Antigenic Evolution Characteristics and Immunological Evaluation of H9N2 Avian Influenza Viruses from 1994–2019 in China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Virus
2.2. Sequence Alignment and Phylogenetic Analysis
2.3. Phylogeographic Analyses
2.4. Structural Modeling of HA Proteins
2.5. Virus Rescue
2.6. HI Assay and Antigenic Cartography
2.7. Immune Protection Experiment
2.8. Statistical Analysis
3. Results
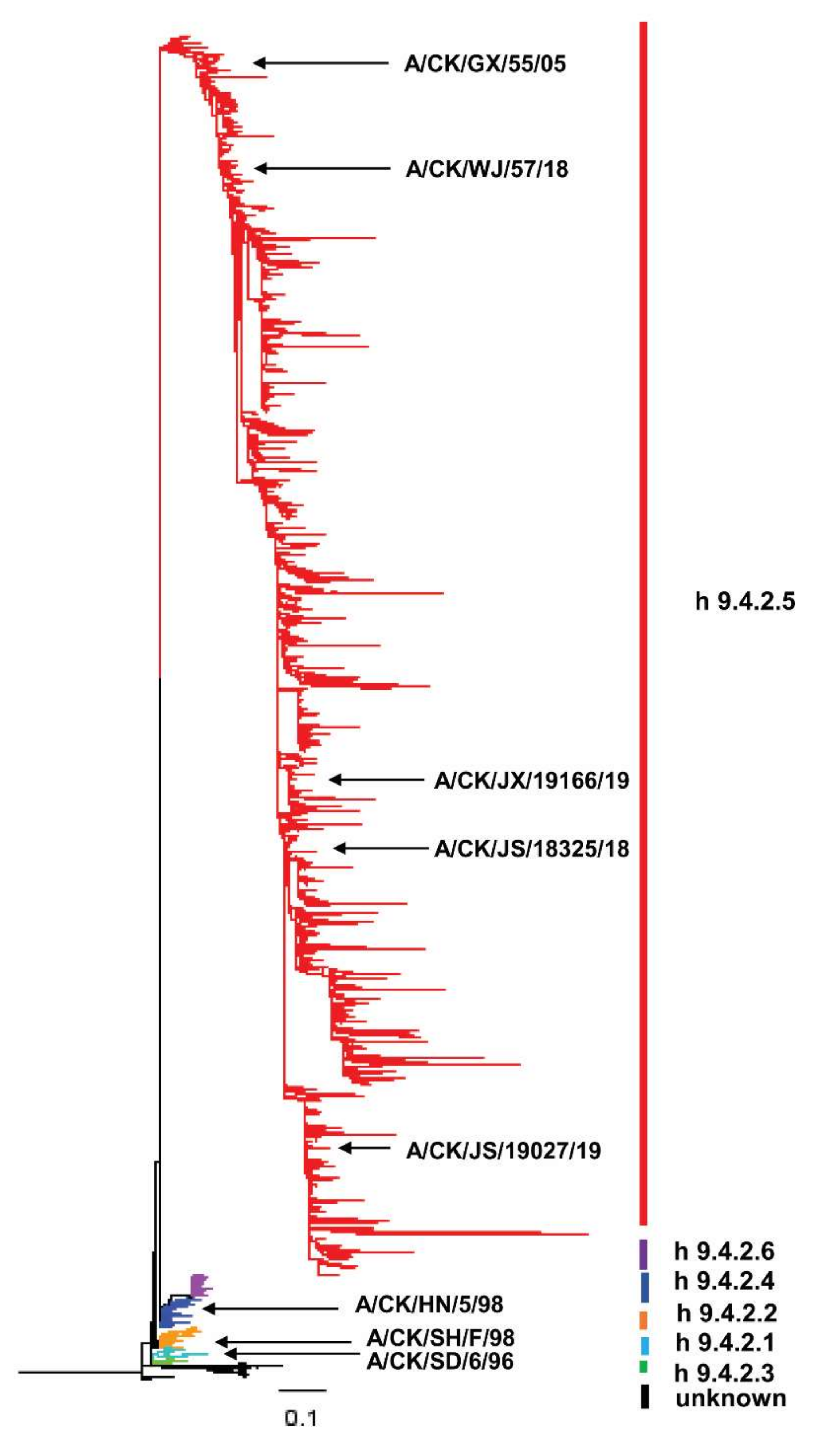
3.1. Genetic Evolution of Domestic H9N2 HA1 Protein from 1994 to 2019
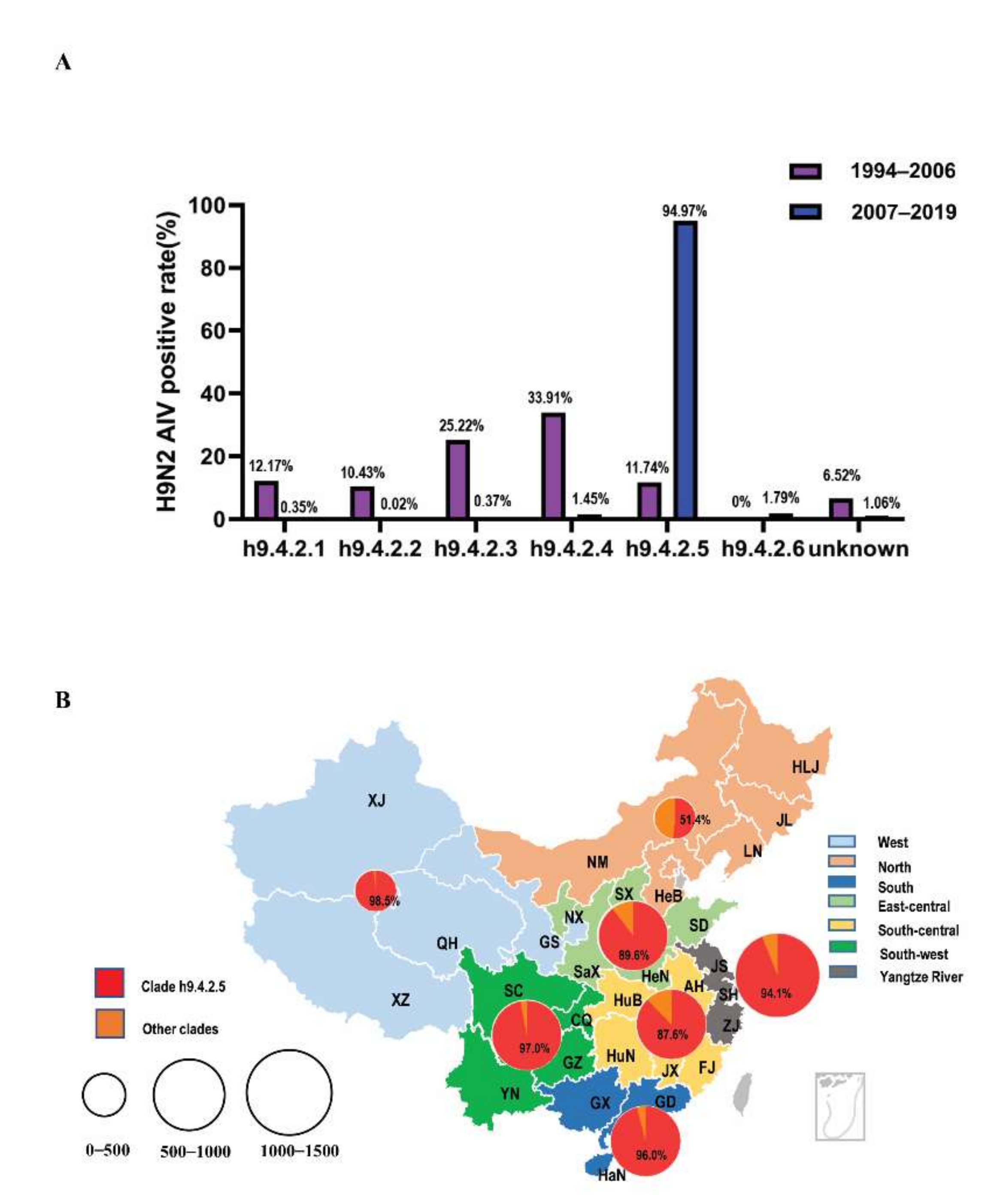
3.2. The Temporal and Spatial Distribution of Genetic Evolution of HA1 Protein
3.3. Screening of 12 Different Mutant Amino Acid Sites from the Sequences around 2006
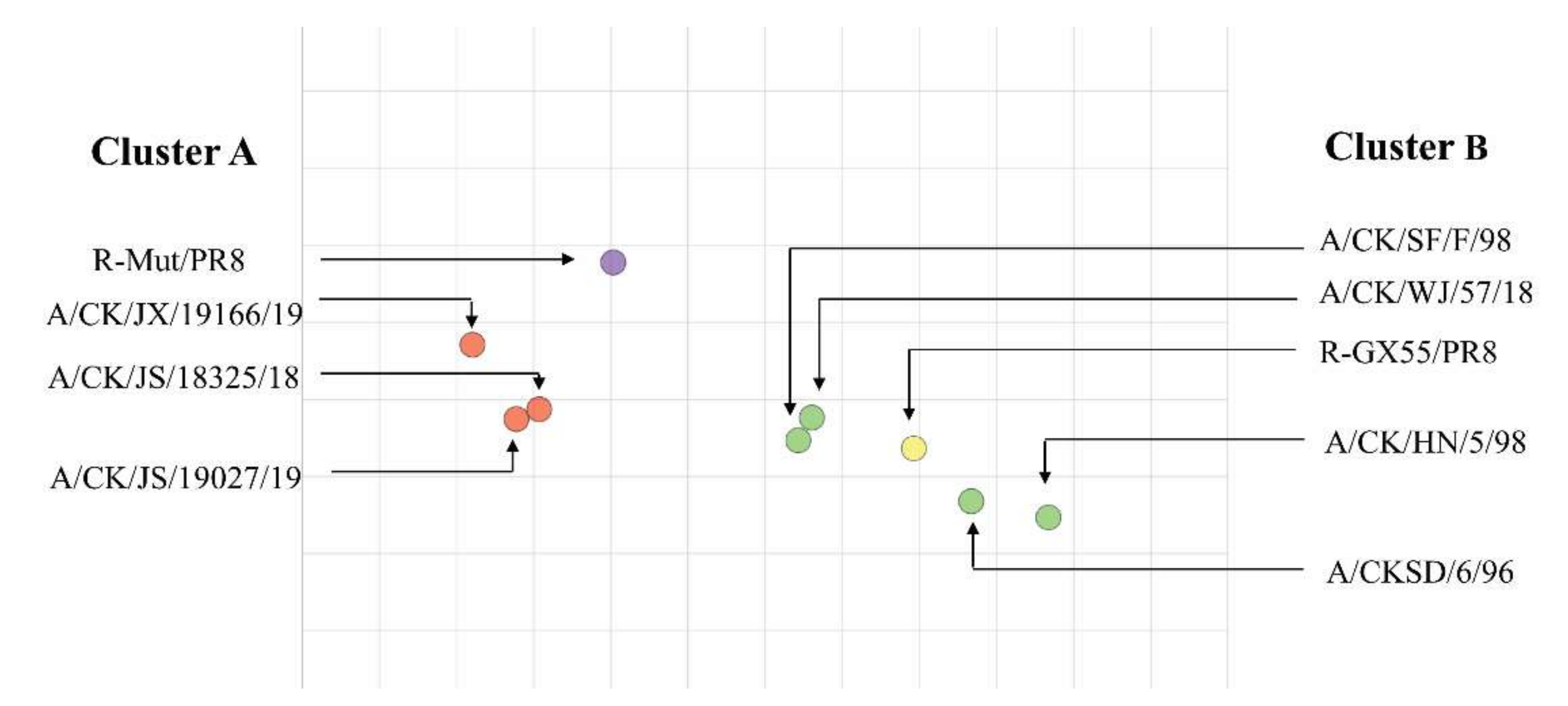
3.4. Antigenicity Analysis of the R-Mut/PR8 and Other Viruses
3.5. R-Mut/PR8 Inactivated Vaccine Provides Adequate Immune Protection against 19166 Strain
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nili, H.; Asasi, K. Natural cases and an experimental study of H9N2 avian influenza in commercial broiler chickens of Iran. Avian Pathol. 2002, 31, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.A.; Cho, S.H.; Kim, H.S.; Seo, S.H. H9N2 influenza viruses isolated from poultry in Korean live bird markets continuously evolve and cause the severe clinical signs in layers. Vet. Microbiol. 2006, 118, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Carnaccini, S.; Perez, D.R. H9 Influenza Viruses: An Emerging Challenge. Cold Spring Harb. Perspect. Med. 2020, 10, a038588. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.M.; Smith, G.J.; Bahl, J.; Duan, L.; Tai, H.; Vijaykrishna, D.; Wang, J.; Zhang, J.X.; Li, K.S.; Fan, X.H.; et al. The genesis and evolution of H9N2 influenza viruses in poultry from southern China, 2000 to 2005. J. Virol. 