Swine Norovirus: Past, Present, and Future
Abstract
1. Introduction
2. Results
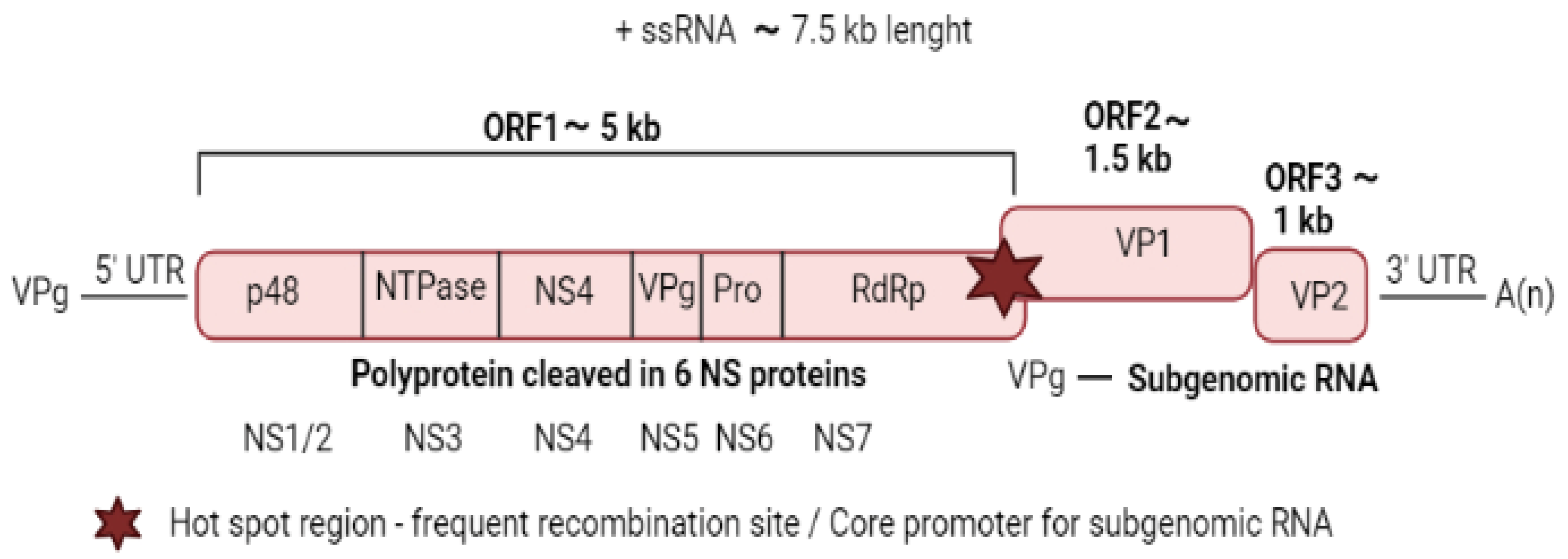
2.1. Viral Structure and Genome Organisation
2.2. Virus Replication
2.3. Studying Swine Norovirus: When and How Frequent
2.4. Where and When: Countries, Years of Detection and Seasonality
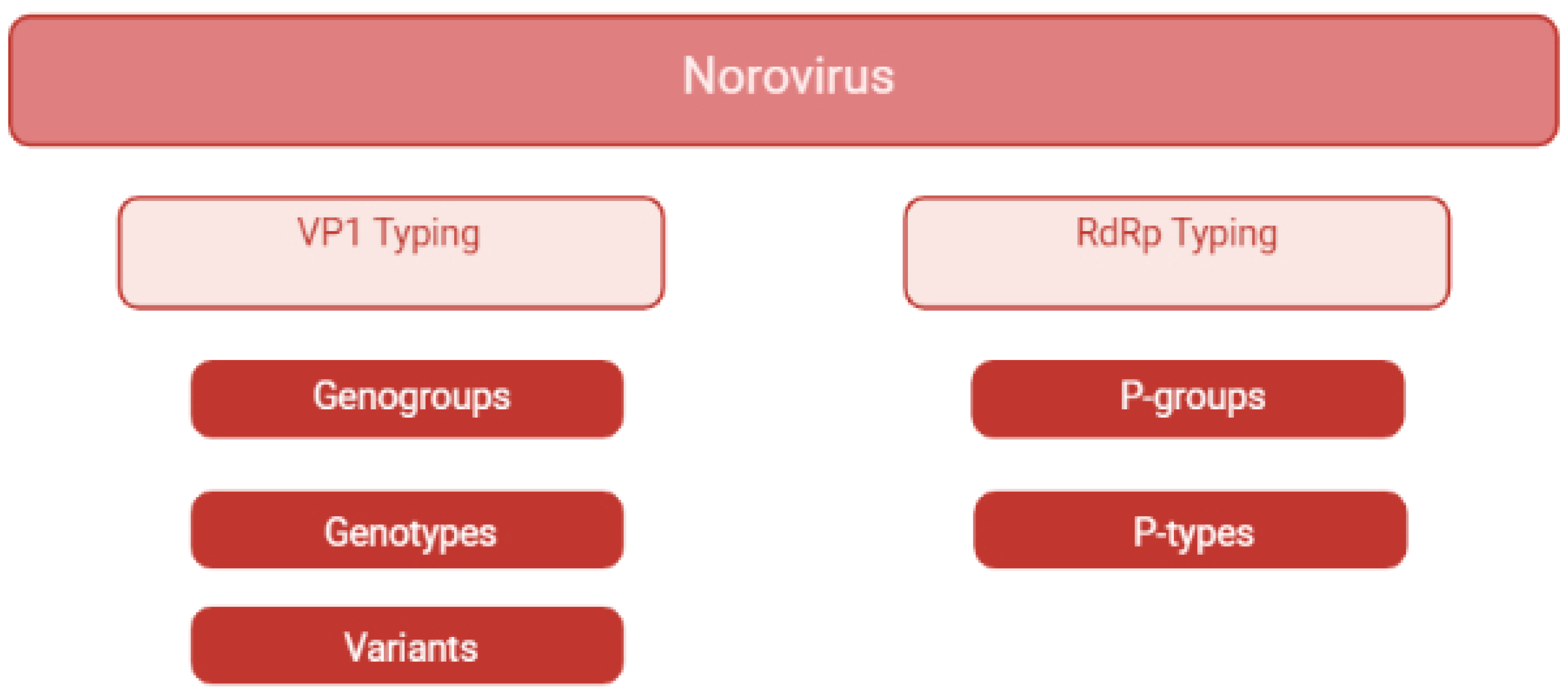
2.5. What: Genetic Diversity
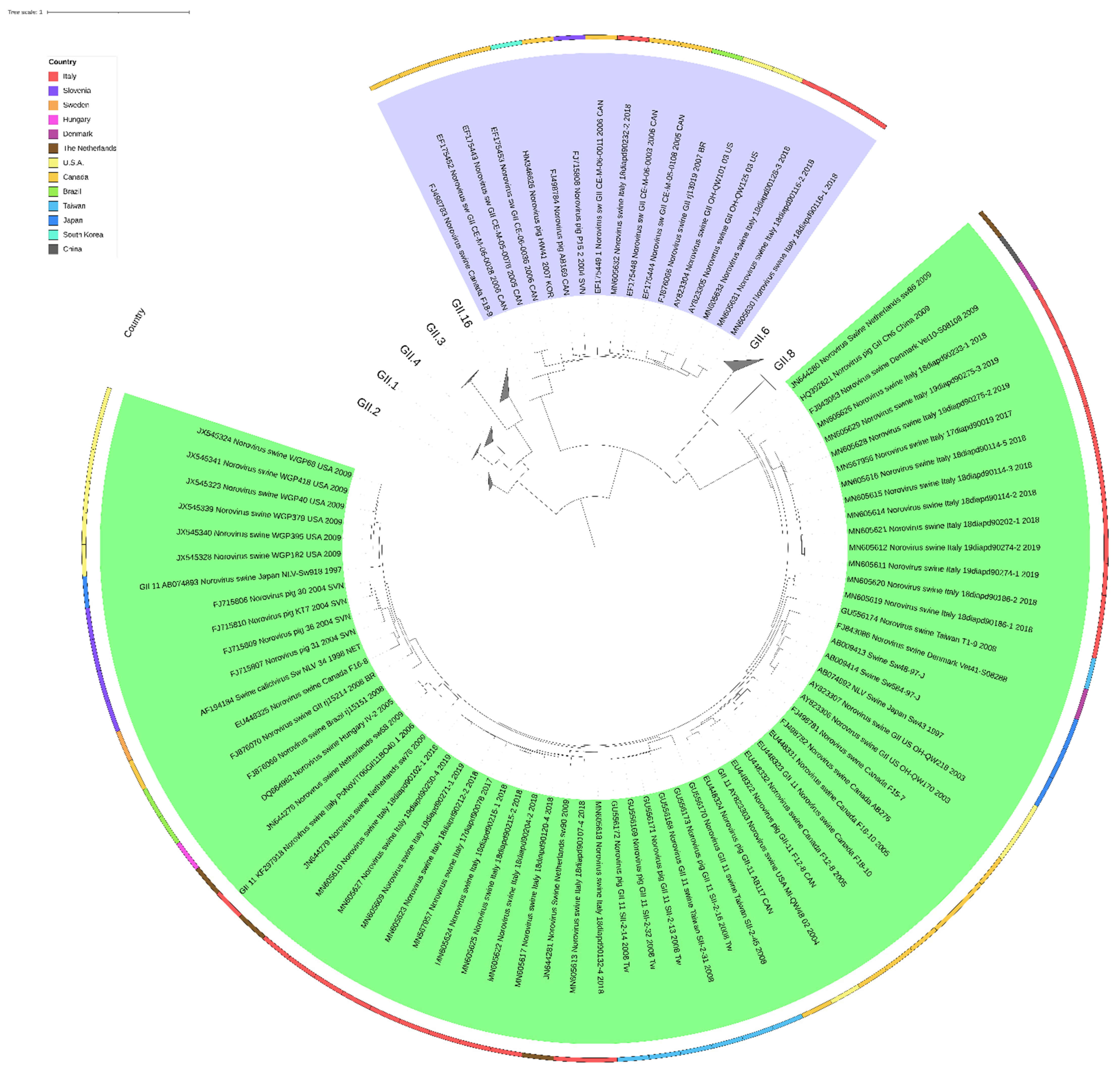
2.6. Phylogeny
2.7. Swine Norovirus in Naturally Infected Swine
2.8. Swine Norovirus in Experimentally Infected Swine
2.9. Host Factors
2.10. Laboratory Methods to Investigate Swine Noroviruses
2.11. Electron Microscopy
2.12. Pre-Treatment of Samples and RNA Extraction for PCR Detection
2.13. RNA Transcription and Amplification Methods
2.14. Whole Genome Sequencing
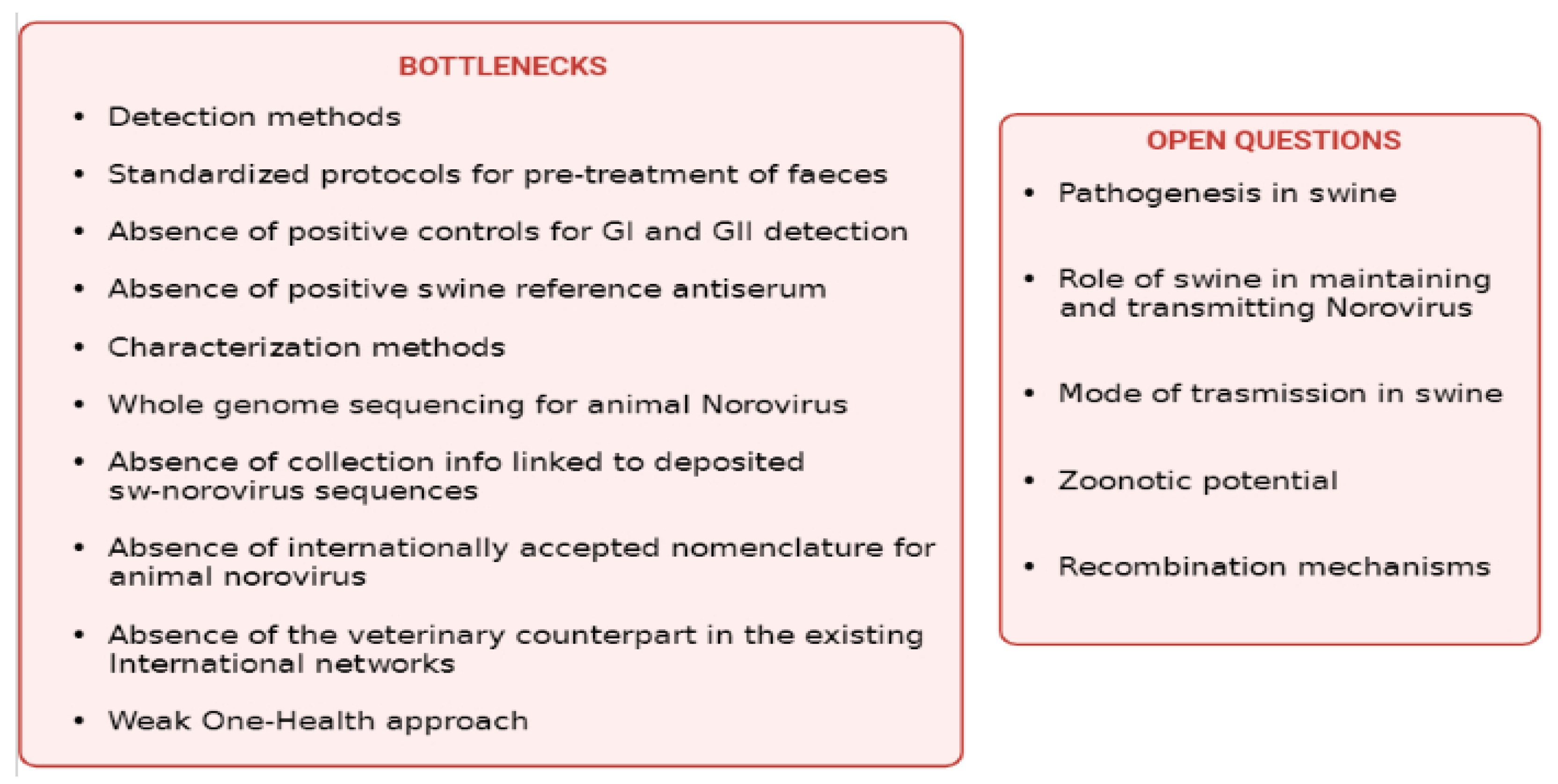
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- DiEuliis, D.; Johnson, K.R.; Morse, S.S.; Schindel, D.E. Opinion: Specimen collections should have a much bigger role in infectious disease research and response. Proc. Natl. Acad. Sci. USA 2016, 113, 4–7. [Google Scholar] [CrossRef] [PubMed]
- Norovirus Worldwide|CDC. 2021. Available online: https://www.cdc.gov/norovirus/trends-outbreaks/worldwide.html (accessed on 29 December 2021).
- European Food Safety Autority (EFSA). The European Union One Health 2020 Zoonoses Report. EFSA J. 2021, 19, e06971. [Google Scholar]
- Gosh, S.; Malik, S.Y.; Kobayashi, N. Animal Caliciviruses. In Animal—Origin Viral Zoonoses, Livestock Diseases Singapore; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Thorne, L.G.; Goodfellow, I. Norovirus gene expression and replication. J. Gen. Virol. 2014, 95 Pt 2, 278–291. [Google Scholar] [CrossRef]
- Olspert, A.; Hosmillo, M.; Chaudhry, Y.; Peil, L.; Truve, E.; Goodfellow, I. Protein-RNA linkage and posttranslational modifications of feline calicivirus and murine norovirus VPg proteins. PeerJ 2016, 4, e2134. [Google Scholar] [CrossRef] [PubMed]
- Bertolotti-Ciarlet, A.; Crawford, S.E.; Hutson, A.M.; Estes, M.K. The 3′ End of Norwalk Virus mRNA Contains Determinants That Regulate the Expression and Stability of the Viral Capsid Protein VP1: A Novel Function for the VP2 Protein. J. Virol. 2003, 77, 11603–11615. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, P.; Karakasiliotis, I.; Bailey, D.; Chaudhry, Y.; Evans, D.J.; Goodfellow, I.G. Bioinformatic and functional analysis of RNA secondary structure elements among different genera of human and animal caliciviruses. Nucleic Acids Res. 2008, 36, 2530–2546. [Google Scholar] [CrossRef] [PubMed]
- Woode, G.N.; Bridger, J.C. Isolation of Small Viruses Resembling Astroviruses and Caliciviruses from Acute Enteritis of Calves. J. Med. Microbiol. 1978, 11, 441–452. [Google Scholar] [CrossRef]
- Bhar, S.; Jones, M.K. In Vitro Replication of Human Norovirus. Viruses 2019, 11, 547. [Google Scholar] [CrossRef]
- Ettayebi, K.; Crawford, S.E.; Murakami, K.; Broughman, J.R.; Karandikar, U.; Tenge, V.; Neill, F.H.; Blutt, S.E.; Zeng, X.-L.; Qu, L.; et al. Replication of human noroviruses in stem cell-derived human enteroids. Science 2016, 353, 1387–1393. [Google Scholar] [CrossRef]
