Identification of Crucial Amino Acids in Begomovirus C4 Proteins Involved in the Modulation of the Severity of Leaf Curling Symptoms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Virus Inoculation
2.2. Plasmid Construction and RCA
2.3. Sequence Analyses
3. Results
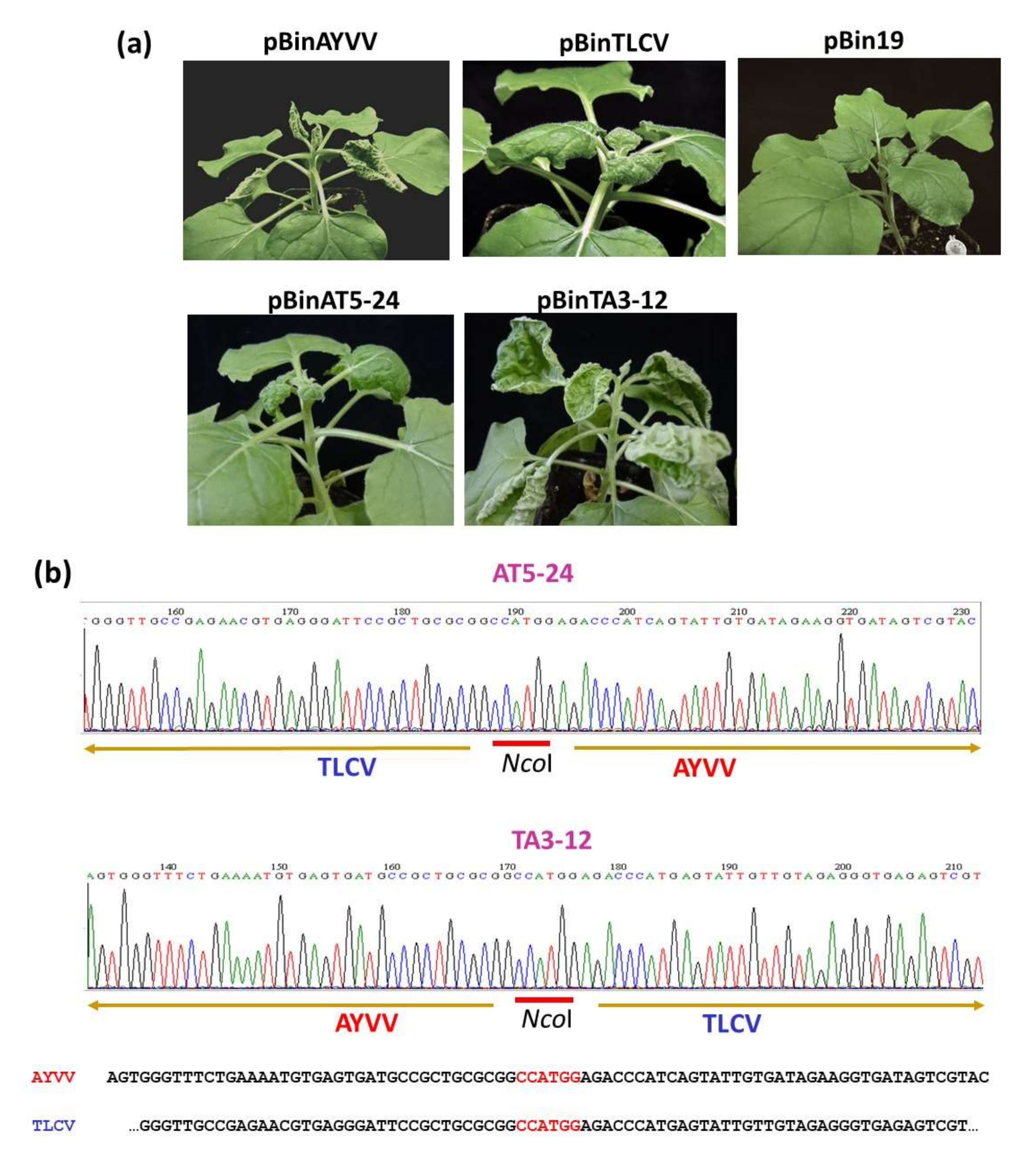
3.1. Identification of the Determinant of Leaf Curling Direction by Genome Recombination
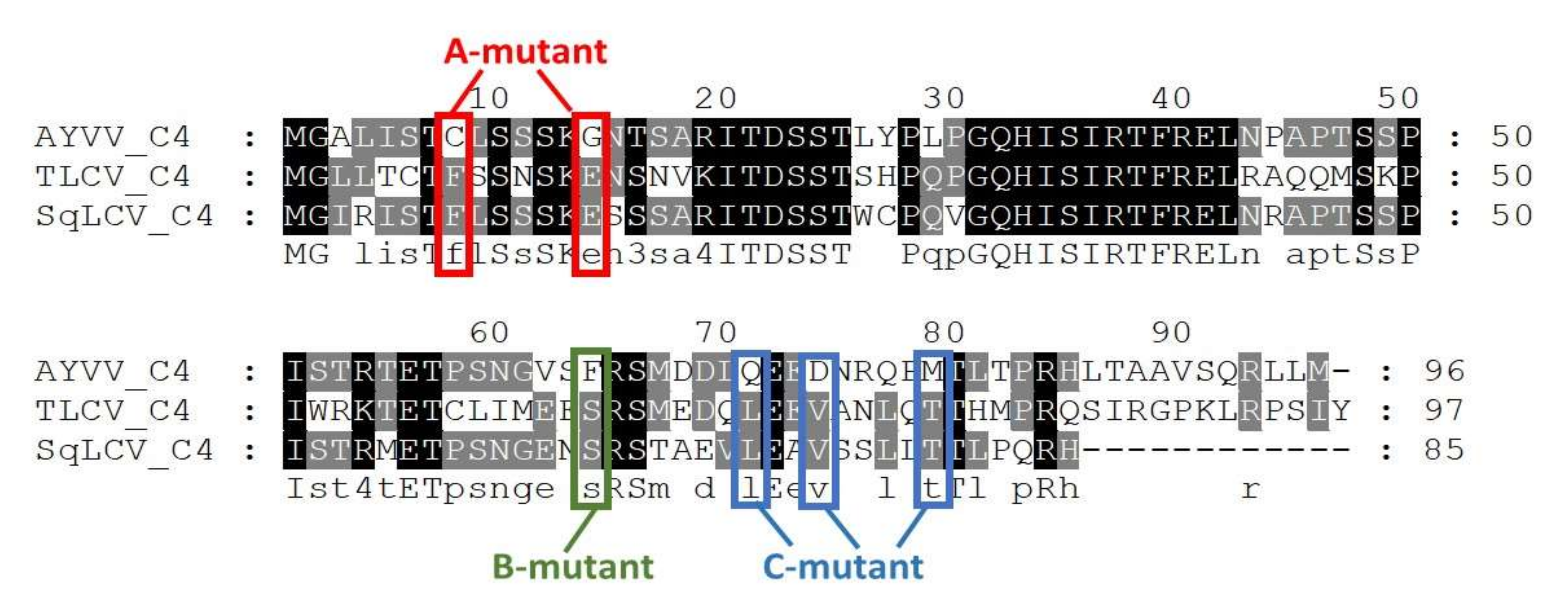
3.2. In Search of Amino Acids Involved in the Modulation of Leaf Curling Symptoms
3.3. Potential Application of A-Mut in Disease Management
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zerbini, F.M.; Briddon, R.W.; Idris, A.; Martin, D.P.; Moriones, E.; Navas-Castillo, J.; Rivera-Bustamante, R.; Roumagnac, P.; Varsani, A.; ICTV Report Consortium. ICTV Virus Taxonomy Profile: Geminiviridae. J. Gen. Virol. 2017, 98, 131–133. [Google Scholar] [CrossRef]
- Rojas, M.R.; Hagen, C.; Lucas, W.J.; Gilbertson, R.L. Exploiting chinks in the plant’s armor: Evolution and emergence of geminiviruses. Annu. Rev. Phytopathol. 2005, 43, 361–394. [Google Scholar] [CrossRef]
- Fauquet, C.M.; Briddon, R.W.; Brown, J.K.; Moriones, E.; Stanley, J.; Zerbini, M.; Zhou, X. Geminivirus strain demarcation and nomenclature. Arch. Virol. 2008, 153, 783–821. [Google Scholar] [CrossRef] [Green Version]
- Leke, W.N.; Mignouna, D.B.; Brown, J.K.; Kvarnheden, A. Begomovirus disease complex: Emerging threat to vegetable production systems of West and Central Africa. Agric. Food Secur. 2015, 4, 1. [Google Scholar] [CrossRef] [Green Version]
- Sattar, M.N.; Kvarnheden, A.; Saeed, M.; Briddon, R.W. Cotton leaf curl disease—An emerging threat to cotton production worldwide. J. Gen. Virol. 2013, 94, 695–710. [Google Scholar] [CrossRef]
- Zeng, R.; Liu, X.; Li, H.; Wu, S.; Huang, W.; Zhai, Z.; Li, M.; Lin, Y.; Xie, Q.; Yang, C.; et al. Danger peptide signaling enhances internalization of a geminivirus symptom determinant in plant cells during infection. J. Exp. Bot. 2020, 71, 2817–2827. [Google Scholar] [CrossRef]
- Mei, Y.; Zhang, F.; Wang, M.; Li, F.; Wang, Y.; Zhou, X. Divergent Symptoms Caused by Geminivirus-Encoded C4 Proteins Correlate with Their Ability to Bind NbSKeta. J. Virol. 2020, 94, e01307-20. [Google Scholar] [CrossRef]
