Characterization of the First SARS-CoV-2 Isolates from Aotearoa New Zealand as Part of a Rapid Response to the COVID-19 Pandemic
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells
2.2. Clinical Specimens
2.3. SARS-CoV-2 Isolation
2.4. SARS-CoV-2 Plaque Assay
2.5. Immunofluorescence Assay
2.6. Western Blot
2.7. SARS-CoV-2 RT-qPCR Assay
2.8. Whole-Genome Sequencing of Patient-Derived SARS-CoV-2 and SARS-CoV-2 Isolates
2.8.1. MiSeq, Illumina
2.8.2. Ion PGM™ System, Ion Torrent™
2.9. Microbiota Analysis
2.10. Phylogenetic Analysis
2.11. Electron Microscopy
2.12. Viral Growth Kinetics Analysis
2.13. Drug Susceptibility
2.14. Statistical Analysis
3. Results
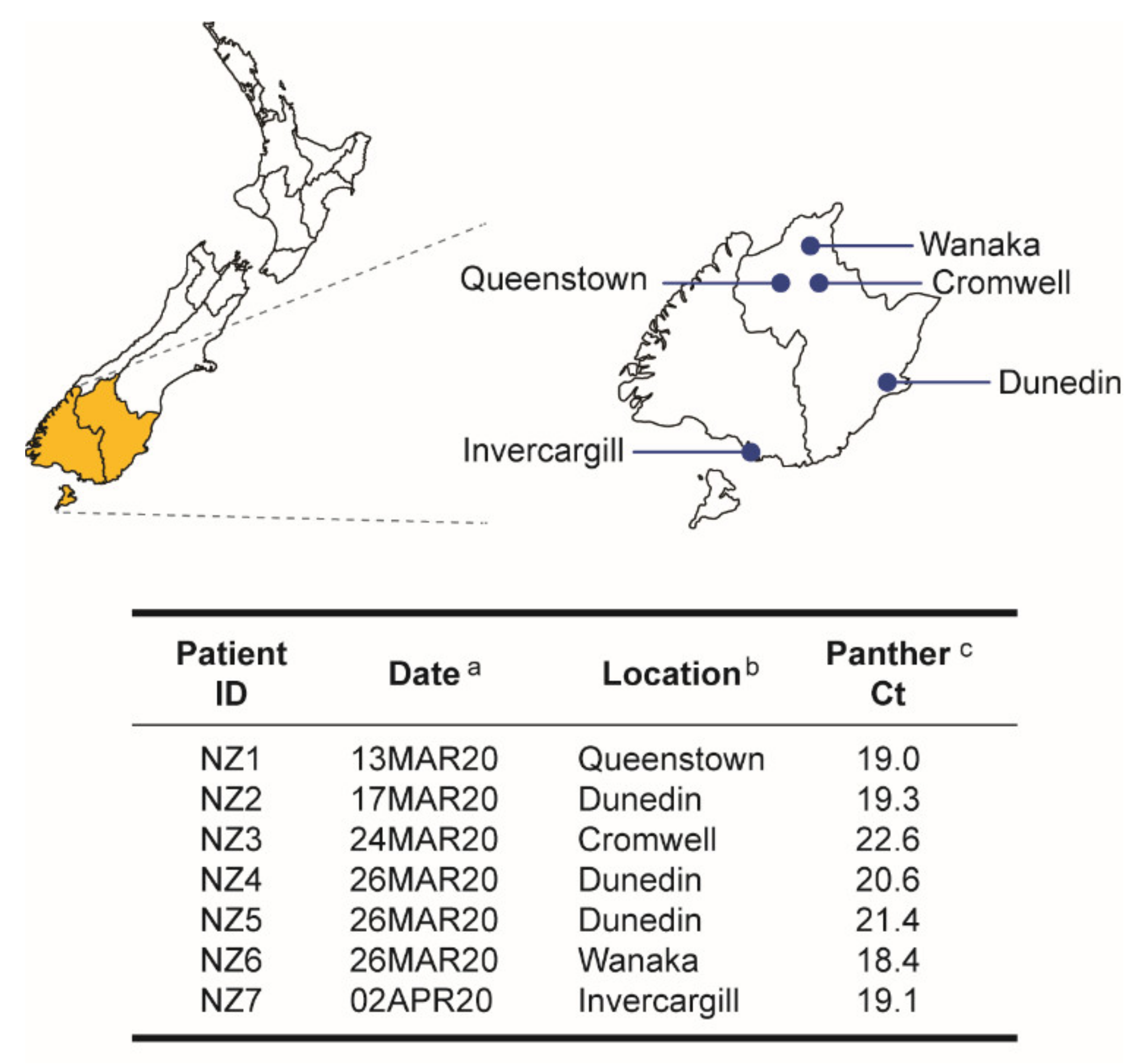
3.1. First COVID-19 Cases in the South Island, New Zealand
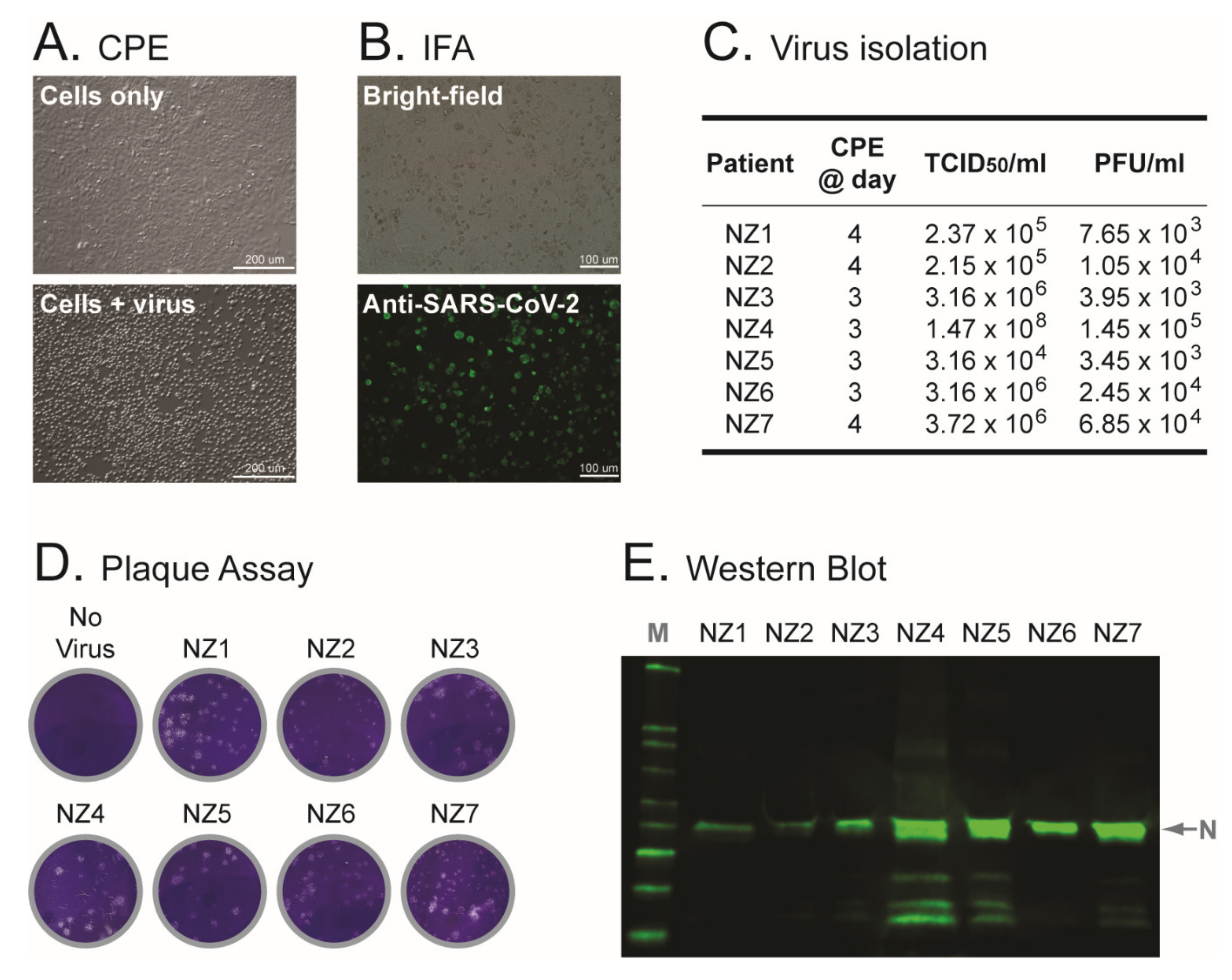
3.2. First Isolation of SARS-CoV-2 in New Zealand
3.3. Electron Microscopy of Vero Cells Infected with SARS-CoV-2 Isolates
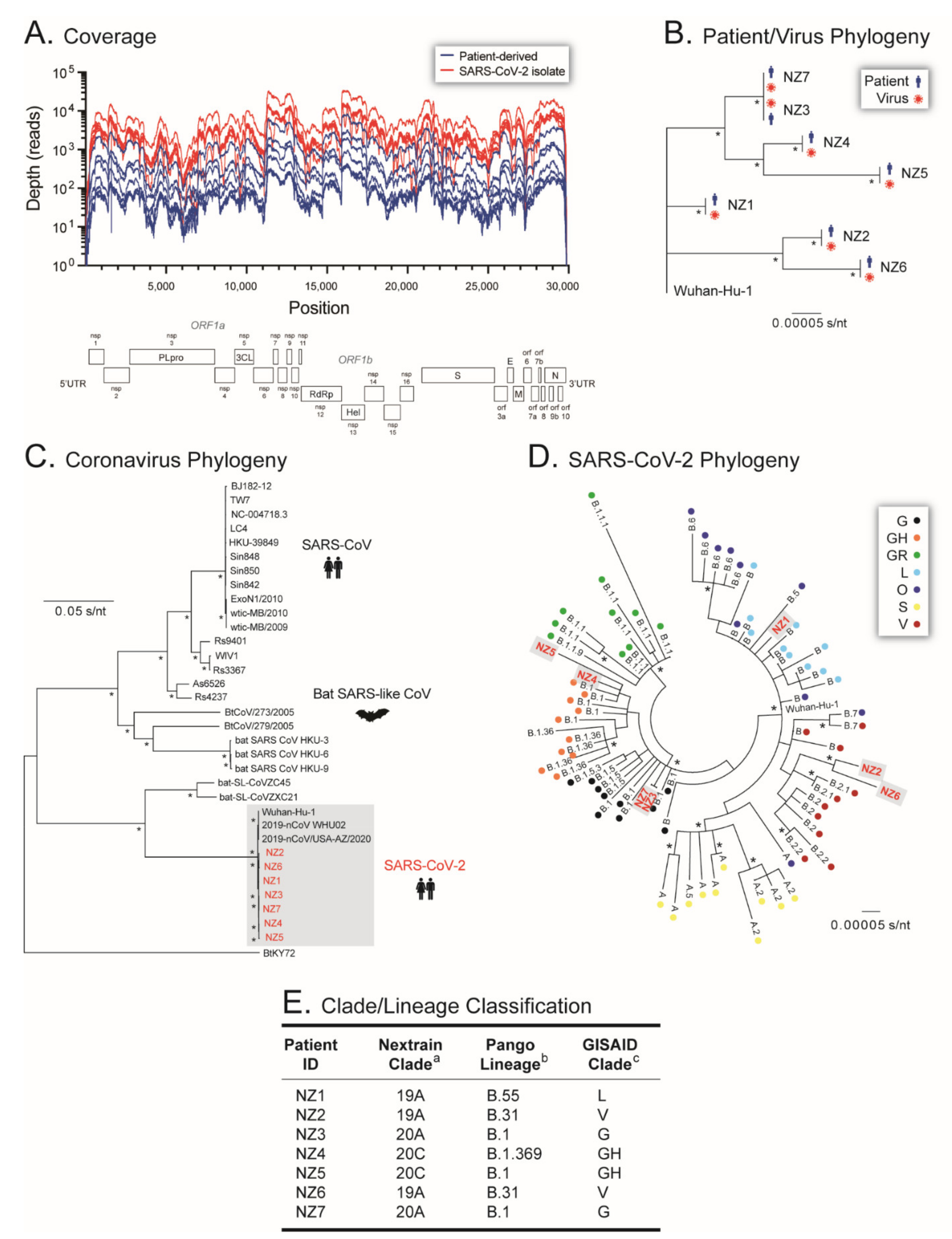
3.4. Whole-Genome Sequencing of Patient-Derived SARS-CoV-2 Samples and SARS-CoV-2 Isolates from Early in the Pandemic in New Zealand
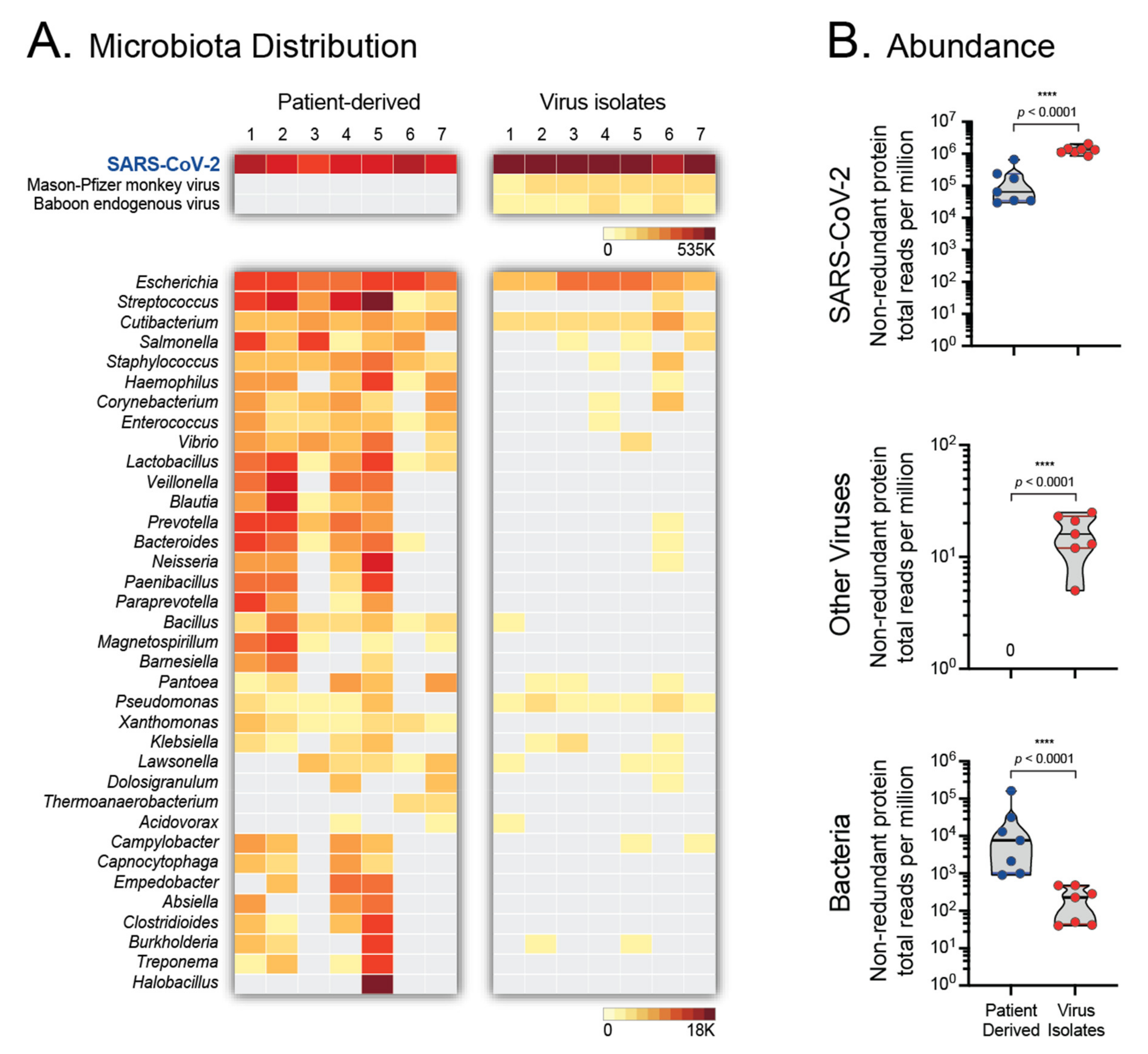
3.5. Microbiota of Patient-Derived Samples and SARS-CoV-2 Isolates
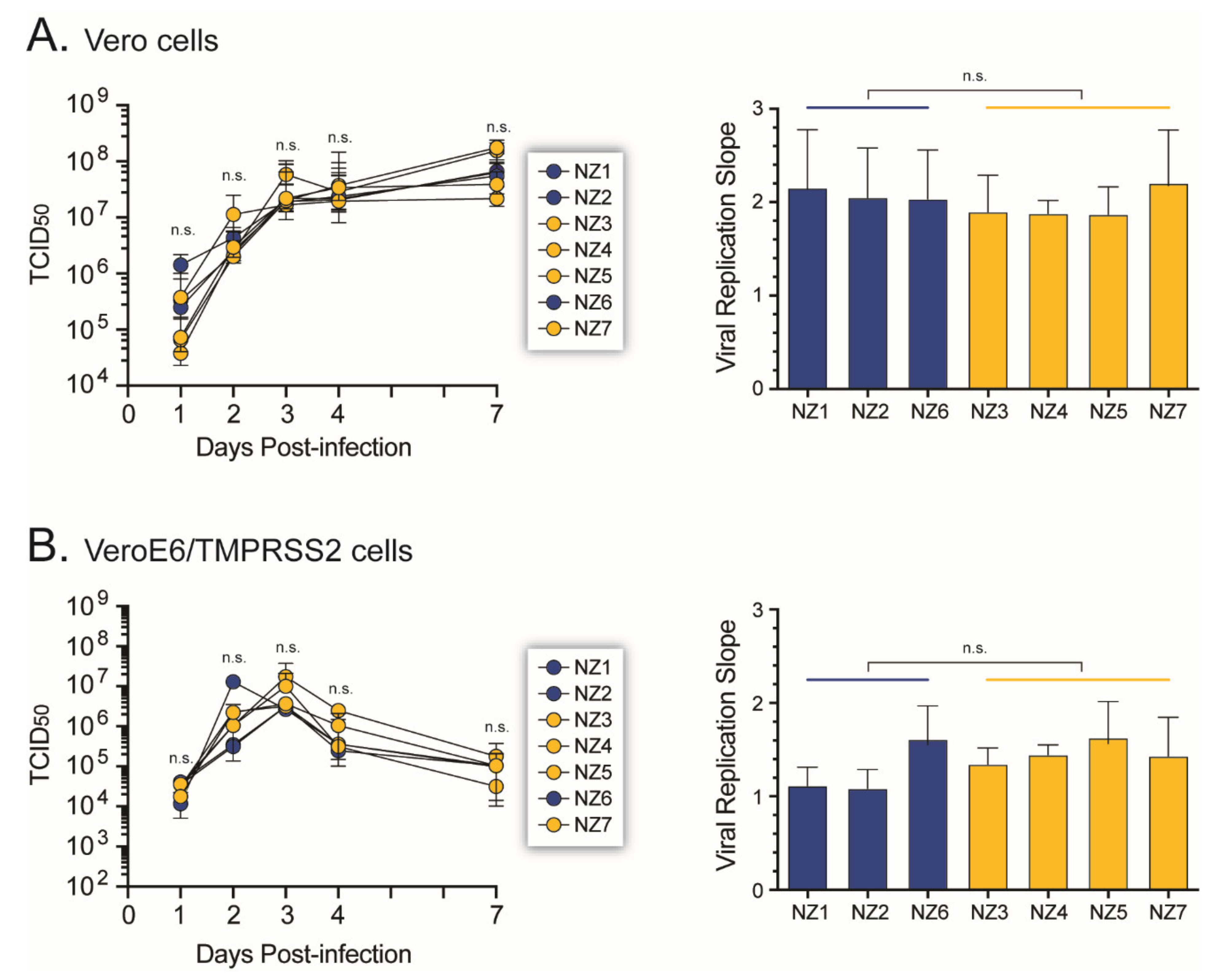
3.6. SARS-CoV-2 Growth Kinetics
3.7. Susceptibility of SARS-CoV-2 Isolates to Antiviral Agents
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Kuiken, T.; Fouchier, R.A.; Schutten, M.; Rimmelzwaan, G.F.; van Amerongen, G.; van Riel, D.; Laman, J.D.; de Jong, T.; van Doornum, G.; Lim, W.; et al. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 2003, 362, 263–270. [Google Scholar] [CrossRef]
- Guery, B.; Poissy, J.; el Mansouf, L.; Séjourné, C.; Ettahar, N.; Lemaire, X.; Vuotto, F.; Goffard, A.; Behillil, S.; Enouf, V.; et al. Clinical features and viral diagnosis of two cases of infection with Middle East Respiratory Syndrome coronavirus: A report of nosocomial transmission. Lancet 2013, 381, 2265–2272. [Google Scholar] [CrossRef]
- Patel, A.; Jernigan, D.B. Initial Public Health Response and Interim Clinical Guidance for the 2019 Novel Coronavirus Outbreak—United States, December 31, 2019–February 4, 2020. MMWR 2020, 69, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Reiche, A.S.; Hernandez, M.M.; Sullivan, M.J.; Ciferri, B.; Alshammary, H.; Obla, A.; Fabre, S.; Kleiner, G.; Polanco, J.; Khan, Z.; et al. Introductions and early spread of SARS-CoV-2 in the New York City area. Science 2020, 17, 297–301. [Google Scholar] [CrossRef]