2007, 81, 10389–10401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Li, C.C.; Diao, Y.X.; Sun, X.Y.; Hao, D.M.; Liu, X.; Ge, P.P. Different outcomes of infection of chickens and ducks with a duck-origin H9N2 influenza A virus. Acta Virol. 2014, 58, 223–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.J.; Krauss, S.; Senne, D.A.; Mo, I.P.; Lo, K.S.; Xiong, X.P.; Norwood, M.; Shortridge, K.F.; Webster, R.G.; Guan, Y. Characterization of the pathogenicity of members of the newly established H9N2 influenza virus lineages in Asia. Virology 2000, 267, 279–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, K.S.; Xu, K.M.; Peiris, J.S.; Poon, L.L.; Yu, K.Z.; Yuen, K.Y.; Shortridge, K.F.; Webster, R.G.; Guan, Y. Characterization of H9 subtype influenza viruses from the ducks of southern China: A candidate for the next influenza pandemic in humans? J. Virol. 2003, 77, 6988–6994. [Google Scholar] [CrossRef] [Green Version]
- Thuy, D.M.; Peacock, T.P.; Bich, V.T.N.; Fabrizio, T.; Hoang, D.N.; Tho, N.D.; Diep, N.T.; Nguyen, M.; Hoa, L.N.M.; Trang, H.T.T.; et al. Prevalence and diversity of H9N2 avian influenza in chickens of Northern Vietnam, 2014. Infect. Genet. Evol. 2016, 44, 530–540. [Google Scholar] [CrossRef] [Green Version]
- Zhu, R.; Xu, D.; Yang, X.; Zhang, J.; Wang, S.; Shi, H. Genetic and biological characterization of H9N2 avian influenza viruses isolated in China from 2011 to 2014. PLoS ONE 2018, 13, e0199260. [Google Scholar] [CrossRef] [Green Version]
- Park, K.J.; Song, M.S.; Kim, E.H.; Kwon, H.I.; Baek, Y.H.; Choi, E.H.; Park, S.J.; Kim, S.M.; Kim, Y.I.; Choi, W.S.; et al. Molecular characterization of mammalian-adapted Korean-type avian H9N2 virus and evaluation of its virulence in mice. J. Microbiol. 2015, 53, 570–577. [Google Scholar] [CrossRef]