- Li, Z.; Du, Z.; Tang, Y.; She, X.; Wang, X.; Zhu, Y.; Yu, L.; Lan, G.; He, Z. C4, the Pathogenic Determinant of Tomato Leaf Curl Guangdong Virus, May Suppress Post-transcriptional Gene Silencing by Interacting with BAM1 Protein. Front. Microbiol. 2020, 11, 851. [Google Scholar] [CrossRef]
- Li, P.; Jing, C.; Ren, H.; Jia, Z.; Ghanem, H.; Wu, G.; Li, M.; Qing, L. Analysis of Pathogenicity and Virulence Factors of Ageratum leaf curl Sichuan virus. Front. Plant Sci. 2020, 11, 527787. [Google Scholar] [CrossRef]
- Jing, C.; Li, P.; Zhang, J.; Wang, R.; Wu, G.; Li, M.; Xie, L.; Qing, L. The Malvastrum Yellow Vein Virus C4 Protein Promotes Disease Symptom Development and Enhances Virus Accumulation in Plants. Front. Microbiol. 2019, 10, 2425. [Google Scholar] [CrossRef] [Green Version]
- Zhan, B.; Zhao, W.; Li, S.; Yang, X.; Zhou, X. Functional Scanning of Apple Geminivirus Proteins as Symptom Determinants and Suppressors of Posttranscriptional Gene Silencing. Viruses 2018, 10, 488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mei, Y.; Yang, X.; Huang, C.; Zhang, X.; Zhou, X. Tomato leaf curl Yunnan virus-encoded C4 induces cell division through enhancing stability of Cyclin D 1.1 via impairing NbSKeta-mediated phosphorylation in Nicotiana benthamiana. PLoS Pathog. 2018, 14, e1006789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Zeng, R.; Chen, Z.; Liu, X.; Cao, Z.; Xie, Q.; Yang, C.; Lai, J. S-acylation of a geminivirus C4 protein is essential for regulating the CLAVATA pathway in symptom determination. J. Exp. Bot. 2018, 69, 4459–4468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rigden, J.E.; Krake, L.R.; Rezaian, M.A.; Dry, I.B. ORF C4 of tomato leaf curl geminivirus is a determinant of symptom severity. Virology 1994, 204, 847–850. [Google Scholar] [CrossRef]
- Stanley, J.; Latham, J.R.; Pinner, M.S.; Bedford, I.; Markham, P.G. Mutational analysis of the monopartite geminivirus beet curly top virus. Virology 1992, 191, 396–405. [Google Scholar] [CrossRef]
- Stanley, J.; Latham, J.R. A symptom variant of beet curly top geminivirus produced by mutation of open reading frame C4. Virology 1992, 190, 506–509. [Google Scholar] [CrossRef]
