Monocyte Gene and Molecular Expression Profiles Suggest Distinct Effector and Regulatory Functions in Beninese HIV Highly Exposed Seronegative Female Commercial Sex Workers
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Socio-Demographic Characteristics of the Study Groups
3.2. Transcriptomic Analyses by RNA-Seq of Total Sorted Blood Monocytes from HESNs Reveal Distinct Effector and Regulatory Capacities
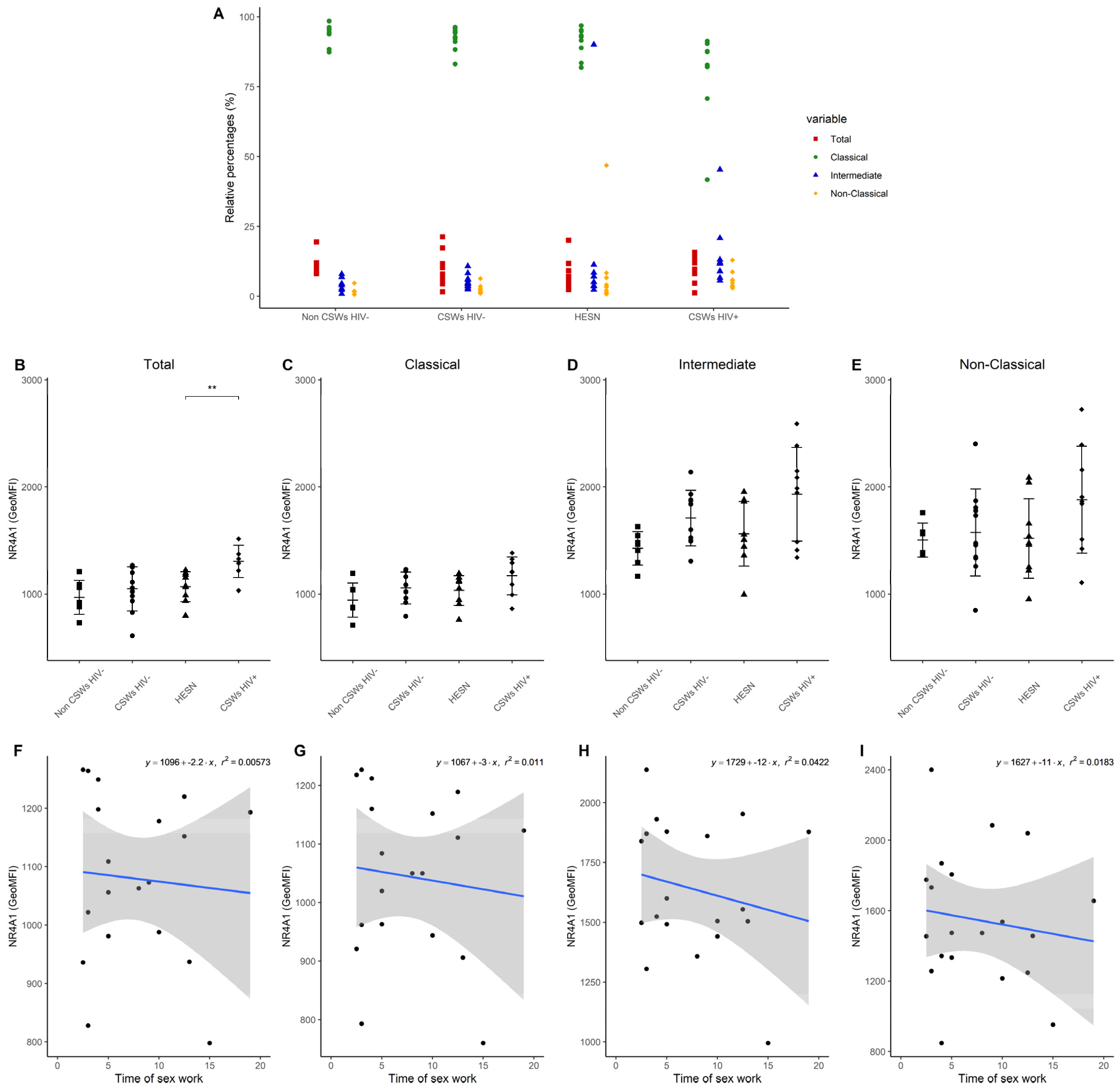
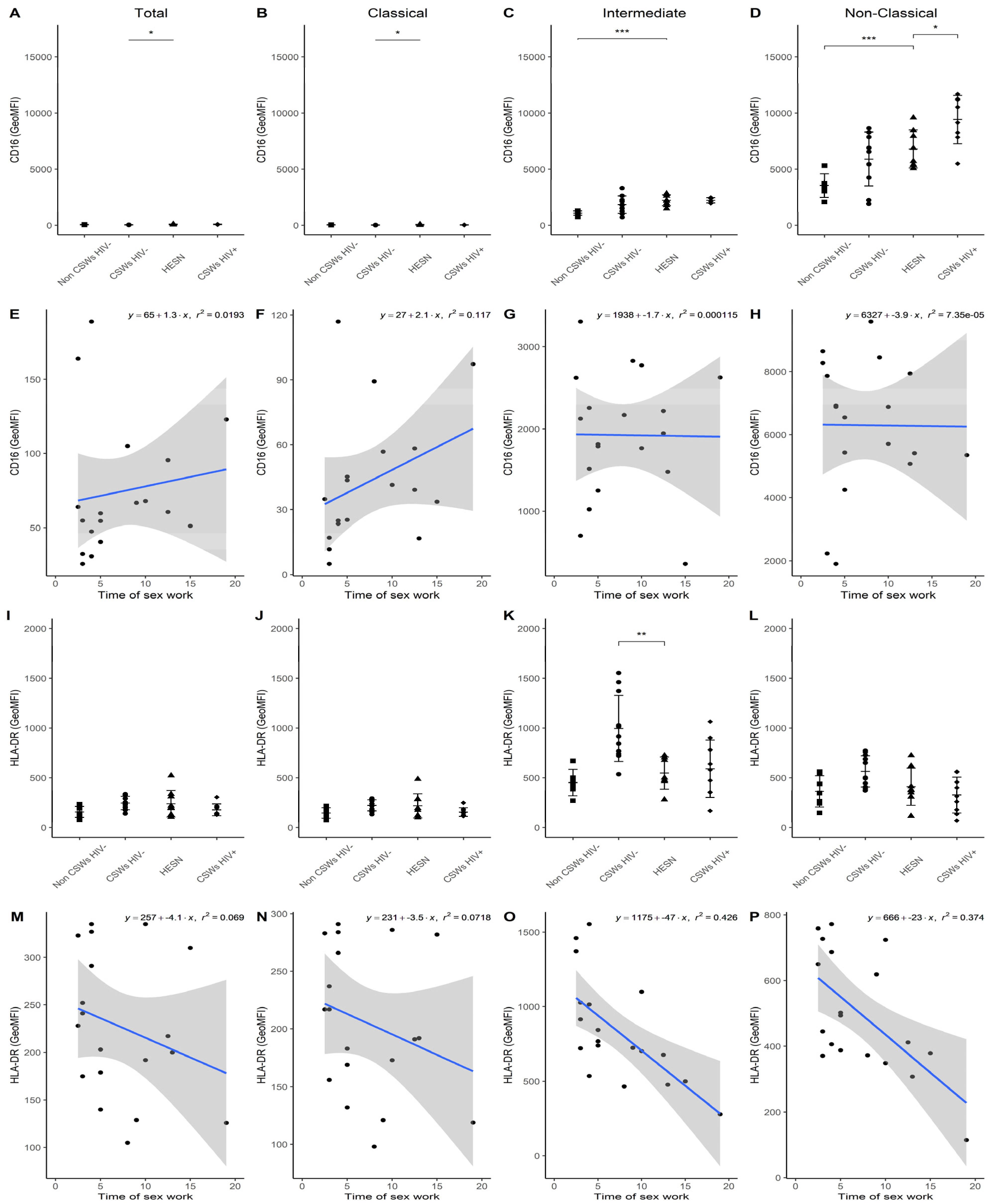
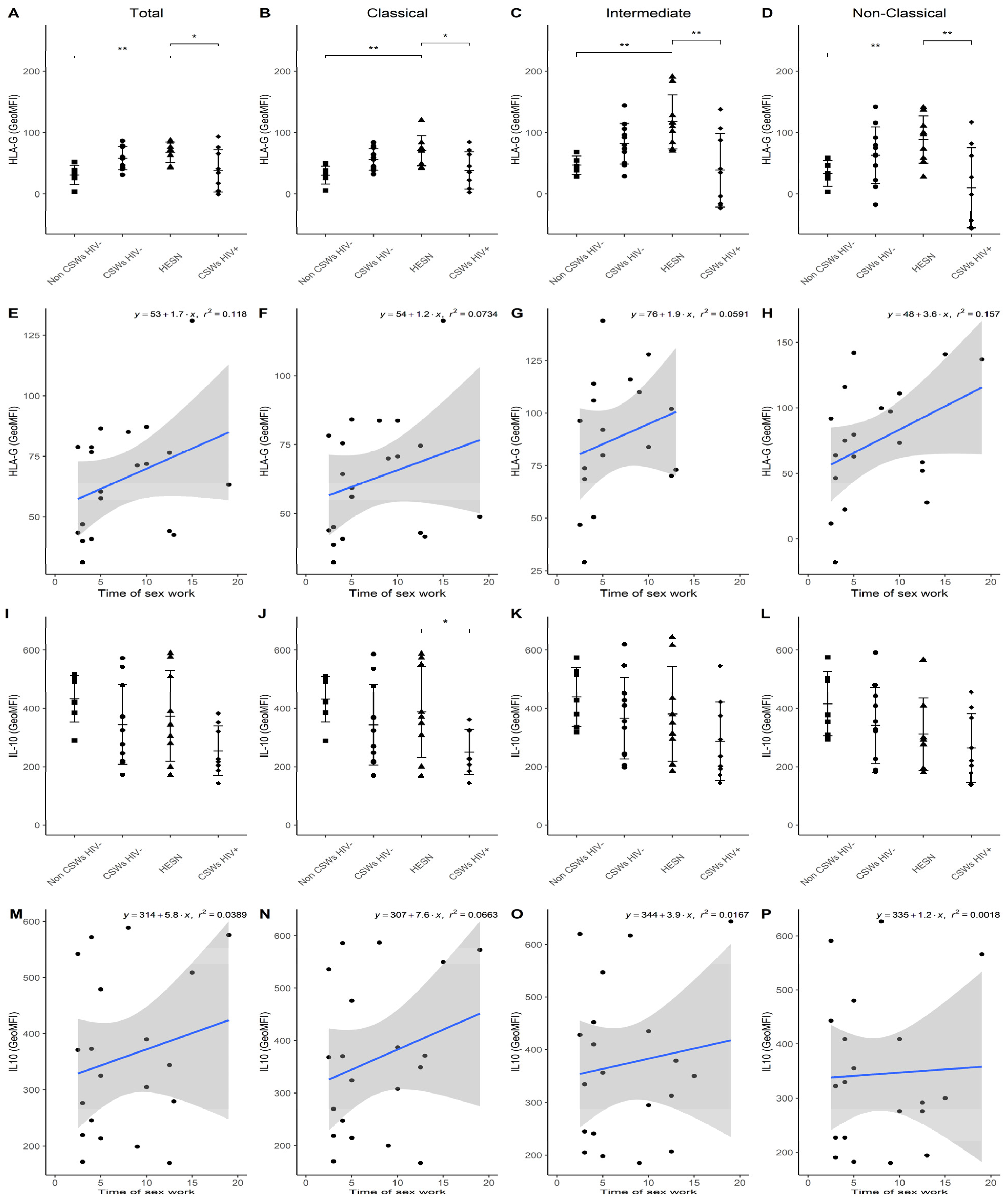
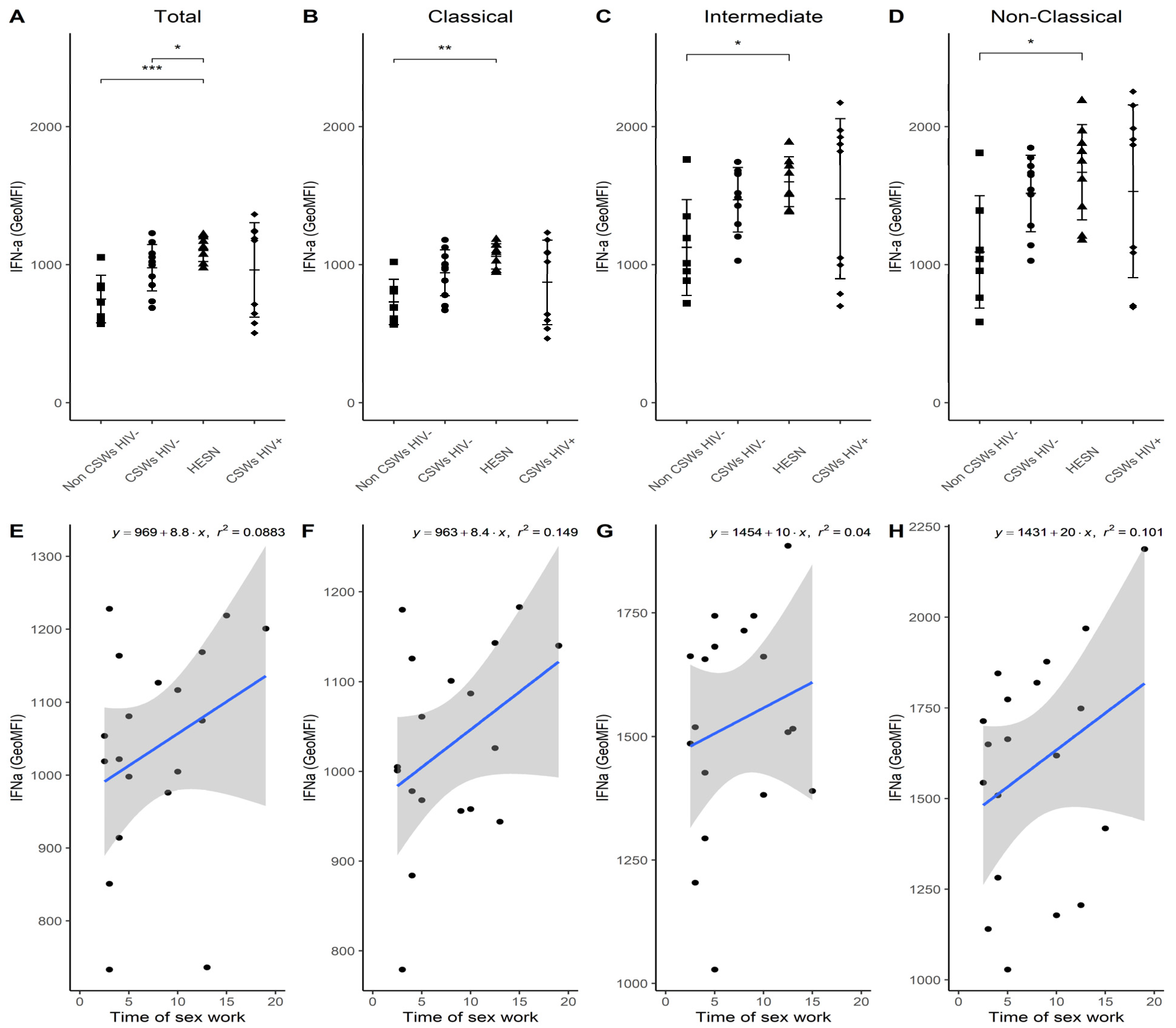
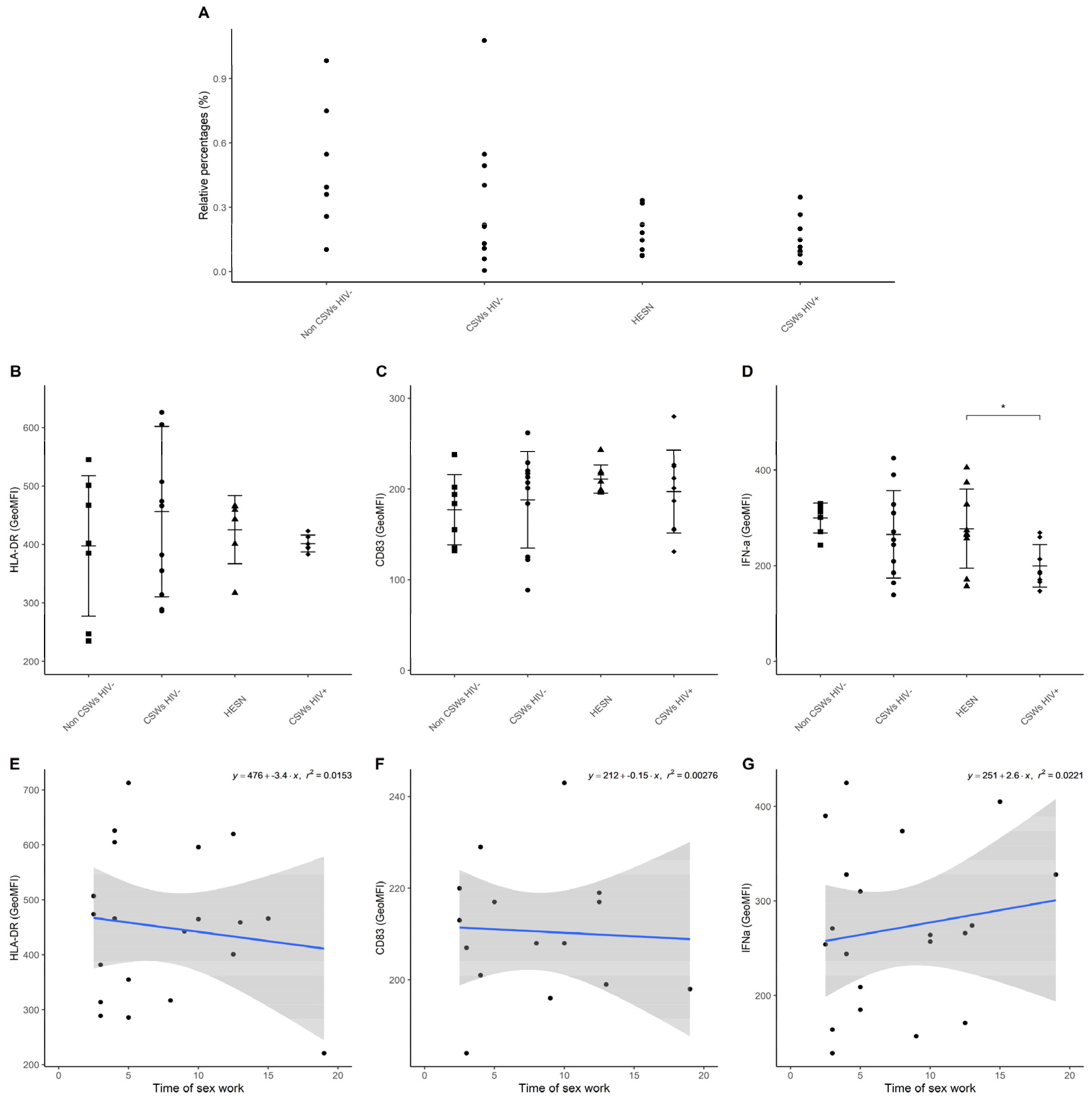
3.3. Multicolor Flow Cytometry Analyses Expose Important Effector and Regulatory Capacities in Blood Monocytes from HESNs
3.4. Flow Cytometry Analyses Suggest That Monocytes and Plasmacytoid DC (pDC) Antiviral Capacities Are Preserved in HESNs When Compared to HIV-Infected CSWs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- UNAIDS. Global HIV & AIDS Statistics 2020 Fact Sheet. Available online: www.unaids.org (accessed on 3 April 2021).
