peptide in the binding groove.
Considering that Mcl-1, KsBcl2 and ORFV125 feature highly similar binding site (Figure 4) it is interesting that they feature significantly differing affinities for Puma. Mcl-1:Puma features the hallmark interaction between the conserved Arg residue of the NWGR motif and the well conserved Asp residue of BH3 motif, whereas in KsBcl-2:Puma the NWGR motif is present but this hallmark interaction with the conserved Asp from the BH3 motif is absent. This may contribute to the four-fold decrease in affinity of KsBcl-2:Puma complex compared to that observed for the Mcl-1:Puma complex. The equivalent BH1 region of ORFV125 features the sequence “SPGR” instead of the conserved NWGR and displays an approximately 250-fold reduced affinity against Puma BH3. Thus, the “SPGR” sequence motif of ORFV125 is directly impacting specificity and binding compared to KsBcl-2.
KsBcl-2 displays a very broad ligand binding profile when compared to its closest homologous proteins, Mcl-1, NRZ, and FPV039, BHRF1 and M11 (Table 3), even when taking into consideration that different methods were used to determine these affinities. However, KsBcl-2 is the only protein that interacts with BH3 motif peptide of pro-apoptotic protein Bok and showed very strong affinity towards Bak, Bax, Bim, Bid, Puma and Bik BH3 motif peptides. Previous studies revealed that overexpression of KsBcl-2 efficiently blocked host cell apoptosis similar to that observed in cellular Bcl-2, Bcl-xL, Mcl-1 or EBV (gammaherpes virus 4) encoded BHRF1 but was not able to heterodimerized with Bax or Bak [13], suggesting that KsBcl-2 may have developed a strategy to escape any negative regulatory effect from cellular Bak and Bax. Overexpression of KsBcl-2 is sufficient to block mitochondrial outer membrane permeabilization and subsequent apoptosis during latent B-cell infection [68]. Overall KsBcl-2 displays a ligand binding profile that most closely resembles Mcl-1, albeit with varied affinities for individual ligands.
Interestingly, KsBcl-2 was shown to be involved in virion assembly via tegument protein (ORF55), with a reported interaction via its N-terminal amino acid residue 11–20 [30]. In our crystal structures and the previously reported NMR structure [73] KsBcl-2 adopts a helical configuration in the N-terminal region proposed as the interacting site, and crucially the identified Glu14 residue is solvent exposed and available for an interaction. However, the precise mode of engagement of ORF55 with KsBcl-2 remains to be clarified and likely requires structure determination of such a complex.
In summary, we report the biochemical and structural analysis of Kaposi sarcoma herpes virus encoded apoptosis inhibitor KsBcl-2, which revealed a broad high affinity binding profile for mammalian pro-apoptotic Bcl-2 proteins. Our findings provide a molecular basis for dissecting the function of KsBcl-2 in KSHV infection and to determine the contribution that abolition of Bcl-2 mediated apoptosis makes to the KSHV life cycle.
Author Contributions
C.D.S.: Acquisition of data; Analysis and interpretation of data; Drafting and revising the article. M.G.H.: Conception and design; Analysis and interpretation of data; Drafting and revising the article. M.K.: Conception and design; Acquisition of data; Analysis and interpretation of data; Drafting and revising the article. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by La Trobe University (Scholarship to C.D.S.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data supporting the findings of this manuscript are available from the corresponding authors upon reasonable request. Coordinate files were deposited at the Protein Data Bank (https://www.rcsb.org/) (accessed on 1 July 2021) using accession codes 7QTW and 7QTX for KsBcl-2: Bid BH3 and KsBcl-2: Puma BH3, respectively. The raw X-ray diffraction data were deposited at the SBGrid Data Bank [41] (https://data.sbgrid.org/data/) using their PDB accession codes 7QTW and 7QTX for KsBcl-2: Bid BH3 and KsBcl-2: Puma BH3, respectively.