- Mei, Y.; Wang, Y.; Hu, T.; He, Z.; Zhou, X. The C4 protein encoded by Tomato leaf curl Yunnan virus interferes with mitogen-activated protein kinase cascade-related defense responses through inhibiting the dissociation of the ERECTA/BKI1 complex. New Phytol. 2021, 231, 747–762. [Google Scholar] [CrossRef]
- Gutierrez, C. DNA replication and cell cycle in plants: Learning from geminiviruses. EMBO J. 2000, 19, 792–799. [Google Scholar] [CrossRef] [Green Version]
- Van Wezel, R.; Liu, H.; Wu, Z.; Stanley, J.; Hong, Y. Contribution of the zinc finger to zinc and DNA binding by a suppressor of posttranscriptional gene silencing. J. Virol. 2003, 77, 696–700. [Google Scholar] [CrossRef] [Green Version]
- Lozano-Duran, R.; Bejarano, E.R. Geminivirus C2 protein might be the key player for geminiviral co-option of SCF-mediated ubiquitination. Plant Signal. Behav. 2011, 6, 999–1001. [Google Scholar] [CrossRef] [Green Version]
- Chandran, S.A.; Jeyabharathy, C.; Usha, R. The C2 protein of Bhendi yellow vein mosaic virus plays an important role in symptom determination and virus replication. Virus Genes 2014, 48, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.; Iwasaki, M.; Machida, C.; Machida, Y.; Zhou, X.; Chua, N.H. betaC1, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses. Genes Dev. 2008, 22, 2564–2577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fondong, V.N. The ever-expanding role of C4/AC4 in geminivirus infection: Punching above its weight? Mol. Plant 2019, 12, 145–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deom, C.M.; Mills-Lujan, K. Toward understanding the molecular mechanism of a geminivirus C4 protein. Plant Signal. Behav. 2015, 10, e1109758. [Google Scholar] [CrossRef]
- Mills-Lujan, K.; Andrews, D.L.; Chou, C.W.; Deom, C.M. The roles of phosphorylation and SHAGGY-like protein kinases in geminivirus C4 protein induced hyperplasia. PLoS ONE 2015, 10, e0122356. [Google Scholar] [CrossRef] [Green Version]
- Dogra, S.C.; Eini, O.; Rezaian, M.A.; Randles, J.W. A novel shaggy-like kinase interacts with the Tomato leaf curl virus pathogenicity determinant C4 protein. Plant Mol. Biol. 2009, 71, 25–38. [Google Scholar] [CrossRef]
- Vanitharani, R.; Chellappan, P.; Pita, J.S.; Fauquet, C.M. Differential roles of AC2 and AC4 of cassava geminiviruses in mediating synergism and suppression of posttranscriptional gene silencing. J. Virol. 2004, 78, 9487–9498. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Q.; Zhou, X.P. The C4 proteins of Ageratum yellow vein China virus and Stachytarpheta leaf curl virus are suppressors of RNA silencing. Wei Sheng Wu Xue Bao = Acta Microbiol. Sin. 2007, 47, 39–43. [Google Scholar]