- Fourcade, L.; Poudrier, J.; Roger, M. Natural Immunity to HIV: A Template for Vaccine Strategies. Viruses 2018, 10, 215. [Google Scholar] [CrossRef] [PubMed]
- Lajoie, J.; Poudrier, J.; Massinga-Loembe, M.; Guédou, F.; Agossa-Gbenafa, C.; Labbé, A.C.; Alary, M.; Roger, M. Differences in immunoregulatory cytokine expression patterns in the systemic and genital tract compartments of HIV-1-infected commercial sex workers in Benin. Mucosal Immunol. 2008, 1, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Lajoie, J.; Poudrier, J.; Massinga Loembe, M.; Guédou, F.; Leblond, F.; Labbé, A.C.; Alary, M.; Roger, M. Chemokine expression patterns in the systemic and genital tract compartments are associated with HIV-1 infection in women from Benin. J. Clin. Immunol. 2010, 30, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Thibodeau, V.; Fourcade, L.; Labbé, A.C.; Alary, M.; Guédou, F.; Poudrier, J.; Roger, M. Highly-Exposed HIV-1 seronegative Female Commercial Sex Workers sustain in their genital mucosa increased frequencies of tolerogenic myeloid and regulatory T-cells. Sci. Rep. 2017, 7, 43857. [Google Scholar] [CrossRef]
- Attanasio, J.; Wherry, E.J. Costimulatory and Coinhibitory Receptor Pathways in Infectious Disease. Immunity 2016, 44, 1052–1068. [Google Scholar] [CrossRef]
- Khaitan, A.; Kravietz, A.; Mwamzuka, M.; Marshed, F.; Ilmet, T.; Said, S.; Ahmed, A.; Borkowsky, W.; Unutmaz, D. FOXP3+Helios+ Regulatory T Cells, Immune Activation, and Advancing Disease in HIV-Infected Children. J. Acquir. Immune Defic. Syndr. 2016, 72, 474–484. [Google Scholar] [CrossRef][Green Version]
- Card, C.M.; McLaren, P.J.; Wachihi, C.; Kimani, J.; Plummer, F.A.; Fowke, K.R. Decreased immune activation in resistance to HIV-1 infection is associated with an elevated frequency of CD4(+)CD25(+)FOXP3(+) regulatory T cells. J. Infect. Dis. 2009, 199, 1318–1322. [Google Scholar] [CrossRef]