- Wang, C.; Wang, Z.; Ren, X.; Wang, L.; Li, C.; Sun, Y.; Wang, M.; Tong, Q.; Sun, H.; Pu, J. Infection of chicken H9N2 influenza viruses in different species of domestic ducks. Vet. Microbiol. 2019, 233, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Ji, K.; Chen, J.; Tai, D.; Jiang, W.; Hou, G.; Chen, J.; Li, J.; Huang, B. Panorama phylogenetic diversity and distribution of Type A influenza virus. PLoS ONE 2009, 4, e5022. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Cao, B.; Hu, Y.; Feng, Z.; Wang, D.; Hu, W.; Chen, J.; Jie, Z.; Qiu, H.; Xu, K.; et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J. Med. 2013, 368, 1888–1897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pu, J.; Wang, S.; Yin, Y.; Zhang, G.; Carter, R.A.; Wang, J.; Xu, G.; Sun, H.; Wang, M.; Wen, C.; et al. Evolution of the H9N2 influenza genotype that facilitated the genesis of the novel H7N9 virus. Proc. Natl. Acad. Sci. USA 2015, 112, 548–553. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Yang, L.; Zhu, W.; Zhang, Y.; Zou, S.; Bo, H.; Gao, R.; Dong, J.; Huang, W.; Guo, J.; et al. Two Outbreak Sources of Influenza A (H7N9) Viruses Have Been Established in China. J. Virol. 2016, 90, 5561–5573. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Tang, Y.; Liu, X.; Peng, D.; Liu, W.; Liu, H.; Lu, S.; Liu, X. Characterization of H9N2 influenza viruses isolated from vaccinated flocks in an integrated broiler chicken operation in eastern China during a 5 year period (1998–2002). J. Gen. Virol. 2008, 89, 3102–3112. [Google Scholar] [CrossRef]
- Park, K.J.; Kwon, H.I.; Song, M.S.; Pascua, P.N.; Baek, Y.H.; Lee, J.H.; Jang, H.L.; Lim, J.Y.; Mo, I.P.; Moon, H.J.; et al. Rapid evolution of low-pathogenic H9N2 avian influenza viruses following poultry vaccination programmes. J. Gen. Virol. 2011, 92, 36–50. [Google Scholar] [CrossRef]
- Wei, Y.; Xu, G.; Zhang, G.; Wen, C.; Anwar, F.; Wang, S.; Lemmon, G.; Wang, J.; Carter, R.; Wang, M.; et al. Antigenic evolution of H9N2 chicken influenza viruses isolated in China during 2009–2013 and selection of a candidate vaccine strain with broad cross-reactivity. Vet. Microbiol. 2016, 182, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Peacock, T.P.; Harvey, W.T.; Sadeyen, J.R.; Reeve, R. The molecular basis of antigenic variation among A(H9N2) avian influenza viruses. Emerg. Microbes Infect. 2018, 7, 176. [Google Scholar] [CrossRef] [Green Version]