- Gopal, P.; Pravin Kumar, P.; Sinilal, B.; Jose, J.; Kasin Yadunandam, A.; Usha, R. Differential roles of C4 and betaC1 in mediating suppression of post-transcriptional gene silencing: Evidence for transactivation by the C2 of Bhendi yellow vein mosaic virus, a monopartite begomovirus. Virus Res. 2007, 123, 9–18. [Google Scholar] [CrossRef]
- Ismayil, A.; Haxim, Y.; Wang, Y.; Li, H.; Qian, L.; Han, T.; Chen, T.; Jia, Q.; Yihao Liu, A.; Zhu, S.; et al. Cotton Leaf Curl Multan virus C4 protein suppresses both transcriptional and post-transcriptional gene silencing by interacting with SAM synthetase. PLoS Pathog. 2018, 14, e1007282. [Google Scholar] [CrossRef]
- Chellappan, P.; Vanitharani, R.; Fauquet, C.M. MicroRNA-binding viral protein interferes with Arabidopsis development. Proc. Natl. Acad. Sci. USA 2005, 102, 10381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, P.; Wang, H.; Xue, H.; Rosas-Diaz, T.; Tang, W.; Zhang, H.; Xu, L.; Lozano-Duran, R. The receptor-like kinases BAM1 and BAM2 promote the cell-to-cell movement of miRNA in the root stele to regulate xylem patterning. bioRxiv 2019. [Google Scholar] [CrossRef]
- Rosas-Diaz, T.; Zhang, D.; Fan, P.; Wang, L.; Ding, X.; Jiang, Y.; Jimenez-Gongora, T.; Medina-Puche, L.; Zhao, X.; Feng, Z.; et al. A virus-targeted plant receptor-like kinase promotes cell-to-cell spread of RNAi. Proc. Natl. Acad. Sci. USA 2018, 115, 1388–1393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mei, Y.; Wang, Y.; Hu, T.; Yang, X.; Lozano-Duran, R.; Sunter, G.; Zhou, X. Nucleocytoplasmic Shuttling of Geminivirus C4 Protein Mediated by Phosphorylation and Myristoylation Is Critical for Viral Pathogenicity. Mol. Plant 2018, 11, 1466–1481. [Google Scholar] [CrossRef] [Green Version]
- Corrales-Gutierrez, M.; Medina-Puche, L.; Yu, Y.; Wang, L.; Ding, X.; Luna, A.P.; Bejarano, E.R.; Castillo, A.G.; Lozano-Duran, R. The C4 protein from the geminivirus Tomato yellow leaf curl virus confers drought tolerance in Arabidopsis through an ABA-independent mechanism. Plant Biotechnol. J. 2020, 18, 1121–1123. [Google Scholar] [CrossRef] [Green Version]