- Amodio, G.; Gregori, S. Human tolerogenic DC-10: Perspectives for clinical applications. Transplant. Res. 2012, 1, 14. [Google Scholar] [CrossRef]
- Gregori, S.; Tomasoni, D.; Pacciani, V.; Scirpoli, M.; Battaglia, M.; Magnani, C.F.; Hauben, E.; Roncarolo, M.G. Differentiation of type 1 T regulatory cells (Tr1) by tolerogenic DC-10 requires the IL-10-dependent ILT4/HLA-G pathway. Blood 2010, 116, 935–944. [Google Scholar] [CrossRef]
- Levings, M.K.; Sangregorio, R.; Galbiati, F.; Squadrone, S.; de Waal Malefyt, R.; Roncarolo, M.G. IFN-alpha and IL-10 induce the differentiation of human type 1 T regulatory cells. J. Immunol. 2001, 166, 5530–5539. [Google Scholar] [CrossRef]
- Wacleche, V.S.; Tremblay, C.L.; Routy, J.P.; Ancuta, P. The Biology of Monocytes and Dendritic Cells: Contribution to HIV Pathogenesis. Viruses 2018, 10, 65. [Google Scholar] [CrossRef] [PubMed]
- Polletti, S.; Natoli, G. Understanding Spontaneous Conversion: The Case of the Ly6C(-) Monocyte. Immunity 2017, 46, 764–766. [Google Scholar] [CrossRef] [PubMed]