- Da Silva Filipe, A.; Shepherd, J.G.; Williams, T.; Hughes, J.; Aranday-Cortes, E.; Asamaphan, P.; Ashraf, S.; Balcazar, C.; Brunker, K.; Campbell, A.; et al. Genomic epidemiology reveals multiple introductions of SARS-CoV-2 from mainland Europe into Scotland. Nat. Microbiol. 2021, 6, 112–122. [Google Scholar] [CrossRef]
- Worobey, M.; Pekar, J.; Larsen, B.B.; Nelson, M.I.; Hill, V.; Joy, J.B.; Rambaut, A.; Suchard, M.A.; Wertheim, J.O.; Lemey, P. The emergence of SARS-CoV-2 in Europe and North America. Science 2020, 370, 564–570. [Google Scholar] [CrossRef]
- Hao, X.; Cheng, S.; Wu, D.; Wu, T.; Lin, X.; Wang, C. Reconstruction of the full transmission dynamics of COVID-19 in Wuhan. Nature 2020, 584, 420–424. [Google Scholar] [CrossRef]
- Li, J.; Lai, S.; Gao, G.F.; Shi, W. The emergence, genomic diversity and global spread of SARS-CoV-2. Nature 2021, 600, 408–418. [Google Scholar] [CrossRef]
- Shu, Y.; McCauley, J. GISAID: Global initiative on sharing all influenza data-from vision to reality. Euro. Surveill 2017, 22, 30494. [Google Scholar] [CrossRef]
- Dong, E.; Du, H.; Gardner, L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020, 20, 533–534. [Google Scholar] [CrossRef]
- Rambaut, A.; Holmes, E.C.; O’Toole, A.; Hill, V.; McCrone, J.T.; Ruis, C.; du Plessis, L.; Pybus, O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020, 5, 1403–1407. [Google Scholar] [CrossRef]
- O’Toole, Á.; Scher, E.; Underwood, A.; Jackson, B.; Hill, V.; McCrone, J.T.; Colquhoun, R.; Ruis, C.; Abu-Dahab, K.; Taylor, B.; et al. Assignment of Epidemiological Lineages in an Emerging Pandemic Using the Pangolin Tool. Virus Evol. 2021, 7, veab064. [Google Scholar] [CrossRef]
- Cui, J.; Li, F.; Shi, Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef]
- Barré-Sinoussi, F.; Chermann, J.C.; Rey, F.; Nugeyre, M.T.; Chamaret, S.; Gruest, J.; Dauguet, C.; Axler-Blin, C.; Vézinet-Brun, F.; Rouzioux, C.; et al. Isolation of a T-Lymphotropic Retroviruses from a Patient at Risk for Acquired Immune Deficiency Syndrome (AIDS). Science 1983, 220, 868–871. [Google Scholar] [CrossRef] [PubMed]
- Brès, P. The epidemic of Ebola haemorrhagic fever in Sudan and Zaire, 1976: Introductory note. Bull. World Health Organ. 1978, 56, 245. [Google Scholar] [PubMed]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.W.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef]