- Ettayebi, K.; Tenge, V.R.; Cortes-Penfield, N.W.; Crawford, S.E.; Neill, F.H.; Zeng, X.-L.; Yu, X.; Ayyar, B.V.; Burrin, D.; Ramani, S.; et al. New Insights and Enhanced Human Norovirus Cultivation in Human Intestinal Enteroids. mSphere 2021, 6, e01136-20. [Google Scholar] [CrossRef]
- Wang, Q.-H.; Costantini, V.; Saif, L.J. Porcine enteric caliciviruses: Genetic and antigenic relatedness to human caliciviruses, diagnosis and epidemiology. Vaccine 2007, 25, 5453–5466. [Google Scholar] [CrossRef] [PubMed]
- Cheetham, S.; Souza, M.; Meulia, T.; Grimes, S.; Han, M.G.; Saif, L.J. Pathogenesis of a Genogroup II Human Norovirus in Gnotobiotic Pigs. J. Virol. 2006, 80, 10372–10381. [Google Scholar] [CrossRef]
- Park, B.; Jung, S.; Choi, C.; Myoung, J.; Ahn, H.; Han, S.; Kim, Y.H.; Go, H.J.; Lee, J.B.; Park, S.Y.; et al. Pathogenesis of Human Norovirus Genogroup II Genotype 4 in Post-Weaning Gnotobiotic Pigs. J. Microbiol. Biotechnol. 2018, 28, 2133–2140. [Google Scholar] [CrossRef] [PubMed]
- Bui, T.; Kocher, J.; Li, Y.; Wen, K.; Li, G.; Liu, F.; Yang, X.; LeRoith, T.; Tan, M.; Xia, M.; et al. Median infectious dose of human norovirus GII.4 in gnotobiotic pigs is decreased by simvastatin treatment and increased by age. J. Gen. Virol. 2013, 94, 2005–2016. [Google Scholar] [CrossRef]
- Souza, M.; Cheetham, S.M.; Azevedo, M.S.P.; Costantini, V.; Saif, L.J. Cytokine and Antibody Responses in Gnotobiotic Pigs after Infection with Human Norovirus Genogroup II.4 (HS66 Strain). J. Virol. 2007, 81, 9183–9192. [Google Scholar] [CrossRef]
- Ludwig-Begall, L.; Mauroy, A.; Thiry, E. Noroviruses—The State of the Art, Nearly Fifty Years after Their Initial Discovery. Viruses 2021, 13, 1541. [Google Scholar] [CrossRef] [PubMed]
- Sugieda, M.; Nagaoka, H.; Kakishima, Y.; Ohshita, T.; Nakamura, S.; Nakajima, S. Detection of Norwalk-like virus genes in the caecum contents of pigs. Arch. Virol. 1998, 143, 1215–1221. [Google Scholar] [CrossRef]
- Farkas, T.; Nakajima, S.; Sugieda, M.; Deng, X.; Zhong, W.; Jiang, X. Seroprevalence of Noroviruses in Swine. J. Clin. Microbiol. 2005, 43, 657–661. [Google Scholar] [CrossRef] [PubMed]
- Martella, V.; Lorusso, E.; Banyai, K.; Decaro, N.; Corrente, M.; Elia, G.; Cavalli, A.; Radogna, A.; Costantini, V.; Saif, L.J.; et al. Identification of a Porcine Calicivirus Related Genetically to Human Sapoviruses. J. Clin. Microbiol. 2008, 46, 1907–1913. [Google Scholar] [CrossRef] [PubMed]
- Martella, V.; Bányai, K.; Lorusso, E.; Bellacicco, A.L.; Decaro, N.; Mari, V.; Saif, L.; Costantini, V.; De Grazia, S.; Pezzotti, G.; et al. Genetic heterogeneity of porcine enteric caliciviruses identified from diarrhoeic piglets. Virus Genes 2008, 36, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Oka, T.; Saif, L.J.; Wang, Q. First Complete Genome Sequence of a Genogroup II Genotype 18 Porcine Norovirus, Strain QW125. Genome Announc. 2013, 1, e00344-13. [Google Scholar] [CrossRef] [PubMed]
- Di Bartolo, I.; Tofani, S.; Angeloni, G.; Ponterio, E.; Ostanello, F.; Ruggeri, F.M. Detection and characterization of porcine caliciviruses in Italy. Arch. Virol. 2014, 159, 2479–2484. [Google Scholar] [CrossRef] [PubMed]
- Silva, P.F.; Alfieri, A.F.; Barry, A.F.; de Arruda Leme, R.; Gardinali, N.R.; van der Poel, W.H.; Alfieri, A.A. High frequency of porcine norovirus infection in finisher units of Brazilian pig-production systems. Trop. Anim. Health Prod. 2015, 47, 237–241. [Google Scholar] [CrossRef]