- Mildner, A.; Schönheit, J.; Giladi, A.; David, E.; Lara-Astiaso, D.; Lorenzo-Vivas, E.; Paul, F.; Chappell-Maor, L.; Priller, J.; Leutz, A.; et al. Genomic Characterization of Murine Monocytes Reveals C/EBPβ Transcription Factor Dependence of Ly6C(-) Cells. Immunity 2017, 46, 849–862. [Google Scholar] [CrossRef] [PubMed]
- Hanna, R.N.; Carlin, L.M.; Hubbeling, H.G.; Nackiewicz, D.; Green, A.M.; Punt, J.A.; Geissmann, F.; Hedrick, C.C. The transcription factor NR4A1 (Nur77) controls bone marrow differentiation and the survival of Ly6C- monocytes. Nat. Immunol. 2011, 12, 778–785. [Google Scholar] [CrossRef]
- Boulet, S.; Daudelin, J.F.; Odagiu, L.; Pelletier, A.N.; Yun, T.J.; Lesage, S.; Cheong, C.; Labrecque, N. The orphan nuclear receptor NR4A3 controls the differentiation of monocyte-derived dendritic cells following microbial stimulation. Proc. Natl. Acad. Sci. USA 2019, 116, 15150–15159. [Google Scholar] [CrossRef]
- Lajoie, J.; Kimani, M.; Plummer, F.A.; Nyamiobo, F.; Kaul, R.; Kimani, J.; Fowke, K.R. Association of sex work with reduced activation of the mucosal immune system. J. Infect. Dis. 2014, 210, 319–329. [Google Scholar] [CrossRef]
- Doyon-Laliberté, K.; Chagnon-Choquet, J.; Byrns, M.; Aranguren, M.; Memmi, M.; Chrobak, P.; Stagg, J.; Poudrier, J.; Roger, M. NR4A Expression by Human Marginal Zone B-Cells. Antibodies 2019, 8, 50. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2020. [Google Scholar]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.A.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Wickham, H. Reshaping Data with the reshape Package. J. Stat. Softw. Artic. 2007, 21, 1–20. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Horikoshi, M.; Tang, Y. ggfortify: Data Visualization Tools for Statistical Analysis Results. 2018. Available online: https://cran.r-project.org/web/packages/ggfortify/citation.html (accessed on 11 November 2021).
