Transcriptomics Reveal Several Novel Viruses from Canegrubs (Coleoptera: Scarabaeidae) in Central Queensland, Australia
Abstract
1. Introduction
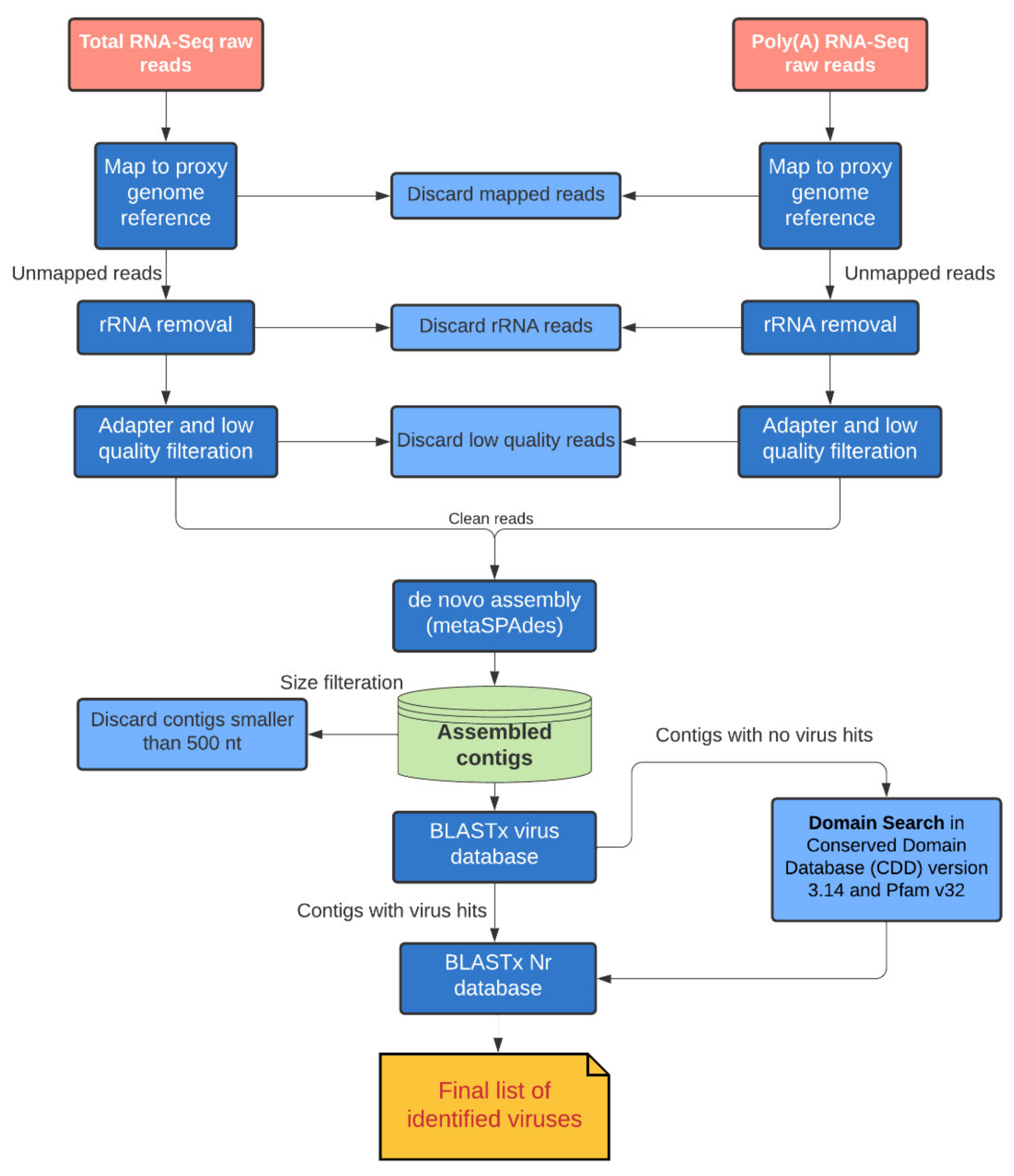
2. Materials and Methods
2.1. Insect Collection and RNA Extraction
2.2. RNA-Seq Data Analysis
2.3. Phylogenetic Analysis
3. Results and Discussion
3.1. Dermolepida Albohirtum Picorna-like Virus (DaPV1)
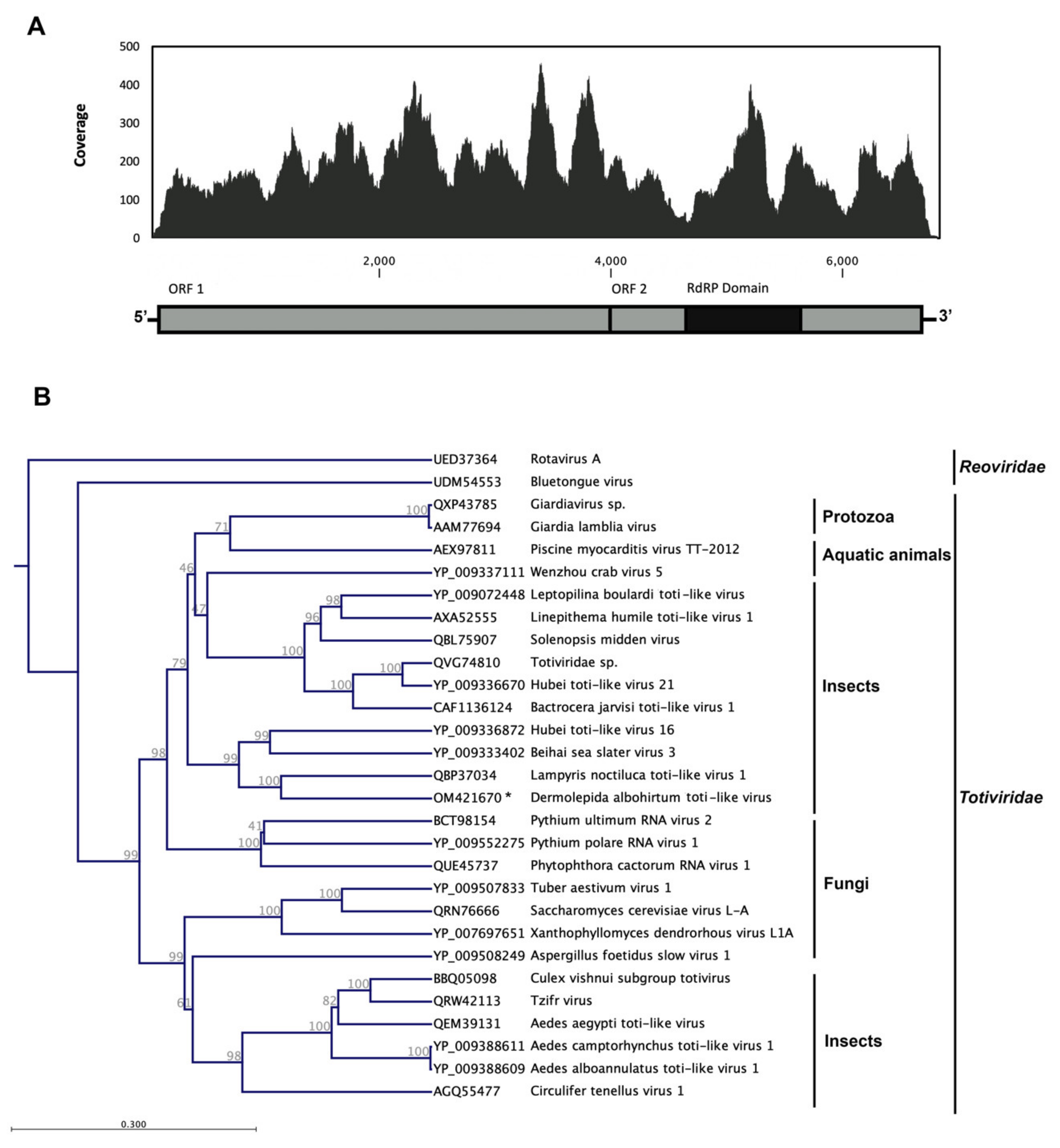
3.2. Dermolepida Albohirtum Toti-like Virus (DaTV1)
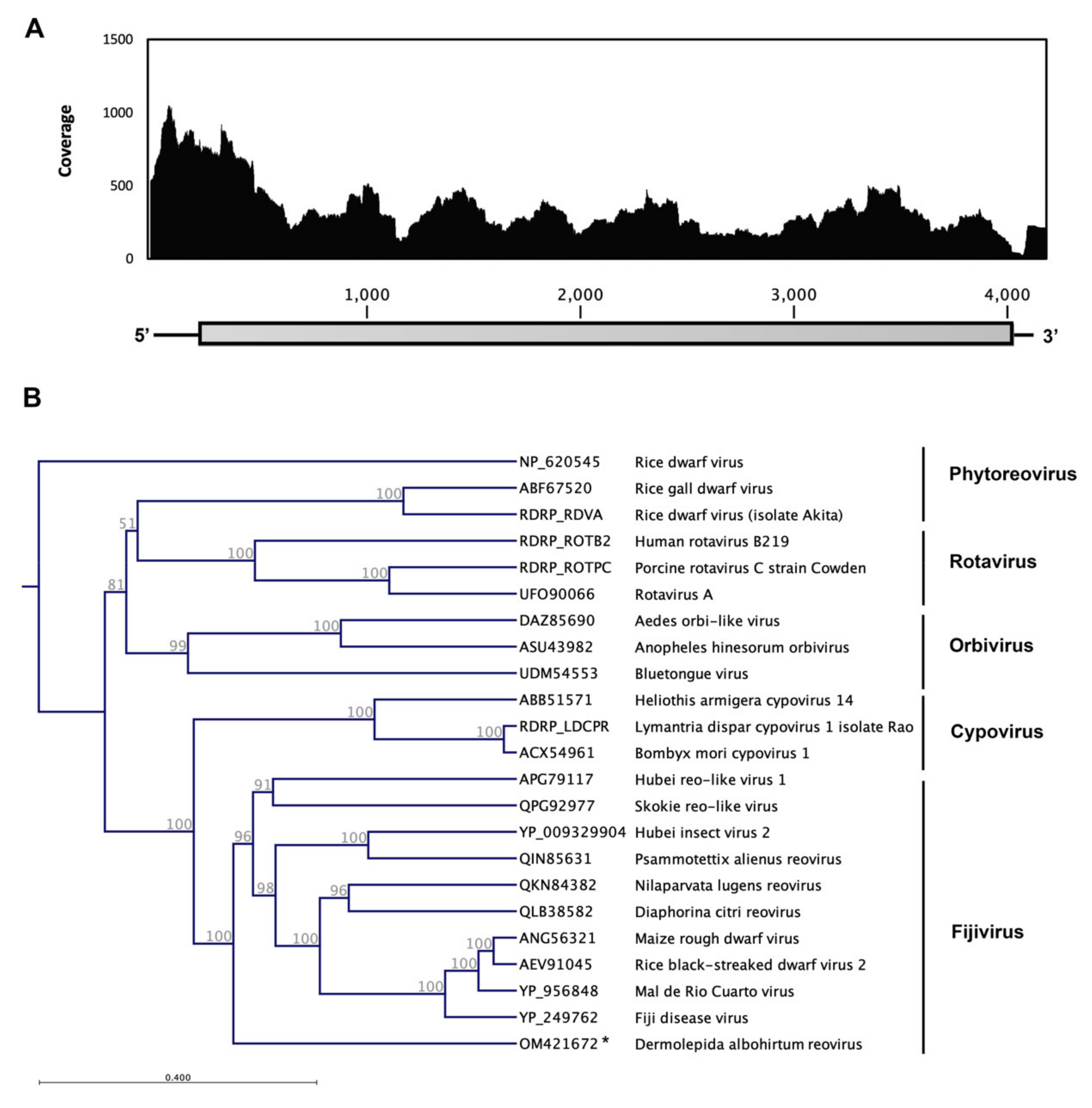
3.3. Dermolepida Albohirtum Reovirus (DaRV)
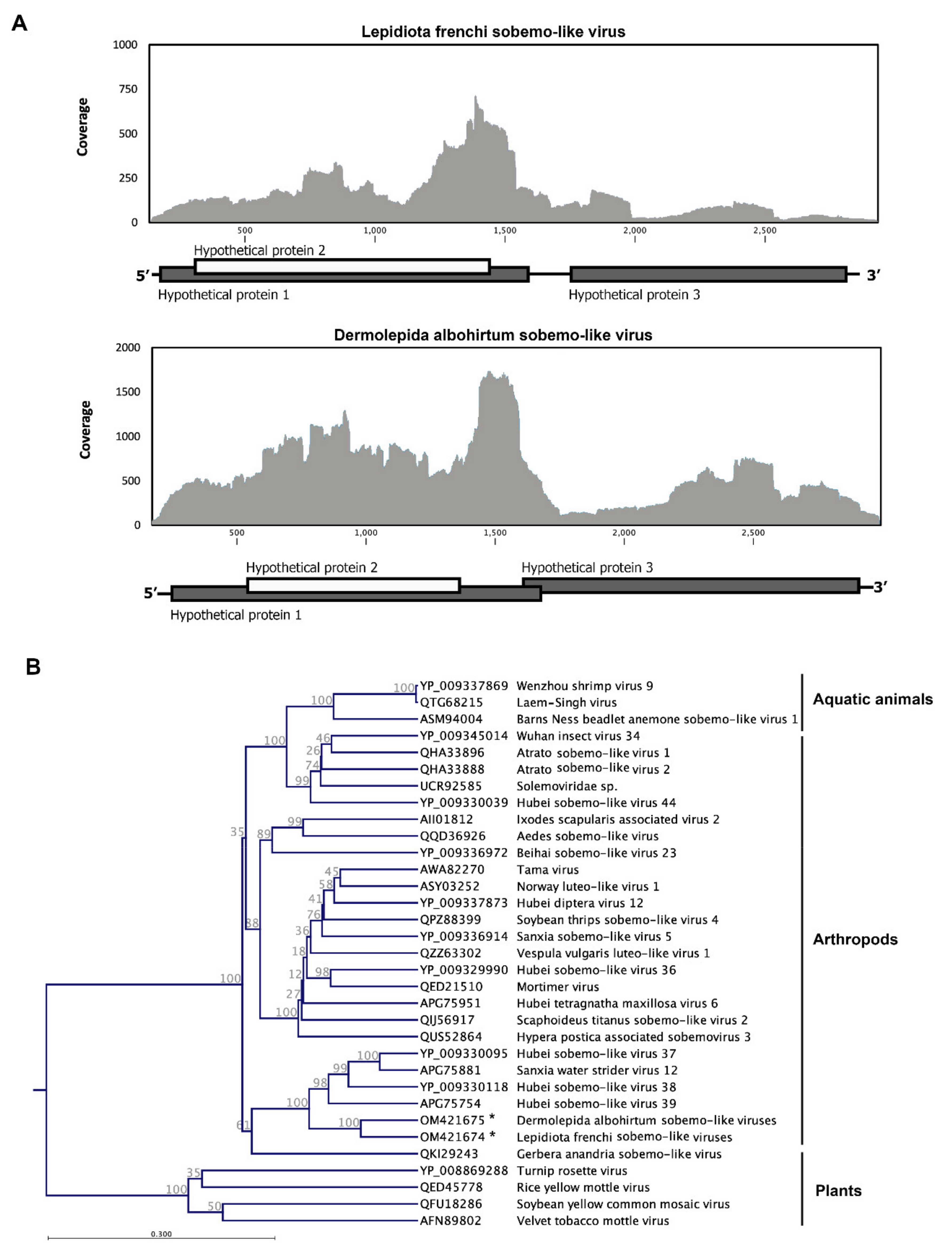
3.4. Canegrubs’ Sobemo-like Viruses
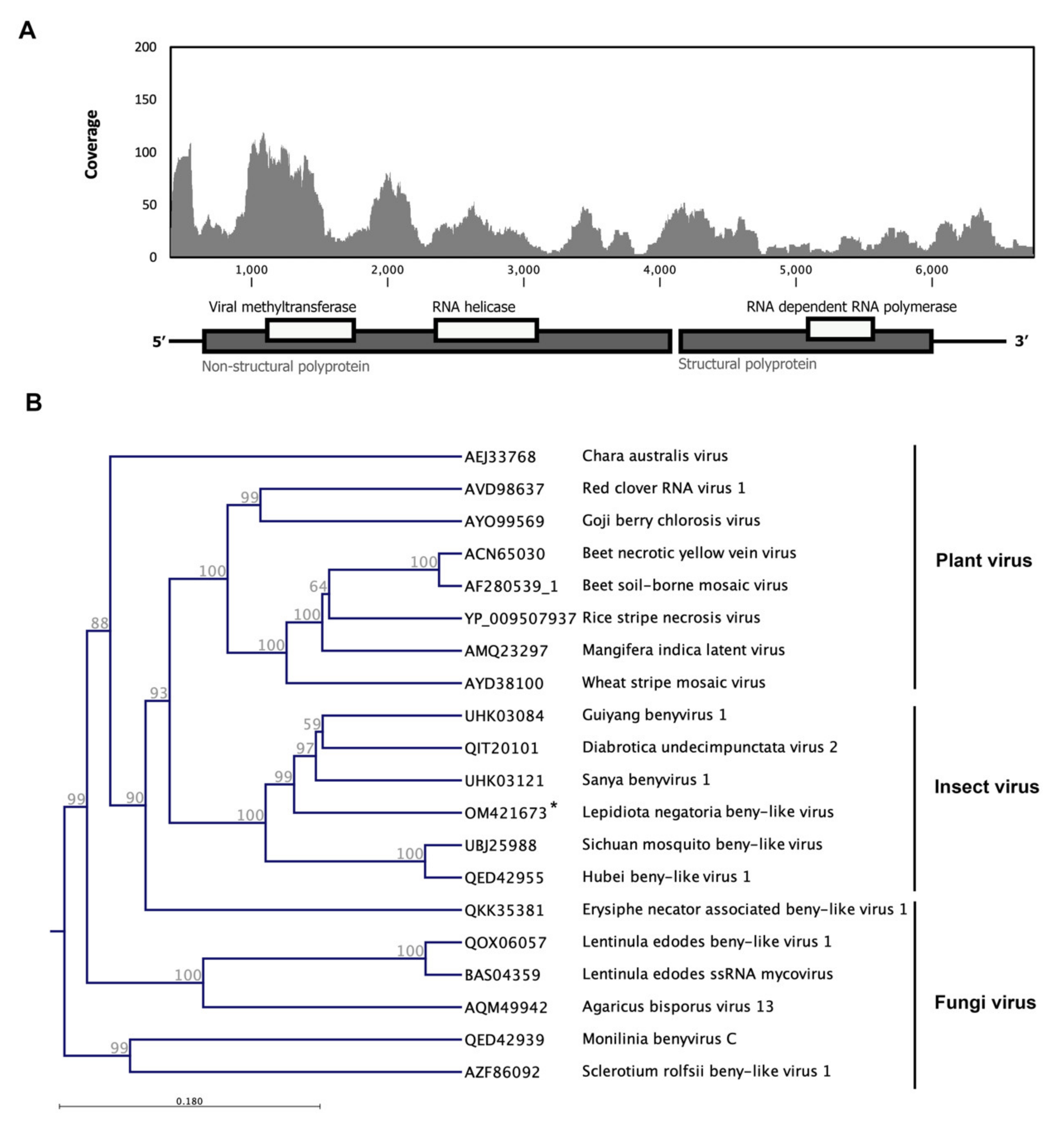
3.5. Lepidiota Negatoria Beny-like Virus (LnBV)
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Long, W.H.; Hensley, S.D. Insect pests of sugar cane. Annu. Rev. Entomol. 1972, 17, 149–176. [Google Scholar] [CrossRef]
- Allsopp, P.G. IPM for whitegrubs in Australian sugarcane: From a continuing success to regressing to the past. Sugar Tech. 2020, 23, 225–238. [Google Scholar] [CrossRef]
- Frew, A.; Barnett, K.; Nielsen, U.N.; Riegler, M.; Johnson, S.N. Belowground ecology of Scarabs feeding on grass roots: Current knowledge and future directions for management in Australasia. Front. Plant. Sci. 2016, 7, 670. [Google Scholar] [CrossRef] [PubMed]
- Miller, L.J.; Allsopp, P.G. Identification of Australian canegrubs (Coleoptera: Scarabaeidae: Melolonthini). Invertebr. Taxon. 2000, 14, 377–409. [Google Scholar] [CrossRef]
- Allsopp, P.G. Integrated management of sugarcane whitegrubs in Australia: An evolving success. Annu. Rev. Entomol. 2010, 55, 329–349. [Google Scholar] [CrossRef]
- Milner, R.J.; Samson, P.R.; Bullard, G.K. FI-1045: A profile of a commercially useful isolate of Metarhizium anisopliae var. anisopliae. Biocontrol Sci. Technol. 2002, 12, 43–58. [Google Scholar] [CrossRef]
- Goulson, D. An overview of the environmental risks posed by neonicotinoid insecticides. J. Appl. Ecol. 2013, 50, 977–987. [Google Scholar] [CrossRef]
- Warne, M.S.; Smith, R.A.; Turner, R.D.R. Analysis of pesticide mixtures discharged to the lagoon of the Great Barrier Reef, Australia. Environ. Pollut. 2020, 265, 114088. [Google Scholar] [CrossRef]
- Sallam, N. Review of current knowledge on the population dynamics of Dermolepida albohirtum (Waterhouse) (Coleoptera: Scarabaeidae). Aust. J. Entomol. 2011, 50, 300–308. [Google Scholar] [CrossRef]
- Goodwin, R.H.; Roberts, R.J. Diagnosis and infectivity of entomopoxviruses from three australian scarab beetle larvae (Coleoptera: Scarabaeidae). J. Invertebr. Pathol. 1975, 25, 47–57. [Google Scholar] [CrossRef]