- Wilhelm, B.; Fazil, A.; Rajić, A.; Houde, A.; McEwen, S.A. Risk Profile of Hepatitis E Virus from Pigs or Pork in Canada. Transbound. Emerg. Dis. 2017, 64, 1694–1708. [Google Scholar] [CrossRef]
- Monini, M.; Di Bartolo, I.; Ianiro, G.; Angeloni, G.; Magistrali, C.F.; Ostanello, F.; Ruggeri, F.M. Detection and molecular characterization of zoonotic viruses in swine fecal samples in Italian pig herds. Arch. Virol. 2015, 160, 2547–2556. [Google Scholar] [CrossRef] [PubMed]
- Cavicchio, L.; Tassoni, L.; Laconi, A.; Cunial, G.; Gagliazzo, L.; Milani, A.; Campalto, M.; Di Martino, G.; Forzan, M.; Monne, I.; et al. Unrevealed genetic diversity of GII Norovirus in the swine population of North East Italy. Sci. Rep. 2020, 10, 9217. [Google Scholar] [CrossRef] [PubMed]
- Cunha, J.; De Mendonça, M.; Miagostovich, M.; Leite, J. First detection of porcine norovirus GII.18 in Latin America. Res. Vet. Sci. 2010, 89, 126–129. [Google Scholar] [CrossRef]
- Cunha, J.B.; De Mendonça, M.C.L.; Miagostovich, M.P.; Leite, J.P.G. Genetic diversity of porcine enteric caliciviruses in pigs raised in Rio de Janeiro State, Brazil. Arch. Virol. 2010, 155, 1301–1305. [Google Scholar] [CrossRef] [PubMed]
- Halaihel, N.; Masía, R.M.; Fernández-Jiménez, M.; Ribes, J.M.; Montava, R.; De Blas, I.; Gironés, O.; Alonso, J.L.; Buesa, J. Enteric calicivirus and rotavirus infections in domestic pigs. Epidemiol. Infect. 2010, 138, 542–548. [Google Scholar] [CrossRef] [PubMed]
- Keum, H.-O.; Moon, H.-J.; Park, S.-J.; Kim, H.-K.; Rho, S.-M.; Park, B.-K. Porcine noroviruses and sapoviruses on Korean swine farms. Arch. Virol. 2009, 154, 1765–1774. [Google Scholar] [CrossRef]
- Machnowska, P.; Ellerbroek, L.; Johne, R. Detection and characterization of potentially zoonotic viruses in faeces of pigs at slaughter in Germany. Vet. Microbiol. 2014, 168, 60–68. [Google Scholar] [CrossRef]
- Mijovski, J.Z.; Poljšak-Prijatelj, M.; Steyer, A.; Barlič-Maganja, D.; Koren, S. Detection and molecular characterisation of noroviruses and sapoviruses in asymptomatic swine and cattle in Slovenian farms. Infect. Genet. Evol. 2010, 10, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Saga, Y.; Iwai, M.; Obara, M.; Horimoto, E.; Hasegawa, S.; Kurata, T.; Okumura, H.; Nagoshi, M.; Takizawa, T. Frequent Detection of Noroviruses and Sapoviruses in Swine and High Genetic Diversity of Porcine Sapovirus in Japan during Fiscal Year 2008. J. Clin. Microbiol. 2010, 48, 1215–1222. [Google Scholar] [CrossRef] [PubMed]
- Okada, A.; Kobayashi, S.; Inoshima, Y. Detection Frequency of Porcine Noroviruses in Healthy Pigs in Japan. Jpn. Agric. Res. Q. JARQ 2019, 53, 305–310. [Google Scholar] [CrossRef]
- Reuter, G.; Bíró, H.; Szücs, G. Enteric caliciviruses in domestic pigs in Hungary. Arch. Virol. 2007, 152, 611–614. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Zhang, W.; Yang, S.; Chen, Y.; Ning, H.; Shan, T.; Liu, J.; Yang, Z.; Cui, L.; Zhu, J.; et al. Molecular detection and prevalence of porcine caliciviruses in eastern China from 2008 to 2009. Arch. Virol. 2009, 154, 1625–1630. [Google Scholar] [CrossRef]
- Shen, Q.; Zhang, W.; Yang, S.; Yang, Z.; Chen, Y.; Cui, L.; Zhu, J.; Hua, X. Recombinant porcine norovirus identified from piglet with diarrhea. BMC Vet. Res. 2012, 8, 155. [Google Scholar] [CrossRef] [PubMed]