Acknowledgments
We thank staff at the MX beamlines at the Australian Synchrotron for help with X-ray data collection. We thank the ACRF for their support of the Eiger MX detector at the Australian Synchrotron MX2 beamline and the Comprehensive Proteomics Platform at La Trobe University for core instrument support. We thank S Caria for helpful discussion and technical support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Jorgensen, I.; Rayamajhi, M.; Miao, E.A. Programmed cell death as a defence against infection. Nat. Rev. Immunol. 2017, 17, 151–164. [Google Scholar] [CrossRef]
- Galluzzi, L.; Brenner, C.; Morselli, E.; Touat, Z.; Kroemer, G. Viral Control of Mitochondrial Apoptosis. PLoS Pathog. 2008, 4, e1000018. [Google Scholar] [CrossRef] [Green Version]
- Kvansakul, M.; Hinds, M.G. Structural biology of the Bcl-2 family and its mimicry by viral proteins. Cell Death Dis. 2013, 4, e909. [Google Scholar] [CrossRef]
- Banjara, S.; Suraweera, C.D.; Hinds, M.G.; Kvansakul, M. The Bcl-2 Family: Ancient Origins, Conserved Structures, and Divergent Mechanisms. Biomolecules 2020, 10, 128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suraweera, C.D.; Hinds, M.G.; Kvansakul, M. Poxviral Strategies to Overcome Host Cell Apoptosis. Pathogens 2021, 10, 6. [Google Scholar] [CrossRef] [PubMed]
- Youle, R.J.; Strasser, A. The BCL-2 protein family: Opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 2008, 9, 47–59. [Google Scholar] [CrossRef]
- Kvansakul, M.; Hinds, M.G. The Structural Biology of BH3-Only Proteins. Methods Enzymol. 2014, 544, 49–74. [Google Scholar] [CrossRef]
- Shamas-Din, A.; Kale, J.; Leber, B.; Andrews, D.W. Mechanisms of Action of Bcl-2 Family Proteins. Cold Spring Harb. Perspect. Biol. 2013, 5, a008714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bellows, D.S.; Chau, B.N.; Lee, P.; Lazebnik, Y.; Burns, W.H.; Hardwick, J.M. Antiapoptotic Herpesvirus Bcl-2 Homologs Escape Caspase-Mediated Conversion to Proapoptotic Proteins. J. Virol. 2000, 74, 5024–5031. [Google Scholar] [CrossRef]
- Lee, M.A.; Yates, J.L. BHRF1 of Epstein-Barr virus, which is homologous to human proto-oncogene bcl2, is not essential for transformation of B cells or for virus replication in vitro. J. Virol. 1992, 66, 1899–1906. [Google Scholar] [CrossRef] [Green Version]
- Fitzsimmons, L.; Cartlidge, R.; Chang, C.; Sejic, N.; Galbraith, L.C.A.; Suraweera, C.; Croom-Carter, D.; Dewson, G.; Tierney, R.J.; Bell, A.; et al. EBV BCL-2 homologue BHRF1 drives chemoresistance and lymphomagenesis by inhibiting multiple cellular pro-apoptotic proteins. Cell Death Differ. 2020, 27, 1554–1568. [Google Scholar] [CrossRef]
- Bellows, D.S.; Howell, M.; Pearson, C.; Hazlewood, S.A.; Hardwick, J.M. Epstein-Barr Virus BALF1 Is a BCL-2-like Antagonist of the Herpesvirus Antiapoptotic BCL-2 Proteins. J. Virol. 2002, 76, 2469–2479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, E.H.-Y.; Nicholas, J.; Bellows, D.S.; Hayward, G.S.; Guo, H.-G.; Reitz, M.S.; Hardwick, J.M. A Bcl-2 homolog encoded by Kaposi sarcoma-associated virus, human herpesvirus 8, inhibits apoptosis but does not heterodimerize with Bax or Bak. Proc. Natl. Acad. Sci. USA 1997, 94, 690–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derfuss, T.; Fickenscher, H.; Kraft, M.S.; Henning, G.; Lengenfelder, D.; Fleckenstein, B.; Meinl, E. Antiapoptotic Activity of the Herpesvirus Saimiri-Encoded Bcl-2 Homolog: Stabilization of Mitochondria and Inhibition of Caspase-3-Like Activity. J. Virol. 1998, 72, 5897–5904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afonso, C.L.; Tulman, E.R.; Lu, Z.; Zsak, L.; Rock, D.L.; Kutish, G.F. The Genome of Turkey Herpesvirus. J. Virol. 2001, 75, 971–978. [Google Scholar] [CrossRef] [Green Version]