- Tang, Y.; Horikoshi, M.; Li, W. ggfortify: Unified Interface to Visualize Statistical Result of Popular R Packages. R J. 2016, 8, 474–485. [Google Scholar] [CrossRef]
- Auguie, B. gridExtra: Miscellaneous Functions for “Grid” Graphics. 2015. Available online: https://cran.rproject.org/web/packages/gridExtra/index.html (accessed on 11 November 2021).
- Kassambara, A. ggpubr: ‘ggplot2’ Based Publication Ready Plots. 2020. Available online: https://cran.r-project.org/web/packages/gridExtra/index.html (accessed on 11 November 2021).
- Wilke, C.; Fox, S.J.; Bates, T.; Manalo, K.; Lang, B.; Barrett, M.; Stoiber, M.; Philipp, A.; Denney, B.; Hesselberth, J.; et al. wilkelab/cowplot: 1.1.1. 2021. Available online: https://zenodo.org/record/4411966#.YgRw-ZYRU2x (accessed on 11 November 2021).
- Ehrenberg, P.K.; Shangguan, S.; Issac, B.; Alter, G.; Geretz, A.; Izumi, T.; Bryant, C.; Eller, M.A.; Wegmann, F.; Apps, R.; et al. A vaccine-induced gene expression signature correlates with protection against SIV and HIV in multiple trials. Sci. Transl. Med. 2019, 11, eaaw4236. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, S.; Ehrenberg, P.K.; Geretz, A.; Yum, L.; Kundu, G.; May, K.; Fourati, S.; Nganou-Makamdop, K.; Williams, L.D.; Sawant, S.; et al. Monocyte-derived transcriptome signature indicates antibody-dependent cellular phagocytosis as the primary mechanism of vaccine-induced protection against HIV-1. bioRxiv 2021. [Google Scholar] [CrossRef]
- Rerks-Ngarm, S.; Pitisuttithum, P.; Nitayaphan, S.; Kaewkungwal, J.; Chiu, J.; Paris, R.; Premsri, N.; Namwat, C.; de Souza, M.; Adams, E.; et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 2009, 361, 2209–2220. [Google Scholar] [CrossRef] [PubMed]
- Gray, G.E.; Huang, Y.; Grunenberg, N.; Laher, F.; Roux, S.; Andersen-Nissen, E.; De Rosa, S.C.; Flach, B.; Randhawa, A.K.; Jensen, R.; et al. Immune correlates of the Thai RV144 HIV vaccine regimen in South Africa. Sci. Transl. Med. 2019, 11. [Google Scholar] [CrossRef]
- Tay, M.Z.; Wiehe, K.; Pollara, J. Antibody-Dependent Cellular Phagocytosis in Antiviral Immune Responses. Front. Immunol. 2019, 10, 332. [Google Scholar] [CrossRef] [PubMed]
- Li, S.S.; Gilbert, P.B.; Tomaras, G.D.; Kijak, G.; Ferrari, G.; Thomas, R.; Pyo, C.W.; Zolla-Pazner, S.; Montefiori, D.; Liao, H.X.; et al. FCGR2C polymorphisms associate with HIV-1 vaccine protection in RV144 trial. J. Clin. Investig. 2014, 124, 3879–3890. [Google Scholar] [CrossRef]
- Lassaunière, R.; Paximadis, M.; Ebrahim, O.; Chaisson, R.E.; Martinson, N.A.; Tiemessen, C.T. The FCGR2C allele that modulated the risk of HIV-1 infection in the Thai RV144 vaccine trial is implicated in HIV-1 disease progression. Genes Immun. 2019, 20, 651–659. [Google Scholar] [CrossRef]
- Zhao, N.Q.; Vendrame, E.; Ferreira, A.M.; Seiler, C.; Ranganath, T.; Alary, M.; Labbé, A.C.; Guédou, F.; Poudrier, J.; Holmes, S.; et al. Natural killer cell phenotype is altered in HIV-exposed seronegative women. PLoS ONE 2020, 15, e0238347. [Google Scholar] [CrossRef]
- Abel, A.M.; Yang, C.; Thakar, M.S.; Malarkannan, S. Natural Killer Cells: Development, Maturation, and Clinical Utilization. Front. Immunol. 2018, 9, 1869. [Google Scholar] [CrossRef]