- Vago, C. A new type of insect virus. J. Invertebr. Pathol. 1963, 5, 275. [Google Scholar]
- Lipa, J.J.; Bartkowski, J. A newly discovered poxlike virus disease of dung beetles, Geotrupes silvaticus (Coleoptera: Scarabaeidae). J. Invertebr. Pathol. 1972, 20, 218–219. [Google Scholar] [CrossRef]
- Sezen, K.; Demirbag, Z. A new isolate of Melolontha melolontha entomopoxvirus in Turkey: Morphology, infectivity and prevalence in the field. Appl. Entomol. Zool. 2006, 41, 471–477. [Google Scholar] [CrossRef][Green Version]
- Katagiri, K.; Kushida, T.; Kasuga, S.; Ohba, M. An entomopoxvirus disease of the cupreous chafer, Anomala cuprea Hope. Jpn. J. Appl. Entomol. Zool. 1975, 19, 243–252. [Google Scholar] [CrossRef]
- Beaudoin, L.; Robert, P.; Lal, S.N. An entomopoxvirus observed in Adoretus versutus Harold (Coleoptera; Scarabaeidae; Rutelinae) in Fiji. Int. J. Pest Manag. 1994, 40, 66–68. [Google Scholar] [CrossRef]
- Scotti, P.D.; Fredericksen, S. Manawatu virus: A nodavirus isolated from Costelytra zealandica (White) (Coleoptera: Scarabaeidae). Arch. Virol. 1987, 97, 85–92. [Google Scholar] [CrossRef]
- Dearing, S.C.; Scotti, P.D.; Wigley, P.J.; Dhana, S.D. A small RNA virus isolated from the grass grub, Costelytra zealandica (Coleoptera: Scarabaeidae). New Zeal. J. Zool. 1980, 7, 267–269. [Google Scholar] [CrossRef]
- Scotti, P.D.; Dearing, S.; Mossop, D.W. Flock House virus: A nodavirus isolated from Costelytra zealandica (White) (Coleoptera: Scarabaeidae). Arch. Virol. 1983, 75, 181–189. [Google Scholar] [CrossRef]
- Longworth, J.F.; Carey, G.P. A small RNA virus with a divided genome from Heteronychus arator (F.) [Coleoptera: Scarabaeidae]. J. Gen. Virol. 1976, 33, 31–40. [Google Scholar] [CrossRef]
- Jenkins, D.A.; Hunter, W.B.; Goenaga, R. Effects of Invertebrate Iridescent Virus 6 in Phyllophaga vandinei and its potential as a biocontrol delivery system. J. Insect Sci. 2011, 11, 44. [Google Scholar] [CrossRef]
- Poprawski, T.J.; Yule, W.N. A new small iridescent virus from grubs of Phyllophaga anxia (LeConte) (Col., Scarabaeidae). J. Appl. Entomol. 1990, 110, 63–67. [Google Scholar] [CrossRef]
- Carey, G.P.; Lescott, T.; Robertson, J.S.; Spencer, L.K.; Kelly, D.C. Three African isolates of small iridescent viruses: Type 21 from Heliothis armigera (Lepidoptera: Noctuidae), type 23 from Heteronychus arator (Coleoptera: Scarabaeidae), and type 28 from Lethocerus columbiae (Hemiptera Heteroptera: Belostomatidae). Virology 1978, 85, 307–309. [Google Scholar] [CrossRef]