- Reddy, V.R.A.P.; Sadigh, Y.; Tang, N.; Yao, Y.; Nair, V.; Jung, J.U. Novel Insights into the Roles of Bcl-2 Homolog Nr-13 (vNr-13) Encoded by Herpesvirus of Turkeys in the Virus Replication Cycle, Mitochondrial Networks, and Apoptosis Inhibition. J. Virol. 2020, 94, e02049-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, D.J.; Ebrahimi, B.C.; Dutia, B.M.; Nash, A.A.; Stewart, J.P. Murine gammaherpesvirus M11 gene product inhibits apoptosis and is expressed during virus persistence. Arch Virol. 2000, 145, 2411–2420. [Google Scholar] [CrossRef] [PubMed]
- Czabotar, P.E.; Westphal, D.; Dewson, G.; Ma, S.; Hockings, C.; Fairlie, W.; Lee, E.; Yao, S.; Robin, A.; Smith, B.; et al. Bax Crystal Structures Reveal How BH3 Domains Activate Bax and Nucleate Its Oligomerization to Induce Apoptosis. Cell 2013, 152, 519–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dorstyn, L.; Akey, C.W.; Kumar, S. New insights into apoptosome structure and function. Cell Death Differ. 2018, 25, 1194–1208. [Google Scholar] [CrossRef] [PubMed]
- Neumann, S.; El Maadidi, S.; Faletti, L.; Haun, F.; Labib, S.; Schejtman, A.; Maurer, U.; Borner, C. How do viruses control mitochondria-mediated apoptosis? Virus Res. 2015, 209, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Renne, R.; Lagunoff, M.; Zhong, W.; Ganem, D. The size and conformation of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) DNA in infected cells and virions. J. Virol. 1996, 70, 8151–8154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russo, J.J.; Bohenzky, R.A.; Chien, M.-C.; Chen, J.; Yan, M.; Maddalena, D.; Parry, J.P.; Peruzzi, D.; Edelman, I.S.; Chang, Y.; et al. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 1996, 93, 14862–14867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neipel, F.; Albrecht, J.C.; Ensser, A.; Huang, Y.Q.; Li, J.J.; Friedman-Kien, A.E.; Fleckenstein, B. Human herpesvirus 8 encodes a homolog of interleukin-6. J. Virol. 1997, 71, 839–842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Epurushothaman, P.; Edabral, P.; Egupta, N.; Esarkar, R.; Verma, S.C. KSHV Genome Replication and Maintenance. Front. Microbiol. 2016, 7, 54. [Google Scholar] [CrossRef]
- Sinha, S.C.; Colbert, C.L.; Becker, N.; Wei, Y.; Levine, B. Molecular basis of the regulation of Beclin 1-dependent autophagy by the γ-herpesvirus 68 Bcl-2 homolog M11. Autophagy 2008, 4, 989–997. [Google Scholar] [CrossRef] [PubMed]
- Ku, B.; Woo, J.-S.; Liang, C.; Lee, K.-H.; Hong, H.-S.; E, X.; Kim, K.-S.; Jung, J.U.; Oh, B.-H. Structural and Biochemical Bases for the Inhibition of Autophagy and Apoptosis by Viral BCL-2 of Murine γ-Herpesvirus 68. PLoS Pathog. 2008, 4, e25. [Google Scholar] [CrossRef]
- Liang, Q.; Chang, B.; Lee, P.; Brulois, K.F.; Ge, J.; Shi, M.; Rodgers, M.; Feng, P.; Oh, B.-H.; Liang, C.; et al. Identification of the Essential Role of Viral Bcl-2 for Kaposi’s Sarcoma-Associated Herpesvirus Lytic Replication. J. Virol. 2015, 89, 5308–5317. [Google Scholar] [CrossRef] [Green Version]