- Lu, X.; Wang, L.; Sakthivel, S.K.; Whitaker, B.; Murray, J.; Kamili, S.; Lynch, B.; Malapati, L.; Burke, S.A.; Harcourt, J.; et al. US CDC Real-Time Reverse Transcription PCR Panel for Detection of Severe Acute Respiratory Syndrome Coronavirus 2. Emerg. Infect. Dis. 2020, 26, 1654–1665. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.; Zinger, T.; Inglima, K.; Woo, K.-M.; Atie, O.; Yurasits, L.; See, B.; Aguero-Rosenfeld, M.E. Performance of Abbott ID Now COVID-19 Rapid Nucleic Acid Amplification Test Using Nasopharyngeal Swabs Transported in Viral Transport Media and Dry Nasal Swabs in a New York City Academic Institution. J. Clin. Microbiol. 2020, 58, 8. [Google Scholar] [CrossRef]
- Kevadiya, B.D.; Machhi, J.; Herskovitz, J.; Oleynikov, M.D.; Blomberg, W.R.; Bajwa, N.; Soni, D.; Das, S.; Hasan, M.; Patel, M.; et al. Diagnostics for SARS-CoV-2 infections. Nat. Mater. 2021, 20, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.-W.; Schmitz, J.E.; Persing, D.H.; Stratton, C.W. The Laboratory Diagnosis of COVID-19 Infection: Current Issues and Challenges. J. Clin. Microbiol. 2020, 58, e00512-20. [Google Scholar] [CrossRef] [PubMed]
- Lawley, B.; Grant, J.; Harfoot, R.; Treece, J.M.; Day, R.; Hernandez, L.C.; Stanton, J.L.; Ussher, J.E.; Quinones-Mateu, M.E. Rapid Response to SARS-CoV-2 in Aotearoa New Zealand: Implementation of a Diagnostic Test and Characterization of the First COVID-19 Cases in the South Island. Viruses 2021, 13, 2222. [Google Scholar] [CrossRef] [PubMed]
- Riva, L.; Yuan, S.; Yin, X.; Martin-Sancho, L.; Matsunaga, N.; Pache, L.; Burgstaller-Muehlbacher, S.; de Jesus, P.D.; Teriete, P.; Hull, M.V.; et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature 2020, 586, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F. SARS-CoV-2 vaccines in development. Nature 2020, 586, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Jefferies, S.; French, N.; Gilkison, C.; Graham, G.; Hope, V.; Marshall, J.; McElnay, C.; McNeill, A.; Muellner, P.; Paine, S.; et al. COVID-19 in New Zealand and the impact of the national response: A descriptive epidemiological study. Lancet Public Health 2020, 5, e612–e623. [Google Scholar] [CrossRef]
- Douglas, J.; Geoghegan, J.L.; Hadfield, J.; Bouckaert, R.; Storey, M.; Ren, X.; de Ligt, J.; French, N.; Welch, D. Real-Time Genomics for Tracking Severe Acute Respiratory Syndrome Coronavirus 2 Border Incursions after Virus Elimination, New Zealand. Emerg. Infect. Dis. 2021, 27, 2361–2368. [Google Scholar] [CrossRef] [PubMed]