- Lacey, L.A.; Adams, J.R. An iridescent virus from Popillia japonica (Col.: Scarabaeidae). Entomophaga 1994, 39, 131–136. [Google Scholar] [CrossRef]
- Moore, S.G.; Kalmakoff, J.; Miles, J.A.R. An iridescent virus and a rickettsia from the grass grub Costelytra zealandica (Coleoptera: Scarabaeidae). New Zeal. J. Zool. 1974, 1, 205–210. [Google Scholar] [CrossRef][Green Version]
- Huger, A.M. A virus disease of the Indian rhinoceros beetle, Oryctes rhinoceros (Linnaeus), caused by a new type of insect virus, Rhabdionvirus oryctes. J. Invertebr. Pathol. 1966, 8, 38–51. [Google Scholar] [CrossRef]
- Lee, S.; Park, K.-H.; Nam, S.-H.; Kwak, K.-W.; Choi, J.-Y. First report of Oryctes rhinoceros nudivirus (Coleoptera: Scarabaeidae) causing severe disease in Allomyrina dichotoma in Korea. J. Insect Sci. 2015, 15, 26. [Google Scholar] [CrossRef] [PubMed]
- Etebari, K.; Shelomi, M.; Furlong, M.J. Identification of a novel Picorna-like Virus in coconut rhinoceros beetles (Oryctes rhinoceros). Virus Res. 2020, 287, 198100. [Google Scholar] [CrossRef]
- Glare, T.R. Further electron microscopic evidence of inapparent viral infections in the midgut of the New Zealand grass grub, Costelytra zealandica White (Coleoptera: Scarabaeidae). J. Invertebr. Pathol. 1992, 59, 206–209. [Google Scholar] [CrossRef]
- Goodwin, R.H.; Filshie, B. Morphology and development of an occluded virus from the black-soil scarab, Othnonius batesi. J. Invertebr. Pathol. 1969, 13, 317–329. [Google Scholar] [CrossRef]
- Poinar, R.H.; Poinar, G.O.; Jackson, T. Electron microscope evidence of a new virus in the grass grub, Costelytra zealandica (Coleoptera: Scarabaeidae). J. Invertebr. Pathol. 1991, 57, 137–140. [Google Scholar] [CrossRef]
- Zelazny, B. Occurrence of the baculovirus disease of the coconut palm rhinoceros beetle in the Philippines and in Indonesia. FAO Plant Prot. Bull. 1977, 25, 73–77. [Google Scholar]
- Etebari, K.; Filipovic, I.; Rasic, G.; Devine, G.J.; Tsatsia, H.; Furlong, M.J. Complete genome sequence of Oryctes rhinoceros nudivirus isolated from the coconut rhinoceros beetle in Solomon Islands. Virus Res. 2020, 278, 197864. [Google Scholar] [CrossRef] [PubMed]
- Caltagirone, L.E. Landmark examples in classical biological control. Annu. Rev. Entomol. 1981, 26, 213–232. [Google Scholar] [CrossRef]
- Reid, S.; Chan, L.C.L.; Matindoost, L.; Pushparajan, C.; Visnovsky, G. Cell culture for production of insecticidal viruses. In Microbial-Based Biopesticides: Methods and Protocols; Glare, T.R., MoranDiez, M.E., Eds.; Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2016; Volume 1477, pp. 95–117. [Google Scholar]