- Gallo, A.; Lampe, M.; Günther, T.; Brune, W. The Viral Bcl-2 Homologs of Kaposi’s Sarcoma-Associated Herpesvirus and Rhesus Rhadinovirus Share an Essential Role for Viral Replication. J. Virol. 2017, 91, e01875-16. [Google Scholar] [CrossRef] [Green Version]
- Gelgor, A.; Kalt, I.; Bergson, S.; Brulois, K.F.; Jung, J.U.; Sarid, R. Viral Bcl-2 Encoded by the Kaposi’s Sarcoma-Associated Herpesvirus Is Vital for Virus Reactivation. J. Virol. 2015, 89, 5298–5307. [Google Scholar] [CrossRef] [Green Version]
- Liang, Q.; Wei, D.; Chung, B.; Brulois, K.F.; Guo, C.; Dong, S.; Gao, S.-J.; Feng, P.; Liang, C.; Jung, J.U. Novel Role of vBcl2 in the Virion Assembly of Kaposi’s Sarcoma-Associated Herpesvirus. J. Virol. 2018, 92, e00914-17. [Google Scholar] [CrossRef] [Green Version]
- Burton, D.R.; Caria, S.; Marshall, B.; Barry, M.; Kvansakul, M. Structural basis of Deerpox virus -mediated inhibition of apoptosis. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015, 71, 1593–1603. [Google Scholar] [CrossRef]
- Chen, L.; Willis, S.N.; Wei, A.; Smith, B.; Fletcher, J.; Hinds, M.; Colman, P.M.; Day, C.; Adams, J.; Huang, D. Differential Targeting of Prosurvival Bcl-2 Proteins by Their BH3-Only Ligands Allows Complementary Apoptotic Function. Mol. Cell 2005, 17, 393–403. [Google Scholar] [CrossRef]
- Kvansakul, M.; Czabotar, P.E. Preparing Samples for Crystallization of Bcl-2 Family Complexes. Methods Mol. Biol. 2016, 1419, 213–229. [Google Scholar] [CrossRef] [PubMed]
- Aragão, D.; Aishima, J.; Cherukuvada, H.; Clarken, R.; Clift, M.; Cowieson, N.P.; Ericsson, D.J.; Gee, C.L.; Macedo, S.; Mudie, N.; et al. MX2: A high-flux undulator microfocus beamline serving both the chemical and macromolecular crystallography communities at the Australian Synchrotron. J. Synchrotron Radiat. 2018, 25, 885–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beilsten-Edmands, J.; Winter, G.; Gildea, R.; Parkhurst, J.; Waterman, D.; Evans, G. Scaling diffraction data in the DIALS software package: Algorithms and new approaches for multi-crystal scaling. Acta Crystallogr. Sect. D Struct. Biol. 2020, 76, 385–399. [Google Scholar] [CrossRef] [Green Version]
- Evans, P. Scaling and assessment of data quality. Acta Crystallogr. Sect. D Biol. Crystallogr. 2005, 62, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 352–367. [Google Scholar] [CrossRef] [Green Version]
- Morin, A.; Eisenbraun, B.; Key, J.; Sanschagrin, P.C.; Timony, M.A.; Ottaviano, M.; Sliz, P. Collaboration gets the most out of software. eLife 2013, 2, e01456. [Google Scholar] [CrossRef]
- Meyer, P.A.; Socias, S.; Key, J.; Ransey, E.; Tjon, E.C.; Buschiazzo, A.; Lei, M.; Botka, C.; Withrow, J.; Neau, D.; et al. Data publication with the structural biology data grid supports live analysis. Nat. Commun. 2016, 7, 10882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krissinel, E.; Henrick, K. Inference of Macromolecular Assemblies from Crystalline State. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef] [PubMed]
- Stewart, M.L.; Fire, E.; Keating, A.E.; Walensky, L.D. The MCL-1 BH3 helix is an exclusive MCL-1 inhibitor and apoptosis sensitizer. Nat. Chem. Biol. 2010, 6, 595–601. [Google Scholar] [CrossRef] [Green Version]
- Kvansakul, M.; Wei, A.; Fletcher, J.; Willis, S.N.; Chen, L.; Roberts, A.; Huang, D.C.S.; Colman, P.M. Structural Basis for Apoptosis Inhibition by Epstein-Barr Virus BHRF1. PLoS Pathog. 2010, 6, e1001236. [Google Scholar] [CrossRef] [Green Version]
