A Novel H-2d Epitope for Influenza A Polymerase Acidic Protein
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Viruses
2.3. In Vivo Depletion of CD8 T Cells
2.4. Influenza MDCK Plaque Assay
2.5. Preparation of Single-Cell Suspensions and Flow Cytometry
2.6. Statistical Analysis
3. Results
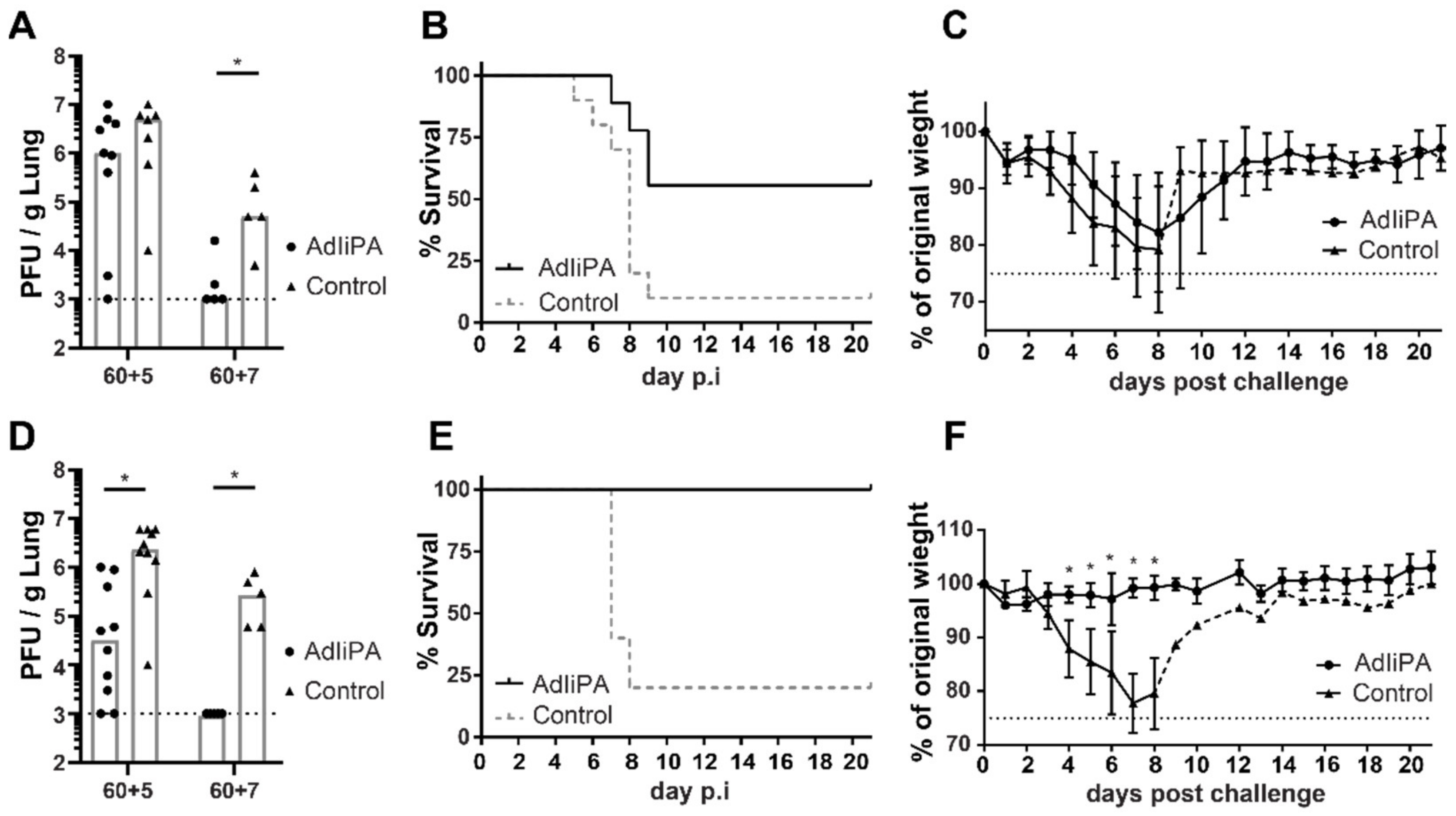
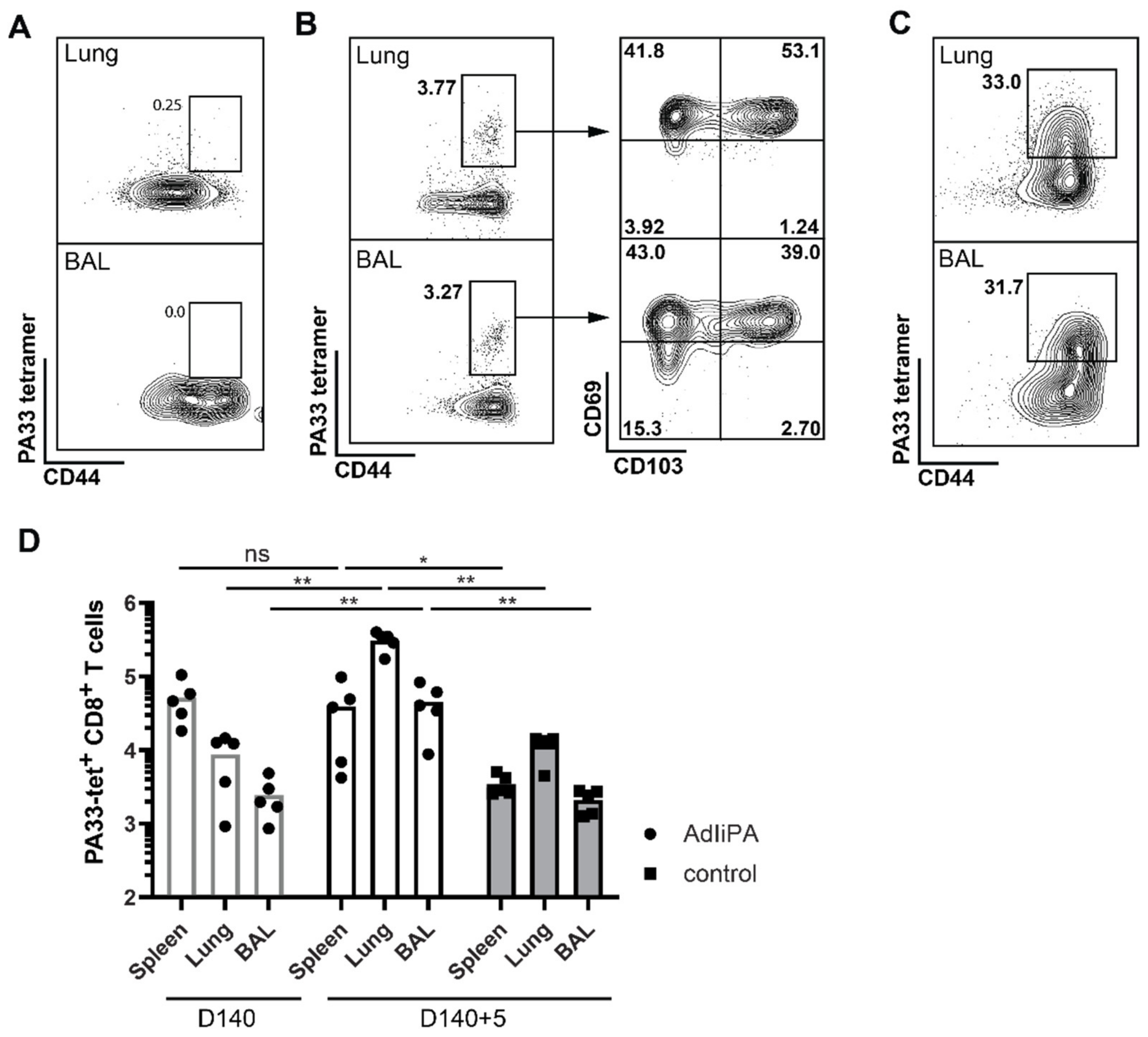
3.1. AdIiPA Vaccination Induced Large Numbers of PA224-Specific CD8 T Cells
3.2. Protection by PA-Specific CD8 T Cells Varied between Mice Strains
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Influenza Virus Infections in Humans. Available online: https://www.who.int/publications/m/item/influenza-virus-infections-in-humans (accessed on 16 February 2022).
- Seasonal Influenza Vaccine Effectiveness, 2004–2018. Available online: https://www.cdc.gov/flu/vaccines-work/past-seasons-estimates.html (accessed on 16 February 2022).