- Yamada, S.; Fukushi, S.; Kinoshita, H.; Ohnishi, M.; Suzuki, T.; Fujimoto, T.; Saijo, M.; Maeda, K. Assessment of SARS-CoV-2 infectivity of upper respiratory specimens from COVID-19 patients by virus isolation using VeroE6/TMPRSS2 cells. BMJ Open Respir. Res. 2021, 8, e000830. [Google Scholar] [CrossRef] [PubMed]
- Reed, L.J.; Muench, H. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 1938, 27, 493–497. [Google Scholar]
- Cleemput, S.; Dumon, W.; Fonseca, V.; Abdool Karim, W.; Giovanetti, M.; Alcantara, L.C.; Deforche, K.; de Oliveira, T. Genome Detective Coronavirus Typing Tool for rapid identification and characterization of novel coronavirus genomes. Bioinformatics 2020, 36, 3552–3555. [Google Scholar] [CrossRef] [PubMed]
- Kalantar, K.L.; Carvalho, T.; A De Bourcy, C.F.; Dimitrov, B.; Dingle, G.; Egger, R.; Han, J.; Holmes, O.B.; Juan, Y.-F.; King, R.; et al. IDseq—An open source cloud-based pipeline and analysis service for metagenomic pathogen detection and monitoring. GigaScience 2020, 9, 10. [Google Scholar] [CrossRef] [PubMed]
- Mercatelli, D.; Triboli, L.; Fornasari, E.; Ray, F.; Giorgi, F.M. Coronapp: A web application to annotate and monitor SARS-CoV-2 mutations. J. Med. Virol. 2021, 93, 3238–3245. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Mastronarde, D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005, 152, 36–51. [Google Scholar] [CrossRef] [PubMed]
- Kremer, J.R.; Mastronarde, D.N.; Mc Intosh, J.R. Computer Visualization of Three-Dimensional Image Data Using IMOD. J. Struct. Biol. 1996, 116, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Goddard, T.D.; Huang, C.C.; Meng, E.C.; Pettersen, E.F.; Couch, G.S.; Morris, J.; Ferrin, T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018, 27, 14–25. [Google Scholar] [CrossRef]
- Weber, J.; Vazquez, A.C.; Winner, D.; Rose, J.D.; Wylie, D.; Rhea, A.M.; Henry, K.; Pappas, J.; Wright, A.; Mohamed, N.; et al. Novel method for simultaneous quantification of phenotypic resistance to maturation, protease, reverse transcriptase, and integrase HIV inhibitors based on 3’Gag(p2/p7/p1/p6)/PR/RT/INT-recombinant viruses: A useful tool in the multitarget era of antiretroviral therapy. Antimicrob. Agents Chemother. 2011, 55, 3729–3742. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.; Rose, J.D.; Vazquez, A.C.; Winner, D.; Margot, N.; McColl, D.J.; Miller, M.D.; Quiñones-Mateu, M.E. Resistance Mutations outside the Integrase Coding Region Have an Effect on Human Immunodeficiency Virus Replicative Fitness but Do Not Affect Its Susceptibility to Integrase Strand Transfer Inhibitors. PLoS ONE 2013, 8, e65631. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Caly, L.; Druce, J.; Roberts, J.; Bond, K.; Tran, T.; Kostecki, R.; Yoga, Y.; Naughton, W.; Taiaroa, G.; Seemann, T.; et al. Isolation and rapid sharing of the 2019 novel coronavirus (SARS -CoV-2) from the first patient diagnosed with COVID-19 in Australia. Med. J. Aust. 2020, 212, 459–462. [Google Scholar] [CrossRef]
- Bushman, D.; Alroy, K.A.; Greene, S.K.; Keating, P.; Wahnich, A.; Weiss, D.; Pathela, P.; Harrison, C.; Rakeman, J.; Langley, G.; et al. Detection and Genetic Characterization of Community-Based SARS-CoV-2 Infections-New York City, March 2020. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 918–922. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, S.; Nao, N.; Shirato, K.; Kawase, M.; Saito, S.; Takayama, I.; Nagata, N.; Sekizuka, T.; Katoh, H.; Kato, F.; et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. USA 2020, 117, 7001–7003. [Google Scholar] [CrossRef] [PubMed]
- Brockwell-Staats, C.; Webster, R.G.; Webby, R.J. Diversity of influenza viruses in swine and the emergence of a novel human pandemic influenza A (H1N1). Influ. Respir. Viruses 2009, 3, 207–213. [Google Scholar] [CrossRef]
- Lycett, S.J.; Duchatel, F.; Digard, P. A brief history of bird flu. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180257. [Google Scholar] [CrossRef] [PubMed]
- Fauci, A.S.; Lane, H.C.; Redfield, R.R. Covid-19-Navigating the Uncharted. N. Engl. J. Med. 2020, 382, 1268–1269. [Google Scholar] [CrossRef] [PubMed]
- Ark, W.B.; Kwon, N.J.; Choi, S.J.; Kang, C.K.; Choe, P.G.; Kim, J.Y.; Yun, J.; Lee, G.W.; Seong, M.W.; Kim, N.J.; et al. Virus Isolation from the First Patient with SARS-CoV-2 in Korea. J. Korean Med. Sci. 2020, 35, e84. [Google Scholar]
- Perera, R.; Tso, E.; Tsang, O.T.Y.; Tsang, D.N.C.; Fung, K.; Leung, Y.W.Y.; Chin, A.W.H.; Chu, D.K.W.; Cheng, S.M.S.; Poon, L.L.M.; et al. SARS-CoV-2 Virus Culture and Subgenomic RNA for Respiratory Specimens from Patients with Mild Coronavirus Disease. Emerg. Infect Dis. 2020, 26, 2701–2704. [Google Scholar] [CrossRef]