- Czabotar, P.E.; Lee, E.F.; van Delft, M.F.; Day, C.L.; Smith, B.J.; Huang, D.C.S.; Fairlie, W.D.; Hinds, M.G.; Colman, P.M. Structural insights into the degradation of Mcl-1 induced by BH3 domains. Proc. Natl. Acad. Sci. USA 2007, 104, 6217–6222. [Google Scholar] [CrossRef] [Green Version]
- Suraweera, C.; Caria, S.; Järvå, M.A.; Hinds, M.G.; Kvansakul, M. A structural investigation of NRZ mediated apoptosis regulation in zebrafish. Cell Death Dis. 2018, 9, 967. [Google Scholar] [CrossRef]
- Holm, L.; Rosenström, P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010, 38, W545–W549. [Google Scholar] [CrossRef]
- Anasir, M.I.; Caria, S.; Skinner, M.; Kvansakul, M.; Anasir, M.I.; Caria, S.; Skinner, M.; Kvansakul, M. Structural basis of apoptosis inhibition by the fowlpox virus protein FPV039. J. Biol. Chem. 2017, 292, 9010–9021. [Google Scholar] [CrossRef] [Green Version]
- Kvansakul, M.; Caria, S.; Hinds, M. The Bcl-2 Family in Host-Virus Interactions. Viruses 2017, 9, 290. [Google Scholar] [CrossRef] [Green Version]
- Kvansakul, M.; van Delft, M.F.; Lee, E.F.; Gulbis, J.M.; Fairlie, W.D.; Huang, D.C.; Colman, P.M. A Structural Viral Mimic of Prosurvival Bcl-2: A Pivotal Role for Sequestering Proapoptotic Bax and Bak. Mol. Cell 2007, 25, 933–942. [Google Scholar] [CrossRef] [PubMed]
- Campbell, S.; Thibault, J.; Mehta, N.; Colman, P.M.; Barry, M.; Kvansakul, M. Structural Insight into BH3 Domain Binding of Vaccinia Virus Antiapoptotic F1L. J. Virol. 2014, 88, 8667–8677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suraweera, C.D.; Anasir, M.I.; Chugh, S.; Javorsky, A.; Impey, R.E.; Zadeh, M.H.; da Costa, T.P.S.; Hinds, M.G.; Kvansakul, M. Structural insight into tanapoxvirus-mediated inhibition of apoptosis. FEBS J. 2020, 287, 3733–3750. [Google Scholar] [CrossRef] [PubMed]
- Banjara, S.; Caria, S.; Dixon, L.K.; Hinds, M.G.; Kvansakul, M. Structural Insight into African Swine Fever Virus A179L-Mediated Inhibition of Apoptosis. J. Virol. 2017, 91, e02228-16. [Google Scholar] [CrossRef] [Green Version]
- Banjara, S.; Mao, J.; Ryan, T.M.; Caria, S.; Kvansakul, M. Grouper iridovirus GIV66 is a Bcl-2 protein that inhibits apoptosis by exclusively sequestering Bim. J. Biol. Chem. 2018, 293, 5464–5477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Q.; Petros, A.M.; Virgin, H.W.; Fesik, S.W.; Olejniczak, E.T. Solution Structure of the BHRF1 Protein From Epstein-Barr Virus, a Homolog of Human Bcl-2. J. Mol. Biol. 2003, 332, 1123–1130. [Google Scholar] [CrossRef]
- Okamoto, T.; Campbell, S.; Mehta, N.; Thibault, J.; Colman, P.M.; Barry, M.; Huang, D.C.S.; Kvansakul, M. Sheeppox Virus SPPV14 Encodes a Bcl-2-Like Cell Death Inhibitor That Counters a Distinct Set of Mammalian Proapoptotic Proteins. J. Virol. 2012, 86, 11501–11511. [Google Scholar] [CrossRef] [Green Version]
- Suraweera, C.D.; Burton, D.R.; Hinds, M.G.; Kvansakul, M. Crystal structures of the sheeppox virus encoded inhibitor of apoptosis SPPV14 bound to the proapoptotic BH3 peptides Hrk and Bax. FEBS Lett. 2020, 594, 2016–2026. [Google Scholar] [CrossRef]
- Banjara, S.; Shimmon, G.L.; Dixon, L.K.; Netherton, C.L.; Hinds, M.G.; Kvansakul, M. Crystal Structure of African Swine Fever Virus A179L with the Autophagy Regulator Beclin. Viruses 2019, 11, 789. [Google Scholar] [CrossRef] [Green Version]