- Tanaka, S.; Harrison, R.L.; Arai, H.; Katayama, Y.; Mizutani, T.; Inoue, M.N.; Miles, J.; Marshall, S.D.G.; Kitalong, C.; Nakai, M. Confirmation of Oryctes rhinoceros nudivirus infections in G-haplotype coconut rhinoceros beetles (Oryctes rhinoceros) from Palauan PCR-positive populations. Sci. Rep. 2021, 11, 18820. [Google Scholar] [CrossRef]
- Etebari, K.; Hereward, J.; Sailo, A.; Ahoafi, E.M.; Tautua, R.; Tsatsia, H.; Jackson, G.V.; Furlong, M.J. Examination of population genetics of the Coconut Rhinoceros Beetle (Oryctes rhinoceros) and the incidence of its biocontrol agent (Oryctes rhinoceros nudivirus) in the South Pacific Islands. Curr. Res. Insect Sci. 2021, 1, 100015. [Google Scholar] [CrossRef]
- Kopylova, E.; Noé, L.; Touzet, H. SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. metaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef]
- Ye, Z.X.; Li, Y.H.; Chen, J.P.; Zhang, C.X.; Li, J.M. Complete genome analysis of a novel iflavirus from a leaf beetle, Aulacophora lewisii. Arch. Virol. 2021, 166, 309–312. [Google Scholar] [CrossRef]
- Liu, Y.; Ren, G.; Jiang, L.; Feng, C.; Liu, D.; Liu, Y.; Xu, P. Sequencing and phylogenetic characterization of a novel RNA virus genome from Harmonia axyridis. Mol. Biol. Rep. 2020, 47, 4015–4019. [Google Scholar] [CrossRef]
- Yu-Juan, H.; Zhuang-Xin, Y.; Jian-Ping, C.; Chuan-Xi, Z.; Gang, L.; Jun-Min, L. Complete genome analysis of a novel picorna-like virus from a ladybird beetle (Cheilomenes sexmaculata). Arch. Virol. 2022, 1–5. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, Y.; Ge, X.; Yuan, J.; Shi, Z. A novel totivirus-like virus isolated from bat guano. Arch. Virol. 2012, 157, 1093–1099. [Google Scholar] [CrossRef]
- Zell, R. Picornaviridae-the ever-growing virus family. Arch. Virol. 2018, 163, 299–317. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Tripp, J.; Bonning, B.C.; Miller, W.A. Challenges associated with research on RNA viruses of insects. Curr. Opin. Insect Sci. 2015, 8, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Carballo, A.; Murillo, R.; Jakubowska, A.; Herrero, S.; Williams, T.; Caballero, P. Co-infection with iflaviruses influences the insecticidal properties of Spodoptera exigua multiple nucleopolyhedrovirus occlusion bodies: Implications for the production and biosecurity of baculovirus insecticides. PLoS ONE 2017, 12, e0177301. [Google Scholar] [CrossRef] [PubMed]