- Sisay, Z.; Wang, Q.; Oka, T.; Saif, L. Prevalence and molecular characterization of porcine enteric caliciviruses and first detection of porcine kobuviruses in US swine. Arch. Virol. 2013, 158, 1583–1588. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sisay, Z.; Djikeng, A.; Berhe, N.; Belay, G.; Abegaz, W.E.; Wang, Q.H.; Saif, L.J. First detection and molecular characterization of sapoviruses and noroviruses with zoonotic potential in swine in Ethiopia. Arch. Virol. 2016, 161, 2739–2747. [Google Scholar] [CrossRef] [PubMed]
- Valkó, A.; Marosi, A.; Cságola, A.; Farkas, R.; Rónai, Z.; Dán, Á. Frequency of diarrhoea-associated viruses in swine of various ages in Hungary. Acta Vet. Hung. 2019, 67, 140–150. [Google Scholar] [CrossRef]
- Wang, Q.-H.; Chang, K.-O.; Han, M.G.; Sreevatsan, S.; Saif, L.J. Development of a new microwell hybridization assay and an internal control RNA for the detection of porcine noroviruses and sapoviruses by reverse transcription-PCR. J. Virol. Methods 2006, 132, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Mauroy, A.; Scipioni, A.; Mathijs, E.; Miry, C.; Ziant, D.; Thys, C.; Thiry, E. Noroviruses and sapoviruses in pigs in Belgium. Arch. Virol. 2008, 153, 1927–1931. [Google Scholar] [CrossRef] [PubMed]
- Stamelou, E.; Giantsis, I.A.; Papageorgiou, K.V.; Petridou, E.; Davidson, I.; Polizopοulou, Z.S.; Papa, A.; Kritas, S.K. Epidemiology of Astrovirus, Norovirus and Sapovirus in Greek pig farms indicates high prevalence of Mamastrovirus suggesting the potential need for systematic surveillance. Porc. Health Manag. 2022, 8, 5. [Google Scholar] [CrossRef] [PubMed]
- Chao, D.-Y.; Wei, J.-Y.; Chang, W.-F.; Wang, J.; Wang, L.-C. Detection of Multiple Genotypes of Calicivirus Infection in Asymptomatic Swine in Taiwan. Zoonoses Public Health 2012, 59, 434–444. [Google Scholar] [CrossRef] [PubMed]
- Martínez, M.A.; Alcala, A.; Carruyo, G.; Botero, L.; Liprandi, F.; Ludert, J.E. Molecular detection of porcine enteric caliciviruses in Venezuelan farms. Vet. Microbiol. 2006, 116, 77–84. [Google Scholar] [CrossRef]
- Laconi, A.; Cavicchio, L.; Tassoni, L.; Cunial, G.; Milani, A.; Ustulin, M.; Di Martino, G.; Forzan, M.; Campalto, M.; Monne, I.; et al. Identification of two divergent swine Noroviruses detected at the slaughterhouse in North East Italy. Porc. Health Manag. 2020, 6, 9. [Google Scholar] [CrossRef]
- Wang, Q.; Scheuer, K.; Zhang, Z.; Gebreyes, W.A.; Molla, B.Z.; Hoet, A.E.; Saif, L.J. Characterization and prevalence of a new porcine calicivirus in Swine, United States. Emerg. Infect. Dis. 2011, 17, 1103–1106. [Google Scholar] [CrossRef]
- Scheuer, K.A.; Oka, T.; Hoet, A.E.; Gebreyes, W.A.; Molla, B.Z.; Saif, L.J.; Wang, Q. Prevalence of Porcine Noroviruses, Molecular Characterization of Emerging Porcine Sapoviruses from Finisher Swine in the United States, and Unified Classification Scheme for Sapoviruses. J. Clin. Microbiol. 2013, 51, 2344–2353. [Google Scholar] [CrossRef]
- Wilhelm, B.; Leblanc, D.; Houde, A.; Brassard, J.; Gagné, M.-J.; Plante, D.; Bellon-Gagnon, P.; Jones, T.H.; Muehlhauser, V.; Janecko, N.; et al. Survey of Canadian retail pork chops and pork livers for detection of hepatitis E virus, norovirus, and rotavirus using real time RT-PCR. Int. J. Food Microbiol. 2014, 185, 33–40. [Google Scholar] [CrossRef]