- Harcourt, J.; Tamin, A.; Lu, X.; Kamili, S.; Sakthivel, S.K.; Murray, J.; Queen, K.; Tao, Y.; Paden, C.R.; Zhang, J.; et al. Severe Acute Respiratory Syndrome Coronavirus 2 from Patient with Coronavirus Disease, United States. Emerg. Infect. Dis. 2020, 26, 1266–1273. [Google Scholar] [CrossRef] [PubMed]
- Licastro, D.; Rajasekharan, S.; dal Monego, S.; Segat, L.; D’Agaro, P.; Marcello, A. Isolation and Full-Length Genome Characterization of SARS-CoV-2 from COVID-19 Cases in Northern Italy. J. Virol. 2020, 94, e00543-20. [Google Scholar] [CrossRef]
- Haveri, A.; Smura, T.; Kuivanen, S.; Osterlund, P.; Hepojoki, J.; Ikonen, N.; Pitkapaasi, M.; Blomqvist, S.; Ronkko, E.; Kantele, A.; et al. Serological and molecular findings during SARS-CoV-2 infection: The first case study in Finland, January to February 2020. Euro. Surveill. 2020, 25, 2000266. [Google Scholar] [CrossRef]
- Snijder, E.J.; Limpens, R.; de Wilde, A.H.; de Jong, A.W.M.; Zevenhoven-Dobbe, J.C.; Maier, H.J.; Faas, F.; Koster, A.J.; Barcena, M. A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis. PLoS Biol. 2020, 18, e3000715. [Google Scholar] [CrossRef]
- Ogando, N.S.; Dalebout, T.J.; Zevenhoven-Dobbe, J.C.; Limpens, R.W.; van der Meer, Y.; Caly, L.; Druce, J.; de Vries, J.J.C.; Kikkert, M.; Bárcena, M.; et al. SARS-coronavirus-2 replication in Vero E6 cells: Replication kinetics, rapid adaptation and cytopathology. J. Gen. Virol. 2020, 101, 925–940. [Google Scholar] [CrossRef] [PubMed]
- Maier, H.J.; Hawes, P.C.; Cottam, E.M.; Mantell, J.; Verkade, P.; Monaghan, P.; Wileman, T.; Britton, P. Infectious Bronchitis Virus Generates Spherules from Zippered Endoplasmic Reticulum Membranes. mBio 2013, 4, e00801-13. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, C.S.; Tatti, K.M.; Ksiazek, T.G.; Rollin, P.; Comer, J.A.; Lee, W.W.; Rota, P.A.; Bankamp, B.; Bellini, W.J.; Zaki, S.R. Ultrastructural Characterization of SARS Coronavirus. Emerg. Infect. Dis. 2004, 10, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Oshiro, L.S.; Schieble, J.H.; Lennette, E.H. Electron Microscopic Studies of Coronavirus. J. Gen. Virol. 1971, 12, 161–168. [Google Scholar] [CrossRef]
- Ulasli, M.; Verheije, M.H.; de Haan, C.A.; Reggiori, F. Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell. Microbiol. 2010, 12, 844–861. [Google Scholar] [CrossRef]