- Jakubowska, A.K.; Murillo, R.; Carballo, A.; Williams, T.; van Lent, J.W.M.; Caballero, P.; Herrero, S. Iflavirus increases its infectivity and physical stability in association with baculovirus. Peerj 2016, 4, e1687. [Google Scholar] [CrossRef] [PubMed]
- King, A.M.Q.; Adams, M.J.; Carstens, E.B.; Lefkowitz, E.J. (Eds.) Family-Totiviridae. In Virus Taxonomy; Elsevier: San Diego, CA, USA, 2012; pp. 639–650. [Google Scholar]
- Shi, M.; Lin, X.D.; Tian, J.H.; Chen, L.J.; Chen, X.; Li, C.X.; Qin, X.C.; Li, J.; Cao, J.P.; Eden, J.S.; et al. Redefining the invertebrate RNA virosphere. Nature 2016, 540, 539–543. [Google Scholar] [CrossRef]
- Viljakainen, L.; Borshagovski, A.-M.; Saarenpää, S.; Kaitala, A.; Jurvansuu, J. Identification and characterisation of common glow-worm RNA viruses. Virus Genes 2020, 56, 236–248. [Google Scholar] [CrossRef]
- Sandlund, L.; Mor, S.K.; Singh, V.K.; Padhi, S.K.; Phelps, N.B.D.; Nylund, S.; Mikalsen, A.B. Comparative molecular characterization of novel and known piscine Toti-Like viruses. Viruses 2021, 13, 1063. [Google Scholar] [CrossRef]
- Zhang, P.; Liu, W.; Cao, M.; Massart, S.; Wang, X. Two novel totiviruses in the white-backed planthopper, Sogatella furcifera. J. Gen. Virol. 2018, 99, 710–716. [Google Scholar] [CrossRef]
- Poimala, A.; Vainio, E.J. Complete genome sequence of a novel toti-like virus from the plant-pathogenic oomycete Phytophthora cactorum. Arch. Virol. 2020, 165, 1679–1682. [Google Scholar] [CrossRef]
- Soo, H.M.; Handley, J.A.; Maugeri, M.M.; Burns, P.; Smith, G.R.; Dale, J.L.; Harding, R.M. Molecular characterization of Fiji disease fijivirus genome segment 9. J. Gen. Virol. 1998, 79, 3155–3161. [Google Scholar] [CrossRef][Green Version]
- Wang, R.Y.; Gergerich, R.C.; Kim, K.S. Entry of ingested plant viruses into the hemocoel of the beetle vector Diabrotica undecimpunctata Howardi. Phytopathology 1994, 84, 147–153. [Google Scholar] [CrossRef]
- King, A.M.Q.; Adams, M.J.; Carstens, E.B.; Lefkowitz, E.J. (Eds.) Family-Reoviridae. In Virus Taxonomy; Elsevier: San Diego, CA, USA, 2012; pp. 541–637. [Google Scholar]
- Nakashima, N.; Noda, H. Nonpathogenic Nilaparvata lugens reovirus is transmitted to the brown planthopper through rice plant. Virology 1995, 207, 303–307. [Google Scholar] [CrossRef]
- Belaganahalli, M.N.; Maan, S.; Maan, N.S.; Tesh, R.; Attoui, H.; Mertens, P.P.C. Umatilla virus genome sequencing and phylogenetic analysis: Identification of stretch lagoon orbivirus as a new member of the Umatilla virus species. PLoS ONE 2011, 6, e23605. [Google Scholar] [CrossRef] [PubMed]