- Shen, Q.; Ren, R.; Zhang, W.; Yang, Z.; Yang, S.; Chen, Y.; Cui, L.; Hua, X. Prevalence of hepatitis E virus and porcine caliciviruses in pig farms of Guizhou province, China. Zahedan J. Res. Med. Sci. 2011, 11, 459–463. [Google Scholar]
- Shen, Q.; Zhang, W.; Yang, S.; Cui, L.; Hua, X. Complete Genome Sequence of a New-Genotype Porcine Norovirus Isolated from Piglets with Diarrhea. J. Virol. 2012, 86, 7015–7016. [Google Scholar] [CrossRef] [PubMed]
- Okada, A.; Inoshima, Y. Near-Complete Genome Sequence of a Swine Norovirus GII.11 Strain Detected in Japan in 2018. Microbiol. Resour. Announc. 2020, 9, e00014-20. [Google Scholar] [CrossRef] [PubMed]
- Sugieda, M.; Nakajima, K. Outbreaks of Norwalk-like virus-associated gastroenteritis traced to shellfish: Coexistence of two genotypes in one specimen. Epidemiol. Infect. 1996, 116, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Sugieda, M.; Nakajima, S. Viruses detected in the caecum contents of healthy pigs representing a new genetic cluster in genogroup II of the genus ‘Norwalk-like viruses’. Virus Res. 2002, 87, 165–172. [Google Scholar] [CrossRef]
- L’Homme, Y.; Sansregret, R.; Plante-Fortier, É.; Lamontagne, A.-M.; Lacroix, G.; Ouardani, M.; Deschamps, J.; Simard, G.; Simard, C. Genetic diversity of porcine Norovirus and Sapovirus: Canada, 2005–2007. Arch. Virol. 2009, 154, 581–593. [Google Scholar] [CrossRef]
- Schuele, L.; Lizarazo-Forero, E.; Strutzberg-Minder, K.; Schütze, S.; Löbert, S.; Lambrecht, C.; Harlizius, J.; Friedrich, A.W.; Peter, S.; Rossen, J.W.A.; et al. Application of shotgun metagenomics sequencing and targeted sequence capture to detect circulating porcine viruses in the Dutch–German border region. Transbound. Emerg. Dis. 2021, 1–14. [Google Scholar] [CrossRef]
- van Der Poel, W.H.; Vinjé, J.; van Der Heide, R.; Herrera, M.I.; Vivo, A.; Koopmans, M.P. Norwalk-like calicivirus genes in farm animals. Emerg. Infect. Dis. 2000, 6, 36–41. [Google Scholar] [CrossRef]
- Rodríguez-Lázaro, D.; Diez-Valcarce, M.; Montes-Briones, R.; Gallego, D.; Hernández, M.; Rovira, J. Presence of pathogenic enteric viruses in illegally imported meat and meat products to EU by international air travelers. Int. J. Food Microbiol. 2015, 209, 39–43. [Google Scholar] [CrossRef]
- Mattison, K.; Shukla, A.; Cook, A.; Pollari, F.; Friendship, R.; Kelton, D.; Bidawid, S.; Farber, J.M. Human Noroviruses in Swine and Cattle. Emerg. Infect. Dis. 2007, 13, 1184–1188. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.-J.; Yu, J.-N.; Nam, H.-M.; Bak, H.-R.; Lee, J.-B.; Park, S.-Y.; Song, C.-S.; Seo, K.-H.; Choi, I.-S. Identification of genetic diversity of porcine Norovirus and Sapovirus in Korea. Virus Genes 2011, 42, 394–401. [Google Scholar] [CrossRef]
- L’Homme, Y.; Sansregret, R.; Simard, C. Broad range RT-PCR assays targeting human noroviruses also detect swine noroviruses. Food Microbiol. 2009, 26, 552–555. [Google Scholar] [CrossRef]
- Wolf, S.; Williamson, W.; Hewitt, J.; Lin, S.; Rivera-Aban, M.; Ball, A.; Scholes, P.; Savill, M.; Greening, G.E. Molecular detection of norovirus in sheep and pigs in New Zealand farms. Vet. Microbiol. 2009, 133, 184–189. [Google Scholar] [CrossRef] [PubMed]