- Wolff, G.; Melia, C.E.; Snijder, E.J.; Bárcena, M. Double-Membrane Vesicles as Platforms for Viral Replication. Trends Microbiol. 2020, 28, 1022–1033. [Google Scholar] [CrossRef]
- Bouhaddou, M.; Memon, D.; Meyer, B.; White, K.M.; Rezelj, V.V.; Correa Marrero, M.; Polacco, B.J.; Melnyk, J.E.; Ulferts, S.; Kaake, R.M.; et al. The global phosphorylation landscape of SARS-CoV-2 infection. Cell 2021, 182, 685–712. [Google Scholar] [CrossRef]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef]
- Boni, M.F.; Lemey, P.; Jiang, X.; Lam, T.T.; Perry, B.W.; Castoe, T.A.; Rambaut, A.; Robertson, D.L. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat. Microbiol. 2020, 5, 1408–1417. [Google Scholar] [CrossRef]
- Harvey, W.T.; Carabelli, A.; Jackson, B.; Gupta, R.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- Sène, M.-A.; Kiesslich, S.; Djambazian, H.; Ragoussis, J.; Xia, Y.; Kamen, A.A. Haplotype-resolved de novo assembly of the Vero cell line genome. NPJ Vaccines 2021, 6, 1–10. [Google Scholar] [CrossRef]
- Geoghegan, J.L.; Ren, X.; Storey, M.; Hadfield, J.; Jelley, L.; Jefferies, S.; Sherwood, J.; Paine, S.; Huang, S.; Douglas, J.; et al. Genomic epidemiology reveals transmission patterns and dynamics of SARS-CoV-2 in Aotearoa New Zealand. Nat. Commun. 2020, 11, 6351. [Google Scholar] [CrossRef] [PubMed]
- Robson, F.; Khan, K.S.; Le, T.K.; Paris, C.; Demirbag, S.; Barfuss, P.; Rocchi, P.; Ng, W.L. Coronavirus RNA Proofreading: Molecular Basis and Therapeutic Targeting. Mol. Cell 2020, 79, 710–727. [Google Scholar] [CrossRef] [PubMed]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.J.; Chiba, S.; Halfmann, P.; Ehre, C.; Kuroda, M.; Dinnon, K.H., 3rd; Leist, S.R.; Schafer, A.; Nakajima, N.; Takahashi, K.; et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 2020, 370, 1464–1468. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Thao, T.T.N.; Hoffmann, D.; Taddeo, A.; Ebert, N.; Labroussaa, F.; Pohlmann, A.; King, J.; Steiner, S.; Kelly, J.N.; et al. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature 2021, 592, 122–127. [Google Scholar] [CrossRef] [PubMed]
- Malin, J.J.; Suárez, I.; Priesner, V.; Fätkenheuer, G.; Rybniker, J. Remdesivir against COVID-19 and Other Viral Diseases. Clin. Microbiol. Rev. 2020, 34, 1. [Google Scholar] [CrossRef]
- Fischer, W.A., 2nd; Eron, J.J., Jr.; Holman, W.; Cohen, M.S.; Fang, L.; Szewczyk, L.J.; Sheahan, T.P.; Baric, R.; Mollan, K.R.; Wolfe, C.R.; et al. Phase 2a clinical trial of Molnupiravir in patients with COVID-19 shows accelerated SARS-CoV-2 RNA clearance and elimination of infectious virus. Sci. Transl. Med. 2022, 14, eabl7430. [Google Scholar] [CrossRef] [PubMed]