- Harvey, W.T.; Benton, D.J.; Gregory, V.; Hall, J.P.; Daniels, R.S.; Bedford, T.; Haydon, D.T.; Hay, A.J.; McCauley, J.W.; Reeve, R. Identification of Low- and High-Impact Hemagglutinin Amino Acid Substitutions That Drive Antigenic Drift of Influenza A(H1N1) Viruses. PLoS Pathog. 2016, 12, e1005526. [Google Scholar] [CrossRef]
- Koel, B.F.; Burke, D.F.; Bestebroer, T.M.; van der Vliet, S.; Zondag, G.C.; Vervaet, G.; Skepner, E.; Lewis, N.S.; Spronken, M.I.; Russell, C.A.; et al. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science 2013, 342, 976–979. [Google Scholar] [CrossRef] [PubMed]
- Hensley, S.E.; Das, S.R.; Bailey, A.L.; Schmidt, L.M.; Hickman, H.D.; Jayaraman, A.; Viswanathan, K.; Raman, R.; Sasisekharan, R.; Bennink, J.R.; et al. Hemagglutinin receptor binding avidity drives influenza A virus antigenic drift. Science 2009, 326, 734–736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abe, Y.; Takashita, E.; Sugawara, K.; Matsuzaki, Y.; Muraki, Y.; Hongo, S. Effect of the addition of oligosaccharides on the biological activities and antigenicity of influenza A/H3N2 virus hemagglutinin. J. Virol. 2004, 78, 9605–9611. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Yu, K.; Tian, G.; Yu, D.; Liu, L.; Jing, B.; Ping, J.; Chen, H. Evolution of H9N2 influenza viruses from domestic poultry in Mainland China. Virology 2005, 340, 70–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iqbal, M.; Yaqub, T.; Reddy, K.; McCauley, J.W. Novel genotypes of H9N2 influenza A viruses isolated from poultry in Pakistan containing NS genes similar to highly pathogenic H7N3 and H5N1 viruses. PLoS ONE 2009, 4, e5788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, W.; Liu, S.; Hou, G.; Li, J.; Zhuang, Q.; Wang, S.; Zhang, P.; Chen, J. Chinese