- Koo, E.S.; Jeong, Y.S. Comparative study of the geographical spread of genogroup II porcine norovirus and human norovirus. J. Microbiol. 2021, 59, 644–650. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, M.F.; López, J.; Ruiz, A.; Osorio, C.; Ulloa, J.C. Identificación de Norovirus Humano (HNoV) en muestras de estiércol de cerdos domésticos. Univ. Sci. 2011, 16, 168. [Google Scholar] [CrossRef]
- Bucardo, F.; González, F.; Reyes, Y.; Blandón, P.; Saif, L.; Nordgren, J. Seroprevalence in Household Raised Pigs Indicate High Exposure to GII Noroviruses in Rural Nicaragua. Zoonoses Public Health 2016, 63, 600–607. [Google Scholar] [CrossRef] [PubMed]
- Farkas, T.; Jiang, X.; Guerrero, M.L.; Zhong, W.; Wilton, N.; Berke, T.S.; Matson, D.O.; Pickering, L.K.; Ruiz-Palacios, G. Prevalence and genetic diversity of human caliciviruses (HuCVs) in Mexican children. J. Med. Virol. 2000, 62, 217. [Google Scholar] [CrossRef]
- Chhabra, P.; de Graaf, M.; Parra, G.I.; Chan, M.C.; Green, K.; Martella, V.; Wang, Q.; White, P.A.; Katayama, K.; Vennema, H.; et al. Updated classification of norovirus genogroups and genotypes. J. Gen. Virol. 2019, 100, 1393–1406. [Google Scholar] [CrossRef]
- Zaczek-Moczydlowska, M.A.; Beizaei, A.; Dillion, M.; Campbell, K. Current state-of-the-art diagnostics for Norovirus detection: Model approaches for point-of-care analysis. Trends Food Sci. Technol. 2021, 114, 684–695. [Google Scholar] [CrossRef]
- L’Homme, Y.; Sansregret, R.; Plante-Fortier, É.; Lamontagne, A.-M.; Ouardani, M.; Lacroix, G.; Simard, C. Genomic characterization of swine caliciviruses representing a new genus of Caliciviridae. Virus Genes 2009, 39, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Reuter, G.; Krisztalovics, K.; Vennema, H.; Koopmans, M.; Szűcs, G. Evidence of the etiological predominance of norovirus in gastroenteritis outbreaks—Emerging new-variant and recombinant noroviruses in Hungary. J. Med. Virol. 2005, 76, 598–607. [Google Scholar] [CrossRef] [PubMed]
- Parra, G.I. Emergence of norovirus strains: A tale of two genes. Virus Evol. 2019, 5, vez048. [Google Scholar] [CrossRef] [PubMed]
- Cheetham, S.; Souza, M.; McGregor, R.; Meulia, T.; Wang, Q.; Saif, L.J. Binding Patterns of Human Norovirus-Like Particles to Buccal and Intestinal Tissues of Gnotobiotic Pigs in Relation to A/H Histo-Blood Group Antigen Expression. J. Virol. 2007, 81, 3535–3544. [Google Scholar] [CrossRef] [PubMed]
- Takanashi, S.; Wang, Q.; Chen, N.; Shen, Q.; Jung, K.; Zhang, Z.; Yokoyama, M.; Lindesmith, L.C.; Baric, R.S.; Saif, L.J. Characterization of Emerging GII.g/GII.12 Noroviruses from a Gastroenteritis Outbreak in the United States in 2010. J. Clin. Microbiol. 2011, 49, 3234–3244. [Google Scholar] [CrossRef]
- Atmar, R.L.; Bernstein, D.I.; Lyon, G.M.; Treanor, J.J.; Al-Ibrahim, M.S.; Graham, D.Y.; Vinje, J.; Jiang, X.; Gregoricus, N.; Frenck, R.W.; et al. Serological Correlates of Protection against a GII.4 Norovirus. Clin. Vaccine Immunol. 2015, 22, 923–929. [Google Scholar] [CrossRef] [PubMed]
- Revilla-Fernández, S.; Wallner, B.; Truschner, K.; Benczak, A.; Brem, G.; Schmoll, F.; Mueller, M.; Steinborn, R. The use of endogenous and exogenous reference RNAs for qualitative and quantitative detection of PRRSV in porcine semen. J. Virol. Methods 2005, 126, 21–30. [Google Scholar] [CrossRef] [PubMed]