- Luna, A.P.; Lozano-Duran, R. Geminivirus-Encoded Proteins: Not All Positional Homologs Are Made Equal. Front. Microbiol. 2020, 11, 878. [Google Scholar] [CrossRef]
- Wu, C.Y.; Lai, Y.C.; Lin, N.S.; Hsu, Y.H.; Tsai, H.T.; Liao, J.Y.; Hu, C.C. A simplified method of constructing infectious clones of begomovirus employing limited restriction enzyme digestion of products of rolling circle amplification. J. Virol. Methods 2008, 147, 355–359. [Google Scholar] [CrossRef]
- Elmer, J.S.; Sunter, G.; Gardiner, W.E.; Brand, L.; Browning, C.K.; Bisaro, D.M.; Rogers, S.G. Agrobacterium-mediated inoculation of plants with tomato golden mosaic virus DNAs. Plant Mol. Biol. 1988, 10, 225–234. [Google Scholar] [CrossRef]
- Grimsley, N.; Hohn, T.; Davies, J.W.; Hohn, B. Agrobacterium-mediated delivery of infectious maize streak virus into maize plants. Nature 1987, 325, 177–179. [Google Scholar] [CrossRef]
- Grimsley, N.; Hohn, B.; Hohn, T.; Walden, R. “Agroinfection,” an alternative route for viral infection of plants by using the Ti plasmid. Proc. Natl. Acad. Sci. USA 1986, 83, 3282–3286. [Google Scholar] [CrossRef] [Green Version]
- Silva, D.; Santos, G.; Barroca, M.; Collins, T. Inverse PCR for Point Mutation Introduction. Methods Mol. Biol. 2017, 1620, 87–100. [Google Scholar] [PubMed]
- Vieira, J.; Messing, J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987, 153, 3–11. [Google Scholar] [PubMed]
- Haible, D.; Kober, S.; Jeske, H. Rolling circle amplification revolutionizes diagnosis and genomics of geminiviruses. J. Virol. Methods 2006, 135, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Nicholas, K.; Nicholas, H. GeneDoc: A Tool for Editing and Annotating Multiple Sequence Alignments; ScienceOpen, Inc.: Burlington, MA, USA, 1997. [Google Scholar]
- Hall, T.A. BioEdit: A User-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Heyraud-Nitschke, F.; Schumacher, S.; Laufs, J.; Schaefer, S.; Schell, J.; Gronenborn, B. Determination of the origin cleavage and joining domain of geminivirus Rep proteins. Nucleic Acids Res. 1995, 23, 910–916. [Google Scholar] [CrossRef] [Green Version]