- Parry, R.; James, M.E.; Asgari, S. Uncovering the worldwide diversity and evolution of the Virome of the Mosquitoes Aedes aegypti and Aedes albopictus. Microorganisms 2021, 9, 1653. [Google Scholar] [CrossRef] [PubMed]
- Colmant, A.M.G.; Etebari, K.; Webb, C.E.; Ritchie, S.A.; Jansen, C.C.; van den Hurk, A.F.; Bielefeldt-Ohmann, H.; Hobson-Peters, J.; Asgari, S.; Hall, R.A. Discovery of new orbiviruses and totivirus from Anopheles mosquitoes in Eastern Australia. Arch. Virol. 2017, 162, 3529–3534. [Google Scholar] [CrossRef]
- Alkhamis, M.A.; Aguilar-Vega, C.; Fountain-Jones, N.M.; Lin, K.; Perez, A.M.; Sanchez-Vizcaino, J.M. Global emergence and evolutionary dynamics of bluetongue virus. Sci. Rep. 2020, 10, 21677. [Google Scholar] [CrossRef]
- Li, Y.; Tan, L.; Li, Y.; Chen, W.; Zhang, J.; Hu, Y. Identification and genome characterization of Heliothis armigera cypovirus types 5 and 14 and Heliothis assulta cypovirus type 14. J. Gen. Virol. 2006, 87, 387–394. [Google Scholar] [CrossRef]
- Hagiwara, K.; Rao, S.J.; Scott, S.W.; Carner, G.R. Nucleotide sequences of segments 1, 3 and 4 of the genome of Bombyx mori cypovirus 1 encoding putative capsid proteins VP1, VP3 and VP4, respectively. J. Gen. Virol. 2002, 83, 1477–1482. [Google Scholar] [CrossRef]
- Tamm, T.; Truve, E. Sobemoviruses. J. Virol. 2000, 74, 6231–6241. [Google Scholar] [CrossRef]
- Sõmera, M.; Sarmiento, C.; Truve, E. Overview on Sobemoviruses and a Proposal for the Creation of the Family Sobemoviridae. Viruses 2015, 7, 3076–3115. [Google Scholar] [CrossRef]
- Taengchaiyaphum, S.; Srisala, J.; Sanguanrut, P.; Ongvarrasopone, C.; Flegel, T.W.; Sritunyalucksana, K. Full genome characterization of Laem Singh virus (LSNV) in shrimp Penaeus monodon. Aquaculture 2021, 538, 736533. [Google Scholar] [CrossRef]
- Kubacki, J.; Flacio, E.; Qi, W.; Guidi, V.; Tonolla, M.; Fraefel, C. Viral metagenomic analysis of aedes albopictus mosquitos from southern switzerland. Viruses 2020, 12, 929. [Google Scholar] [CrossRef] [PubMed]
- Sanborn, M.A.; Klein, T.A.; Kim, H.-C.; Fung, C.K.; Figueroa, K.L.; Yang, Y.; Asafo-adjei, E.A.; Jarman, R.G.; Hang, J. Metagenomic analysis reveals three novel and prevalent mosquito viruses from a single pool of Aedes vexans nipponii Collected in the Republic of Korea. Viruses 2019, 11, 222. [Google Scholar] [CrossRef] [PubMed]
- Thongsripong, P.; Chandler, J.A.; Kittayapong, P.; Wilcox, B.A.; Kapan, D.D.; Bennett, S.N. Metagenomic shotgun sequencing reveals host species as an important driver of virome composition in mosquitoes. Sci. Rep. 2021, 11, 8448. [Google Scholar] [CrossRef] [PubMed]