and global distribution of H9 subtype avian influenza viruses. PLoS ONE 2012, 7, e52671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shanmuganatham, K.; Feeroz, M.M.; Jones-Engel, L.; Smith, G.J.; Fourment, M.; Walker, D.; McClenaghan, L.; Alam, S.M.; Hasan, M.K.; Seiler, P.; et al. Antigenic and molecular characterization of avian influenza A(H9N2) viruses, Bangladesh. Emerg. Infect. Dis. 2013, 19, 1393–1402. [Google Scholar] [CrossRef]
- Shanmuganatham, K.; Feeroz, M.M.; Jones-Engel, L.; Walker, D.; Alam, S.; Hasan, M.; McKenzie, P.; Krauss, S.; Webby, R.J.; Webster, R.G. Genesis of avian influenza H9N2 in Bangladesh. Emerg. Microbes Infect. 2014, 3, e88. [Google Scholar] [CrossRef]
- Song, H.; Nieto, G.R.; Perez, D.R. A new generation of modified live-attenuated avian influenza viruses using a two-strategy combination as potential vaccine candidates. J. Virol. 2007, 81, 9238–9248. [Google Scholar] [CrossRef] [Green Version]
- Cai, Y.; Song, H.; Ye, J.; Shao, H.; Padmanabhan, R.; Sutton, T.C.; Perez, D.R. Improved hatchability and efficient protection after in ovo vaccination with live-attenuated H7N2 and H9N2 avian influenza viruses. Virol. J. 2011, 8, 31. [Google Scholar] [CrossRef] [Green Version]
- Lin, W.; Fan, H.; Cheng, X.; Ye, Y.; Chen, X.; Ren, T.; Qi, W.; Liao, M. A baculovirus dual expression system-based vaccine confers complete protection against lethal challenge with H9N2 avian influenza virus in mice. Virol. J. 2011, 8, 273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Xue, L.; Hu, S.; Cheng, H.; Deng, Y.; Hu, Z.; Wang, X.; Liu, X. Chimeric Newcastle disease virus-vectored vaccine protects chickens against H9N2 avian influenza virus in the presence of pre-existing NDV immunity. Arch. Virol. 2018, 163, 3365–3371. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Zhao, Q.; Zhao, K.; Wang, X.; Zhao, G.; Li, Q.; Gu, M.; Peng, D.; Liu, X. The antigenic drift molecular basis of the H5N1 influenza viruses in a novel branch of clade 2.3.4. Vet. Microbiol. 