- van Elden, L.J.; Nijhuis, M.; Schipper, P.; Schuurman, R.; van Loon, A.M. Simultaneous detection of influenza viruses A and B using real-time quantitative PCR. J. Clin. Microbiol. 2001, 39, 196–200. [Google Scholar] [CrossRef] [PubMed]
- Laconi, A.; Fortin, A.; Bedendo, G.; Shibata, A.; Sakoda, Y.; Awuni, J.A.; Go-Maro, E.; Arafa, A.; Maken Ali, A.S.; Terregino, C.; et al. Detection of avian influenza virus: A comparative study of the in silico and in vitro performances of current RT-qPCR assays. Sci. Rep. 2020, 10, 8441. [Google Scholar] [CrossRef]
- Ludert, J.E.; Alcalá, A.C.; Liprandi, F. Primer Pair p289–p290, Designed to Detect Both Noroviruses and Sapoviruses by Reverse Transcription-PCR, Also Detects Rotaviruses by Cross-Reactivity. J. Clin. Microbiol. 2004, 42, 835–836. [Google Scholar] [CrossRef][Green Version]
| Country | Sampling Years | Genotype | Paper |
|---|---|---|---|
| Japan | 1997 | GII.11 | Sugieda et al., 1998 [19] |
| The Netherlands | 1998 | GII.11 | Van der Poel et al., 2000 [56] |
| USA | 2003 | GII.18 | Oka et al., 2013 [23] |
| Japan | 2002–2003 | GII | Farkas et al., 2005 [20] |
| USA | 2003–2005 | GII.18 | Wang et al., 2006 [43] |
| Slovenia | 2004–2005 | GII.11, GII.18 | Mijovski et al., 2010 [34] |
| Canada | 2005–2007 | GII.18, GII.4, GII.11 | Mattison et al., 2007; L’Homme et al., 2009 [58,59] |
| Hungary | 2005 | NA | Reuter et al., 2007 [37] |
| Italy | 2006–2007 | GII.11 | Di bartolo et al., 2014 [24] |
| New Zealand | 2006–2007 | GII | Wolf et al., 2009 [60] |
| Brazil | 2007 | GII.18 | Cunha et al., 2010, Cunha et al., 2010 [29,30] |
| South Korea | 2007–2009 | GII.11, GII.18 | Keum et al., 2009 [32] |
| Belgium | 2007 | GII.19 | Mauroy et al., 2008 [44] |
| China | 2008–2009 | GII.19 | Shen et al., 2012 [39] |
| Japan | 2008–2009 | GII.11, GII.18, GII.19, GII.3, GII.4, GII.13 | Shen et al., 2009 [38] |
| Brazil | 2008–2009 | GII.11 | Silva et al., 2015 [25] |
| USA | 2009 | GII.18, GII.11 | Scheuer et al., 2013 [57] |
| Ethiopia | 2013 | GII.1 | Sisay et al., 2016 [41] |
| Italy | 2017–2018 | GII.11, GII.18 | Laconi et al., 2020, Cavicchio et al., 2020 [28,61] |
| Dutch–German border region | 2017–2018 | GII.2 | Scheule et al., 2021 [55] |
| Japan | 2017–2018 | GII.11 | Okada et al., 2020 [52] |
| Italy | 2019 | GII.11 | Cavicchio et al., 2020 [28] |
| Primer Name | Sequence | Paper |
|---|---|---|
| For 35 | 5′-CTTGTTGGTTTGAGGCCATAT-3′ | Sugieda et al., 1998 * [19] |
| Rev36 | 5′-ATAAAAGTTGGCATGAACA-3′ | |
| For SMA82 | 5′-CCACTATGATGCAGATTA-3′ | |
| REV NV81 | 5′-ATCTCATCATCACCATA-3′ | |
| ReVNV82 | 5′-CACTATGATGCAGATTA-3′ | |
| For JV12 | 5′-ATACCACTATGATGCAGATTA-3′ | Van der poel et al., 2000 *, Mauroy et al., 2008nd [44,56] |
| Rev JV13 | 5′-TCATCATCACCATAGAAAGAG-3′ | |
| For P290 | 5′-GATTACTCCAAGTGGGACTCCAC-3′ | Wang et al., 2006 *†Δ, Martinezet al., 2006nd, Reuter et al., 2007 ◊, Martella et al., 2008nd, Mauroy et al., 2008nd, Schen et al., 2009 †, L’homme et al., 2009 *Δ, Halaihel et al., 2010nd, Nakamura et al., 2010 *†Δ, Chao et al., 2012 *, Shen et al., 2012 *, Scheur et al., 2013 *Δ, Sisay et al., 2013nd, Silva et al., 2014 *, Di Bartolo et al., 2014 *, Sisay et al., 2016 ◊, Valko et al., 2019 ◊, Stamelou et al., 2020nd [21,22,24,25,31,35,38,39,41,42,43,44,45,46,47,57,58] |