- Le Lay, C.; Shi, M.; Buček, A.; Bourguignon, T.; Lo, N.; Holmes, E.C. Unmapped RNA virus diversity in termites and their symbionts. Viruses 2020, 12, 1145. [Google Scholar] [CrossRef]
- Liu, S.; Valencia-Jiménez, A.; Darlington, M.; Vélez, A.M.; Bonning, B.C. Diabrotica undecimpunctata virus 2, a novel small RNA virus discovered from Southern corn rootworm, Diabrotica undecimpunctata howardi Barber (Coleoptera: Chrysomelidae). Microbiol. Resour. Announc. 2020, 9, e00380-20. [Google Scholar] [CrossRef]
- Gilmer, D.; Ratti, C.; Consortium, I.R. ICTV virus taxonomy profile: Benyviridae. J. Gen. Virol. 2017, 98, 1571–1572. [Google Scholar] [CrossRef]
- Kondo, H.; Hirano, S.; Chiba, S.; Andika, I.B.; Hirai, M.; Maeda, T.; Tamada, T. Characterization of burdock mottle virus, a novel member of the genus Benyvirus, and the identification of benyvirus-related sequences in the plant and insect genomes. Virus Res. 2013, 177, 75–86. [Google Scholar] [CrossRef]

| Virus Name | Taxonomy | Host Common Name | Scientific Name | Location | Reference |
|---|---|---|---|---|---|
| Poxi-like virus | Poxviridae | Cockchafer (maybug) | Melolontha spp. | Poland | [11] |
| Poxi-like virus | Poxviridae | Dung beetles | Geotrupes spp. | Poland | [12] |
| Entomopoxvirus | Poxviridae | Cockchafer (maybug) | Melolontha melolontha | Turkey | [13] |
| Entomopoxvirus | Poxviridae | Cupreous chafer | Anomala cuprea | Japan | [14] |
| Entomopoxvirus | Poxviridae | Rose beetle | Adoretus versutus | Fiji | [15] |
| Manawatu virus | Nodaviridae | Grass grub | Costelytra zealandica | New Zealand | [16] |
| Flock house virus | Nodaviridae | Grass grub | Costelytra zealandica | New Zealand | [17,18] |
| Black beetle virus | Nodaviridae | African black beetle | Heteronychus arator | New Zealand | [19] |
| Iridescent virus 6 | Iridoviridae | June bugs | Phyllophaga vandinei | USA | [20] |
| Small iridescent virus | Iridoviridae | Cranberry white grub | Phyllophaga anxia | Canada | [21] |
| Small iridescent virus | Iridoviridae | African black beetle | Heteronychus arator | South Africa | [22] |
| Blue iridovirus | Iridoviridae | Japanese beetle | Popillia japonica | Terceira Island, Portugal | [23] |
| Small iridescent virus | Iridoviridae | Grass grub | Costelytra zealandica | New Zealand | [24] |
| Small iridescent virus | Iridoviridae | African black beetle | Heteronychus arator | South Africa | [22] |
| Oryctes rhinoceros nudivirus | Nudiviridae | Coconut rhinoceros beetle | Oryctes rhinoceros | Asia, Pacific | [25] |