2014, 171, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Neumann, G.; Watanabe, T.; Ito, H.; Watanabe, S.; Goto, H.; Gao, P.; Hughes, M.; Perez, D.R.; Donis, R.; Hoffmann, E.; et al. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 1999, 96, 9345–9350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edwards, S. OIE laboratory standards for avian influenza. Dev. Biol. 2006, 124, 159–162. [Google Scholar]
- Németh, B.; Fasseeh, A.; Molnár, A.; Bitter, I.; Horváth, M.; Kóczián, K.; Götze, Á.; Nagy, B. A systematic review of health economic models and utility estimation methods in schizophrenia. Expert Rev. Pharm. Outcomes Res. 2018, 18, 267–275. [Google Scholar] [CrossRef]
- Kaverin, N.V.; Rudneva, I.A.; Ilyushina, N.A.; Lipatov, A.S.; Krauss, S.; Webster, R.G. Structural differences among hemagglutinins of influenza A virus subtypes are reflected in their antigenic architecture: Analysis of H9 escape mutants. J. Virol. 2004, 78, 240–249. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.N.; Lee, D.H.; Park, J.K.; Lim, T.H.; Youn, H.N.; Yuk, S.S.; Lee, Y.J.; Mo, I.P.; Sung, H.W.; Lee, J.B.; et al. Isolation and characterization of a novel H9N2 influenza virus in Korean native chicken farm. Avian Dis. 2011, 55, 724–727. [Google Scholar] [CrossRef]
- Okamatsu, M.; Sakoda, Y.; Kishida, N.; Isoda, N.; Kida, H. Antigenic structure of the hemagglutinin of H9N2 influenza viruses. Arch. Virol. 2008, 153, 2189–2195. [Google Scholar] [CrossRef] [Green Version]
- Wan, Z.; Ye, J.; Xu, L.; Shao, H.; Jin, W.; Qian, K.; Wan, H.; Qin, A. Antigenic mapping of the hemagglutinin of an H9N2 avian influenza virus reveals novel critical amino acid positions in antigenic sites. J. Virol. 2014, 88, 3898–3901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peacock, T.; Reddy, K.; James, J.; Adamiak, B.; Barclay, W.; Shelton, H.; Iqbal, M. Antigenic mapping of an H9N2 avian influenza virus reveals two discrete antigenic sites and a novel mechanism of immune escape. Sci. Rep. 2016, 6, 18745. [Google Scholar] [CrossRef] [PubMed]