- Gaglani, M.; Pruszynski, J.; Murthy, K.; Clipper, L.; Robertson, A.; Reis, M.; Chung, J.R.; Piedra, P.A.; Avadhanula, V.; Nowalk, M.P.; et al. Influenza Vaccine Effectiveness Against 2009 Pandemic Influenza A(H1N1) Virus Differed by Vaccine Type during 2013–2014 in the United States. J. Infect. Dis. 2016, 213, 1546–1556. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, Y.; Lee, Y.T.; Bouchard, K.R.; Benechet, A.; Khanna, K.; Cauley, L.S. Lung-resident memory CD8 T cells (TRM) are indispensable for optimal cross-protection against pulmonary virus infection. J. Leukoc. Biol. 2014, 95, 215–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sridhar, S.; Begom, S.; Bermingham, A.; Hoschler, K.; Adamson, W.; Carman, W.; Bean, T.; Barclay, W.; Deeks, J.J.; Lalvani, A. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat. Med. 2013, 19, 1305–1312. [Google Scholar] [CrossRef]
- Piet, B.; de Bree, G.J.; Smids-Dierdorp, B.S.; van der Loos, C.M.; Remmerswaal, E.B.; von der Thusen, J.H.; van Haarst, J.M.; Eerenberg, J.P.; ten Brinke, A.; van der Bij, W.; et al. CD8(+) T cells with an intraepithelial phenotype upregulate cytotoxic function upon influenza infection in human lung. J. Clin. Investig. 2011, 121, 2254–2263. [Google Scholar] [CrossRef] [Green Version]
- Uddback, I.E.; Pedersen, L.M.; Pedersen, S.R.; Steffensen, M.A.; Holst, P.J.; Thomsen, A.R.; Christensen, J.P. Combined local and systemic immunization is essential for durable T-cell mediated heterosubtypic immunity against influenza A virus. Sci. Rep. 2016, 6, 20137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uddbäck, I.; Cartwright, E.K.; Schøller, A.S.; Wein, A.N.; Hayward, S.L.; Lobby, J.; Takamura, S.; Thomsen, A.R.; Kohlmeier, J.E.; Christensen, J.P. Long-term maintenance of lung resident memory T cells is mediated by persistent antigen. Mucosal Immunol. 2021, 14, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Uddback, I.E.; Steffensen, M.A.; Pedersen, S.R.; Nazerai, L.; Thomsen, A.R.; Christensen, J.P. PB1 as a potential target for increasing the breadth of T-cell mediated immunity to Influenza A. Sci. Rep. 2016, 6, 35033. [Google Scholar] [CrossRef] [Green Version]
- Powell, T.J.; Strutt, T.; Reome, J.; Hollenbaugh, J.A.; Roberts, A.D.; Woodland, D.L.; Swain, S.L.; Dutton, R.W. Priming with cold-adapted influenza A does not prevent infection but elicits long-lived protection against supralethal challenge with heterosubtypic virus. J. Immunol. 2007, 178, 1030–1038. [Google Scholar] [CrossRef] [Green Version]
- Holst, P.J.; Sorensen, M.R.; Mandrup Jensen, C.M.; Orskov, C.; Thomsen, A.R.; Christensen, J.P. MHC class II-associated invariant chain linkage of antigen dramatically improves cell-mediated immunity induced by adenovirus vaccines. J. Immunol. 2008, 180, 3339–3346. [Google Scholar] [CrossRef]
- Becker, T.C.; Noel, R.J.; Coats, W.S.; Gómez-Foix, A.M.; Alam, T.; Gerard, R.D.; Newgard, C.B. Use of recombinant adenovirus for metabolic engineering of mammalian cells. Methods Cell Biol. 1994, 43, 161–189. [Google Scholar] [CrossRef]
- Cobbold, S.P.; Jayasuriya, A.; Nash, A.; Prospero, T.D.; Waldmann, H. Therapy with monoclonal antibodies by elimination of T-cell subsets in vivo. Nature 1984, 312, 548–551. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.G.; Mayer-Barber, K.; Sung, H.; Beura, L.; James, B.R.; Taylor, J.J.; Qunaj, L.; Griffith, T.S.; Vezys, V.; Barber, D.L.; et al. Intravascular staining for discrimination of vascular and tissue leukocytes. Nat. Protoc. 2014, 9, 209–222. [Google Scholar] [CrossRef] [Green Version]
- Anderson, K.G.; Sung, H.; Skon, C.N.; Lefrancois, L.; Deisinger, A.; Vezys, V.; Masopust, D. Cutting edge: Intravascular staining redefines lung CD8 T cell responses. J. Immunol. 2012, 189, 2702–2706. [Google Scholar] [CrossRef] [Green Version]