- Yurkovetskiy, L.; Wang, X.; Pascal, K.E.; Tomkins-Tinch, C.; Nyalile, T.P.; Wang, Y.; Baum, A.; Diehl, W.E.; Dauphin, A.; Carbone, C.; et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell 2020, 183, 739–751.e8. [Google Scholar] [CrossRef]
- Weissman, D.; Alameh, M.-G.; de Silva, T.; Collini, P.; Hornsby, H.; Brown, R.; LaBranche, C.C.; Edwards, R.J.; Sutherland, L.; Santra, S.; et al. D614G Spike Mutation Increases SARS CoV-2 Susceptibility to Neutralization. Cell Host Microbe 2020, 29, 23–31.e4. [Google Scholar] [CrossRef]
- Zou, J.; Xie, X.; Fontes-Garfias, C.R.; Swanson, K.A.; Kanevsky, I.; Tompkins, K.; Cutler, M.; Cooper, D.; Dormitzer, P.R.; Shi, P.Y. The effect of SARS-CoV-2 D614G mutation on BNT162b2 vaccine-elicited neutralization. NPJ Vaccines 2021, 6, 44. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harfoot, R.; Lawley, B.; Hernández, L.C.; Kuang, J.; Grant, J.; Treece, J.M.; LeQueux, S.; Day, R.; Jack, S.; Stanton, J.-A.L.; et al. Characterization of the First SARS-CoV-2 Isolates from Aotearoa New Zealand as Part of a Rapid Response to the COVID-19 Pandemic. Viruses 2022, 14, 366. https://doi.org/10.3390/v14020366
Harfoot R, Lawley B, Hernández LC, Kuang J, Grant J, Treece JM, LeQueux S, Day R, Jack S, Stanton J-AL, et al. Characterization of the First SARS-CoV-2 Isolates from Aotearoa New Zealand as Part of a Rapid Response to the COVID-19 Pandemic. Viruses. 2022; 14(2):366. https://doi.org/10.3390/v14020366
Chicago/Turabian StyleHarfoot, Rhodri, Blair Lawley, Leonor C. Hernández, Joanna Kuang, Jenny Grant, Jackson M. Treece, Sharon LeQueux, Robert Day, Susan Jack, Jo-Ann L. Stanton, and et al. 2022. "Characterization of the First SARS-CoV-2 Isolates from Aotearoa New Zealand as Part of a Rapid Response to the COVID-19 Pandemic" Viruses 14, no. 2: 366. https://doi.org/10.3390/v14020366
APA StyleHarfoot, R., Lawley, B., Hernández, L. C., Kuang, J., Grant, J., Treece, J. M., LeQueux, S., Day, R., Jack, S., Stanton, J.-A. L., Bostina, M., Ussher, J. E., & Quiñones-Mateu, M. E. (2022). Characterization of the First SARS-CoV-2 Isolates from Aotearoa New Zealand as Part of a Rapid Response to the COVID-19 Pandemic. Viruses, 14(2), 366. https://doi.org/10.3390/v14020366