| Vaccines | HI Titers(log2) | Challenge Virus | Positive Viral Isolation Chickens/Total Chickens | |||
|---|---|---|---|---|---|---|
| Day 3 | Day 5 | Day 7 | Day 9 | |||
| R-WT/PR8 | 10.67 ± 0.82 | GX/55 19166 | 10/10 10/10 | 2/10 6/10 | 0/10 0/10 | 0/10 0/10 |
| 19166 | 11.25 ± 0.66 | GX/55 19166 | 10/10 10/10 | 4/10 2/10 | 0/10 0/10 | 0/10 0/10 |
| R-Mut/PR8 | 11.4 ± 0.66 | GX/55 19166 | 10/10 10/10 | 4/10 4/10 | 0/10 0/10 | 0/10 0/10 |
| PBS | 0 | GX/55 19166 | 10/10 10/10 | 10/10 10/10 | 0/10 1/10 | 0/10 0/10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Q.; Zhao, L.; Guo, Y.; Zhao, Y.; Li, Y.; Chen, N.; Lu, Y.; Yu, M.; Deng, L.; Ping, J. Antigenic Evolution Characteristics and Immunological Evaluation of H9N2 Avian Influenza Viruses from 1994–2019 in China. Viruses 2022, 14, 726. https://doi.org/10.3390/v14040726
Liu Q, Zhao L, Guo Y, Zhao Y, Li Y, Chen N, Lu Y, Yu M, Deng L, Ping J. Antigenic Evolution Characteristics and Immunological Evaluation of H9N2 Avian Influenza Viruses from 1994–2019 in China. Viruses. 2022; 14(4):726. https://doi.org/10.3390/v14040726
Chicago/Turabian StyleLiu, Qingzheng, Lingcai Zhao, Yanna Guo, Yongzhen Zhao, Yingfei Li, Na Chen, Yuanlu Lu, Mengqi Yu, Lulu Deng, and Jihui Ping. 2022. "Antigenic Evolution Characteristics and Immunological Evaluation of H9N2 Avian Influenza Viruses from 1994–2019 in China" Viruses 14, no. 4: 726. https://doi.org/10.3390/v14040726
APA StyleLiu, Q., Zhao, L., Guo, Y., Zhao, Y., Li, Y., Chen, N., Lu, Y., Yu, M., Deng, L., & Ping, J. (2022). Antigenic Evolution Characteristics and Immunological Evaluation of H9N2 Avian Influenza Viruses from 1994–2019 in China. Viruses, 14(4), 726. https://doi.org/10.3390/v14040726