- Argüello-Astorga, G.R.; Ruiz-Medrano, R. An iteron-related domain is associated to Motif 1 in the replication proteins of geminiviruses: Identification of potential interacting amino acid-base pairs by a comparative approach. Arch. Virol. 2001, 146, 1465–1485. [Google Scholar] [CrossRef]
- Chen, T.H.; Hu, C.C.; Liao, J.T.; Lee, Y.L.; Huang, Y.W.; Lin, N.S.; Lin, Y.L.; Hsu, Y.H. Production of Japanese Encephalitis Virus Antigens in Plants Using Bamboo Mosaic Virus-Based Vector. Front. Microbiol. 2017, 8, 788. [Google Scholar] [CrossRef]
- Liou, M.R.; Huang, Y.W.; Hu, C.C.; Lin, N.S.; Hsu, Y.H. A dual gene-silencing vector system for monocot and dicot plants. Plant Biotechnol. J. 2014, 12, 330–343. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Lee, H.J.; Cheon, C.I.; Kim, S.H.; Hur, Y.S.; Auh, C.K.; Im, K.H.; Yun, D.J.; Lee, S.; Davis, K.R. The Arabidopsis thaliana homeobox gene ATHB12 is involved in symptom development caused by geminivirus infection. PLoS ONE 2011, 6, e20054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, T.; Wang, Y.; Teotia, S.; Wang, Z.; Shi, C.; Sun, H.; Gu, Y.; Zhang, Z.; Tang, G. The interaction between miR160 and miR165/166 in the control of leaf development and drought tolerance in Arabidopsis. Sci. Rep. 2019, 9, 2832. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Wang, Y.; Teotia, S.; Zhang, Z.; Tang, G. The Making of Leaves: How Small RNA Networks Modulate Leaf Development. Front. Plant Sci. 2018, 9, 824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koyama, T.; Sato, F.; Ohme-Takagi, M. Roles of miR319 and TCP Transcription Factors in Leaf Development. Plant Physiol. 2017, 175, 874–885. [Google Scholar] [CrossRef] [Green Version]
- Chitwood, D.H.; Sinha, N.R. Evolutionary and Environmental Forces Sculpting Leaf Development. Curr. Biol. 2016, 26, R297–R306. [Google Scholar] [CrossRef] [Green Version]
- Kalve, S.; De Vos, D.; Beemster, G.T. Leaf development: A cellular perspective. Front. Plant Sci. 2014, 5, 362. [Google Scholar] [CrossRef] [Green Version]
- Palatnik, J.F.; Allen, E.; Wu, X.; Schommer, C.; Schwab, R.; Carrington, J.C.; Weigel, D. Control of leaf morphogenesis by microRNAs. Nature 2003, 425, 257–263. [Google Scholar] [CrossRef] [Green Version]



| Inoculation Efficiency 2 | ||||||||
|---|---|---|---|---|---|---|---|---|
| Experiment I | Experiment II | |||||||
| Inoculum | Leaf Curling 1 | 14 dpi | 21 dpi | 28 dpi | 14 dpi | 21 dpi | 28 dpi | Infectivity 3 |