| Rev p289 | 5′-TGACAATGTAATCATCACCATA-3′ | |
| For PNV7 | 5′-AGGTGGTGGCCGAGGAYCTCCT-3′ | Wang et al., 2006 ◊, Scheur et al., 2013 *Δ, Sisay et al., 2013nd, Di Bartolo et al., 2014 *, Monini et al., 2015nd, Valko et al., 2019 ◊ [24,27,40,42,43,50] |
| Rev PNV8 | 5′-CACCATAGAAGGARAAGCA-3′ | |
| Rev p289H,I- | 5′-TGACGATTTCATCATCACCATA-3′ 5′-TGACGATTTCATCATCCCCGTA-3′ | Wolf et al., 2009nd, Farkas et al., 2005 ◊, Martella et al., 2008nd, Cunha et al., 2010 *†Δ [20,21,22,29,30,60] |
| For p290H,I,J,K, | 5′-GATTACTCCAGGTGGGACTCCAC-3′ 5′-GATTACTCCAGGTGGGACTCAAC-3′ 5′-GATTACTCCACCTGGGATTCAAC-3′ 5′-GATTACTCCACCTGGGATTCCAC-3′ | |
| Monroe region B 431/433 For | 5′-TGGACIAGRGGICCYAAYCA-3′ 5′-GAAYCTCATCCAYCTGAACAT-3′ | Mattison et al., 2007 *Δ◊, Cunha et al., 2010 *†Δ, Gutierrez et al., 2011 ◊ [30,64,67] |
| Monroe region B 432/434 Rev | 5′-TGGACICGYGGICCYAAYCA-3′ 5′-GGAYCGCATCCARCGGAACAT-3′ | |
| Ando region A For G-2 SR46/G-1 SR48/G-1 SR50/G-1 SR52 | 5′-TGGAATTCCATCGCCCACTGG-3′ 5′-GTGAACAGCATAAATCACTGG-3′ 5′-GTGAACAGTATAAACCACTGG-3′ 5′-GTGAACAGTATAAACCATTGG-3′ | Mattison et al., 2007 *Δ◊ [59] Mattison et al., 2007 *Δ◊ [59] |
| Ando region A rev G-1, G-2 SR33 | 5′-TGTCACGATCTCATCATCACC-3′ | |
| Region C for MR3/Yuri22F | 5′-CCGTCAGAGTGGGTATGAA-3′ 5′-ATGAATGAGGATGGACCCAT-3′ | Wolf et al., 2009nd [60] |
| Region C rev MR4/Yuri22R | 5′-AGTGGGTTTGAGGCCGTA-3′ 5′-CATCATCCCCGTAGAAAGAT-3′ | |
| For swNo F | 5′-AGGCAGCTCTATTGGACTAG-3′ | Mauroy et al., 2008 † [44] |
| Rev swNo R | 5′-GGTCTCATTATTGACCTCTGG-3′ | |
| Rev P289N | 5′-TCACGATTTCATCATCACCATA-3′ | L’homme et al., 2009 *Δ [58] |
| For P290N | 5′-GACTATTCACGGTGGGACTCCAC-3′ | |
| For JV12Y | 5′-ATACCACTATGATGCAGAYTA-3′ | Mijovsky et al., 2010 *Δ, Halaihel et al., 2010nd [17,21] |
| Rev JV13I | 5′-TCATCATCACCATAGAAIGAG-3′ | |
| For P290 | 5′-GATTACTCCAAGTGGGACTCCAC-3′ | Mijovsky et al., 2010 *Δ, Di Bartolo et al., 2014 *, Cavicchio et al., 2020 *Δ, Laconi et al., 2020 * [24,28,34,48] |
| Rev P110 | 5′-ACDATYTCATCATCACCATA-3′ | |
| For COG2F | 5′-CARGARBCIATGTTYAGRTGGATGAG-3′ | L’homme et al., 2009nd [64] |
| Rev COG2R | 5′-TCGACGCCATCTTCATTCACA-3′ | |
| Probe RING2 | 5′-TGGGAGGGCGATCGCAATCT-3′ | |
| For GIIF1 RT F RT | 5′-GGGAGGGCGATCGCAATCT-3′ | Keum et al., 2009 *◊ [32] |
| Rev GIIR RT, semi-nested R | 5′-CCRCCIGCATRICCRTTRTACAT-3′ | |
| For Cap GIIF2 Semi-nested F: | 5′-TTGTGAATGAAGATGGCGTCGA-3′ | |
| Song For | 5′-GATTACTCCAGTGGACTTCCAAC-3′ | Song et al., 2011 ◊ [69] |
| Song Rev | 5′-TGACGATTTCATCATCACCCAGTA-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cavicchio, L.; Laconi, A.; Piccirillo, A.; Beato, M.S. Swine Norovirus: Past, Present, and Future. Viruses 2022, 14, 537. https://doi.org/10.3390/v14030537
Cavicchio L, Laconi A, Piccirillo A, Beato MS. Swine Norovirus: Past, Present, and Future. Viruses. 2022; 14(3):537. https://doi.org/10.3390/v14030537
Chicago/Turabian StyleCavicchio, Lara, Andrea Laconi, Alessandra Piccirillo, and Maria Serena Beato. 2022. "Swine Norovirus: Past, Present, and Future" Viruses 14, no. 3: 537. https://doi.org/10.3390/v14030537
APA StyleCavicchio, L., Laconi, A., Piccirillo, A., & Beato, M. S. (2022). Swine Norovirus: Past, Present, and Future. Viruses, 14(3), 537. https://doi.org/10.3390/v14030537