- Caria, S.; Hinds, M.G.; Kvansakul, M. Structural insight into an evolutionarily ancient programmed cell death regulator—The crystal structure of marine sponge BHP2 bound to LB-Bak-2. Cell Death Dis. 2017, 8, e2543. [Google Scholar] [CrossRef]
- Liu, Q.; Moldoveanu, T.; Sprules, T.; Matta-Camacho, E.; Mansur-Azzam, N.; Gehring, K. Apoptotic Regulation by MCL-1 through Heterodimerization. J. Biol. Chem. 2010, 285, 19615–19624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suraweera, C.D.; Hinds, M.G.; Kvansakul, M. Structural Investigation of Orf Virus Bcl-2 Homolog ORFV125 Interactions with BH3-Motifs from BH3-Only Proteins Puma and Hrk. Viruses 2021, 13, 1374. [Google Scholar] [CrossRef] [PubMed]
- Suraweera, C.D.; Hinds, M.G.; Kvansakul, M. Crystal structures of ORFV125 provide insight into orf virus-mediated inhibition of apoptosis. Biochem. J. 2020, 477, 4527–4541. [Google Scholar] [CrossRef] [PubMed]
- Marshall, B.; Puthalakath, H.; Caria, S.; Chugh, S.S.; Doerflinger, M.; Colman, P.M.; Kvansakul, M. Variola virus F1L is a Bcl-2-like protein that unlike its vaccinia virus counterpart inhibits apoptosis independent of Bim. Cell Death Dis. 2015, 6, e1680. [Google Scholar] [CrossRef] [PubMed]
- Day, C.; Smits, C.; Fan, F.C.; Lee, E.; Fairlie, W.; Hinds, M.G. Structure of the BH3 Domains from the p53-Inducible BH3-Only Proteins Noxa and Puma in Complex with Mcl-1. J. Mol. Biol. 2008, 380, 958–971. [Google Scholar] [CrossRef] [PubMed]
- Borrero, L.J.H.; El-Deiry, W.S. Tumor suppressor p53: Biology, signaling pathways, and therapeutic targeting. Biochim. Biophys. Acta 2021, 1876, 188556. [Google Scholar] [CrossRef]
- Rautureau, G.J.P.; Yabal, M.; Yang, H.; Huang, D.; Kvansakul, M.; Hinds, M. The restricted binding repertoire of Bcl-B leaves Bim as the universal BH3-only prosurvival Bcl-2 protein antagonist. Cell Death Dis. 2012, 3, e443. [Google Scholar] [CrossRef]
- Burrer, C.M.; Foight, G.W.; Keating, A.E.; Chan, G.C. Selective peptide inhibitors of antiapoptotic cellular and viral Bcl-2 proteins lead to cytochrome c release during latent Kaposi’s sarcoma-associated herpesvirus infection. Virus Res. 2015, 211, 86–88. [Google Scholar] [CrossRef] [Green Version]
- Merino, D.; Kelly, G.L.; Lessene, G.; Wei, A.H.; Roberts, A.W.; Strasser, A. BH3-Mimetic Drugs: Blazing the Trail for New Cancer Medicines. Cancer Cell 2018, 34, 879–891. [Google Scholar] [CrossRef] [Green Version]
- Sattler, M.; Liang, H.; Nettesheim, D.; Meadows, R.P.; Harlan, J.E.; Eberstadt, M.; Yoon, H.S.; Shuker, S.B.; Chang, B.S.; Minn, A.J.; et al. Structure of Bcl-x L -Bak Peptide Complex: Recognition Between Regulators of Apoptosis. Science 1997, 275, 983–986. [Google Scholar] [CrossRef]
- Popgeorgiev, N.; Sa, J.D.; Jabbour, L.; Banjara, S.; Nguyen, T.T.M.; Akhavan-E-Sabet, A.; Gadet, R.; Ralchev, N.; Manon, S.; Hinds, M.G.; et al. Ancient and conserved functional interplay between Bcl-2 family proteins in the mitochondrial pathway of apoptosis. Sci. Adv. 2020, 6, eabc4149. [Google Scholar] [CrossRef] [PubMed]
- Campbell, S.; Carlson, K.J.; Buchholz, C.J.; Helmers, M.R.; Ghosh, I. Mapping the BH3 Binding Interface of Bcl-xL, Bcl-2, and Mcl-1 Using Split-Luciferase Reassembly. Biochemistry 2015, 54, 2632–2643. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Petros, A.M.; Virgin, H.W.; Fesik, S.W.; Olejniczak, E.T. Solution structure of a Bcl-2 homolog from Kaposi sarcoma virus. Proc. Natl. Acad. Sci. USA 2002, 99, 3428–3433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).