| pBin19 | - | 0/4 | 0/4 | 0/4 | 0/10 | 0/10 | 0/10 | 0 |
| pBinAYVV | Up | 2/2 | 2/2 | 2/2 | 10/10 | 10/10 | 10/10 | 100 |
| pBinTLCV | Dw | 2/2 | 2/2 | 2/2 | 10/10 | 10/10 | 10/10 | 100 |
| pBinTA3-12 | Up | 0/3 | 2/3 | 2/3 | 0/8 | 4/8 | 7/8 | 82 |
| pBinAT5-20 | Dw | 2/3 | 2/3 | 2/3 | 6/8 | 6/8 | 6/8 | 73 |
| pBinAT5-24 | Up | 2/3 | 2/3 | 2/3 | 78 | 7/8 | 7/8 | 82 |
| Infectivity 1 | ||||
|---|---|---|---|---|
| Inoculum | Experiment I | Experiment II | Experiment III | Leaf Curling Symptom |
| Buffer | 0/1 | 0/1 | 0/1 | - |
| pBin19 | 0/1 | 0/1 | 0/1 | - |
| pBinAYVV | 1/1 | 1/1 | 4/4 | Severe upward |
| A-mut | 4/10 | 9/9 | 4/4 | Mild upward |
| B-mut | ND 2 | 14/14 | 5/5 | Severe upward |
| C-mut | ND | ND | 4/4 | Severe upward |
| Experiment I | Experiment II | Experiment III | ||||
|---|---|---|---|---|---|---|
| Inoculum | Infectivity 1 | Attenuation 2 | Infectivity | Attenuation | Infectivity | Attenuation |
| pBinAYVV | 4/4 | 0/4 | 4/4 | 0/4 | 4/4 | 0/4 |
| A-mut | 4/4 | 4/4 | 4/4 | 4/4 | 3/4 | 3/4 |
| pBinAYVV → A-mut | 2/4 | 0/4 | ND 3 | ND | 4/4 | 0/4 |
| A-mut → pBinAYVV | 3/4 | 3/4 | 3/4 | 3/4 | 3/4 | 3/4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, K.-W.; Tsai, Y.-T.; Wu, C.-Y.; Lai, Y.-C.; Lin, N.-S.; Hu, C.-C. Identification of Crucial Amino Acids in Begomovirus C4 Proteins Involved in the Modulation of the Severity of Leaf Curling Symptoms. Viruses 2022, 14, 499. https://doi.org/10.3390/v14030499
Dai K-W, Tsai Y-T, Wu C-Y, Lai Y-C, Lin N-S, Hu C-C. Identification of Crucial Amino Acids in Begomovirus C4 Proteins Involved in the Modulation of the Severity of Leaf Curling Symptoms. Viruses. 2022; 14(3):499. https://doi.org/10.3390/v14030499
Chicago/Turabian StyleDai, Kao-Wei, Yu-Ting Tsai, Chia-Ying Wu, Yi-Chin Lai, Na-Sheng Lin, and Chung-Chi Hu. 2022. "Identification of Crucial Amino Acids in Begomovirus C4 Proteins Involved in the Modulation of the Severity of Leaf Curling Symptoms" Viruses 14, no. 3: 499. https://doi.org/10.3390/v14030499
APA StyleDai, K.-W., Tsai, Y.-T., Wu, C.-Y., Lai, Y.-C., Lin, N.-S., & Hu, C.-C. (2022). Identification of Crucial Amino Acids in Begomovirus C4 Proteins Involved in the Modulation of the Severity of Leaf Curling Symptoms. Viruses, 14(3), 499. https://doi.org/10.3390/v14030499