- Zens, K.D.; Chen, J.K.; Farber, D.L. Vaccine-generated lung tissue-resident memory T cells provide heterosubtypic protection to influenza infection. JCI Insight 2016, 1, e85832. [Google Scholar] [CrossRef]
- Schenkel, J.M.; Fraser, K.A.; Vezys, V.; Masopust, D. Sensing and alarm function of resident memory CD8(+) T cells. Nat. Immunol. 2013, 14, 509–513. [Google Scholar] [CrossRef] [Green Version]
- Hayward, S.L.; Scharer, C.D.; Cartwright, E.K.; Takamura, S.; Li, Z.T.; Boss, J.M.; Kohlmeier, J.E. Environmental cues regulate epigenetic reprogramming of airway-resident memory CD8(+) T cells. Nat. Immunol. 2020, 21, 309–320. [Google Scholar] [CrossRef]
- Tatsis, N.; Fitzgerald, J.C.; Reyes-Sandoval, A.; Harris-McCoy, K.C.; Hensley, S.E.; Zhou, D.; Lin, S.W.; Bian, A.; Xiang, Z.Q.; Iparraguirre, A.; et al. Adenoviral vectors persist in vivo and maintain activated CD8+ T cells: Implications for their use as vaccines. Blood 2007, 110, 1916–1923. [Google Scholar] [CrossRef] [Green Version]
- Cupovic, J.; Ring, S.S.; Onder, L.; Colston, J.M.; Lutge, M.; Cheng, H.W.; De Martin, A.; Provine, N.M.; Flatz, L.; Oxenius, A.; et al. Adenovirus vector vaccination reprograms pulmonary fibroblastic niches to support protective inflating memory CD8(+) T cells. Nat. Immunol. 2021, 22, 1042–1051. [Google Scholar] [CrossRef]
- Crowe, S.R.; Miller, S.C.; Shenyo, R.M.; Woodland, D.L. Vaccination with an acidic polymerase epitope of influenza virus elicits a potent antiviral T cell response but delayed clearance of an influenza virus challenge. J. Immunol. 2005, 174, 696–701. [Google Scholar] [CrossRef] [Green Version]
- Crowe, S.R.; Turner, S.J.; Miller, S.C.; Roberts, A.D.; Rappolo, R.A.; Doherty, P.C.; Ely, K.H.; Woodland, D.L. Differential antigen presentation regulates the changing patterns of CD8+ T cell immunodominance in primary and secondary influenza virus infections. J. Exp. Med. 2003, 198, 399–410. [Google Scholar] [CrossRef]
- Belz, G.T.; Xie, W.; Altman, J.D.; Doherty, P.C. A previously unrecognized H-2D(b)-restricted peptide prominent in the primary influenza A virus-specific CD8(+) T-cell response is much less apparent following secondary challenge. J. Virol. 2000, 74, 3486–3493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yewdell, J.W. Confronting complexity: Real-world immunodominance in antiviral CD8+ T cell responses. Immunity 2006, 25, 533–543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.; Zanker, D.; Valkenburg, S.; Tan, B.; Kedzierska, K.; Zou, Q.M.; Doherty, P.C.; Chen, W. Systematic identification of immunodominant CD8+ T-cell responses to influenza A virus in HLA-A2 individuals. Proc. Natl. Acad. Sci. USA 2011, 108, 9178–9183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinones-Parra, S.; Grant, E.; Loh, L.; Nguyen, T.H.; Campbell, K.A.; Tong, S.Y.; Miller, A.; Doherty, P.C.; Vijaykrishna, D.; Rossjohn, J.; et al. Preexisting CD8+ T-cell immunity to the H7N9 influenza A virus varies across ethnicities. Proc. Natl. Acad. Sci. USA 2014, 111, 1049–1054. [Google Scholar] [CrossRef] [Green Version]


| Position | Restriction | Sequence |
|---|---|---|
| PA33–41 | Kd | KFAAICTHL |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uddbäck, I.; Kohlmeier, J.E.; Thomsen, A.R.; Christensen, J.P. A Novel H-2d Epitope for Influenza A Polymerase Acidic Protein. Viruses 2022, 14, 601. https://doi.org/10.3390/v14030601
Uddbäck I, Kohlmeier JE, Thomsen AR, Christensen JP. A Novel H-2d Epitope for Influenza A Polymerase Acidic Protein. Viruses. 2022; 14(3):601. https://doi.org/10.3390/v14030601
Chicago/Turabian StyleUddbäck, Ida, Jacob E. Kohlmeier, Allan R. Thomsen, and Jan P. Christensen. 2022. "A Novel H-2d Epitope for Influenza A Polymerase Acidic Protein" Viruses 14, no. 3: 601. https://doi.org/10.3390/v14030601
APA StyleUddbäck, I., Kohlmeier, J. E., Thomsen, A. R., & Christensen, J. P. (2022). A Novel H-2d Epitope for Influenza A Polymerase Acidic Protein. Viruses, 14(3), 601. https://doi.org/10.3390/v14030601