| Allomyrina dichotoma nudivirus | Nudiviridae | Korean horn beetle | Allomyrina dichotoma | Korea | [26] |
| Oryctes rhinoceros picorna-like virus | Picornavirales | Coconut rhinoceros beetle | Oryctes rhinoceros | Taiwan | [27] |
| Unknown virus | Unknown | Grass grub | Costelytra zealandica | New Zealand | [28] |
| Spheroidosis viruses | Unknown | Black soil scrab | Othnonius batesi | Australia | [29] |
| Lake Grasmere virus | Unknown | Grass grub | Costelytra zealandica | New Zealand | [30] |
| Samples | Raw Reads | Clean Reads | Genome 1 (Oryctes rhinoceros) | Genome 2 (Onthophagus taurus) * | Assembly Overview | ||||
|---|---|---|---|---|---|---|---|---|---|
| Mapped (%) | Unmapped Reads | Mapped (%) | Unmapped Reads | Contigs >500 bp | N50 | L50 (bp) | |||
| Dermolepida albohirtum | |||||||||
| Total RNA-Seq | 90,875,308 | 90,823,228 | 73.19% | 24,333,795 | 28.47% | 17,405,199 | 31,911 | 1373 | 7905 |
| mRNA-Seq | 73,406,632 | 70,531,633 | 24.19% | 51,654,119 | 6.64% | 48,224,340 | |||
| Lepidiota negatoria | |||||||||
| Total RNA-Seq | 83,850,940 | 83,784,724 | 85.17% | 12,418,859 | 13.34% | 10,762,711 | 30,155 | 1503 | 7428 |
| mRNA-Seq | 75,752,788 | 72,943,712 | 18.87% | 57,271,175 | 4.94% | 54,442,653 | |||
| Lepidiota frenchi | |||||||||
| Total RNA-Seq | 81,565,214 | 81,520,706 | 88.98% | 8,978,364 | 7.45% | 8,309,559 | 26,973 | 1403 | 6889 |
| mRNA-Seq | 69,835,996 | 67,249,833 | 18.95% | 52,746,565 | 5.2% | 49,958,261 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Etebari, K.; Lenancker, P.; Powell, K.S.; Furlong, M.J. Transcriptomics Reveal Several Novel Viruses from Canegrubs (Coleoptera: Scarabaeidae) in Central Queensland, Australia. Viruses 2022, 14, 649. https://doi.org/10.3390/v14030649
Etebari K, Lenancker P, Powell KS, Furlong MJ. Transcriptomics Reveal Several Novel Viruses from Canegrubs (Coleoptera: Scarabaeidae) in Central Queensland, Australia. Viruses. 2022; 14(3):649. https://doi.org/10.3390/v14030649
Chicago/Turabian StyleEtebari, Kayvan, Pauline Lenancker, Kevin S. Powell, and Michael J. Furlong. 2022. "Transcriptomics Reveal Several Novel Viruses from Canegrubs (Coleoptera: Scarabaeidae) in Central Queensland, Australia" Viruses 14, no. 3: 649. https://doi.org/10.3390/v14030649
APA StyleEtebari, K., Lenancker, P., Powell, K. S., & Furlong, M. J. (2022). Transcriptomics Reveal Several Novel Viruses from Canegrubs (Coleoptera: Scarabaeidae) in Central Queensland, Australia. Viruses, 14(3), 649. https://doi.org/10.3390/v14030649