- Yeap, W.H.; Wong, K.L.; Shimasaki, N.; Teo, E.C.; Quek, J.K.; Yong, H.X.; Diong, C.P.; Bertoletti, A.; Linn, Y.C.; Wong, S.C. CD16 is indispensable for antibody-dependent cellular cytotoxicity by human monocytes. Sci. Rep. 2016, 6, 34310. [Google Scholar] [CrossRef]
- Haynes, B.F.; Gilbert, P.B.; McElrath, M.J.; Zolla-Pazner, S.; Tomaras, G.D.; Alam, S.M.; Evans, D.T.; Montefiori, D.C.; Karnasuta, C.; Sutthent, R.; et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 2012, 366, 1275–1286. [Google Scholar] [CrossRef]
- Kim, J.H.; Excler, J.L.; Michael, N.L. Lessons from the RV144 Thai phase III HIV-1 vaccine trial and the search for correlates of protection. Annu. Rev. Med. 2015, 66, 423–437. [Google Scholar] [CrossRef] [PubMed]
- Batraville, L.A.; Richard, J.; Veillette, M.; Labbé, A.C.; Alary, M.; Guédou, F.; Kaufmann, D.E.; Poudrier, J.; Finzi, A.; Roger, M. Short communication: Anti-HIV-1 envelope immunoglobulin Gs in blood and cervicovaginal samples of Beninese commercial sex workers. AIDS Res. Hum. Retrovir. 2014, 30, 1145–1149. [Google Scholar] [CrossRef] [PubMed]
- Fourcade, L.; Sabourin-Poirier, C.; Perraud, V.; Faucher, M.C.; Chagnon-Choquet, J.; Labbé, A.C.; Alary, M.; Guédou, F.; Poudrier, J.; Roger, M. Natural Immunity to HIV is associated with Low BLyS/BAFF levels and low frequencies of innate marginal zone like CD1c+ B-cells in the genital tract. PLoS Pathog. 2019, 15, e1007840. [Google Scholar] [CrossRef] [PubMed]
- Williams, W.B.; Liao, H.X.; Moody, M.A.; Kepler, T.B.; Alam, S.M.; Gao, F.; Wiehe, K.; Trama, A.M.; Jones, K.; Zhang, R.; et al. HIV-1 VACCINES. Diversion of HIV-1 vaccine-induced immunity by gp41-microbiota cross-reactive antibodies. Science 2015, 349, aab1253. [Google Scholar] [CrossRef]
- Mayr, L.M.; Su, B.; Moog, C. Non-Neutralizing Antibodies Directed against HIV and Their Functions. Front. Immunol. 2017, 8, 1590. [Google Scholar] [CrossRef] [PubMed]
- Tomaras, G.D.; Yates, N.L.; Liu, P.; Qin, L.; Fouda, G.G.; Chavez, L.L.; Decamp, A.C.; Parks, R.J.; Ashley, V.C.; Lucas, J.T.; et al. Initial B-cell responses to transmitted human immunodeficiency virus type 1: Virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J. Virol. 2008, 82, 12449–12463. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, N.K.; Ribeiro, R.M.; Liu, P.; Haynes, B.F.; Tomaras, G.D.; Perelson, A.S. Correlation Between Anti-gp41 Antibodies and Virus Infectivity Decay During Primary HIV-1 Infection. Front. Microbiol. 2018, 9, 1326. [Google Scholar] [CrossRef] [PubMed]
- Wills, S.; Hwang, K.K.; Liu, P.; Dennison, S.M.; Tay, M.Z.; Shen, X.; Pollara, J.; Lucas, J.T.; Parks, R.; Rerks-Ngarm, S.; et al. HIV-1-Specific IgA Monoclonal Antibodies from an HIV-1 Vaccinee Mediate Galactosylceramide Blocking and Phagocytosis. J. Virol. 2018, 92, e01552-17. [Google Scholar] [CrossRef]
- Duchemin, M.; Khamassi, M.; Xu, L.; Tudor, D.; Bomsel, M. IgA Targeting Human Immunodeficiency Virus-1 Envelope gp41 Triggers Antibody-Dependent Cellular Cytotoxicity Cross-Clade and Cooperates with gp41-Specific IgG to Increase Cell Lysis. Front. Immunol. 2018, 9, 244. [Google Scholar] [CrossRef] [PubMed]
- Duchemin, M.; Tudor, D.; Cottignies-Calamarte, A.; Bomsel, M. Antibody-Dependent Cellular Phagocytosis of HIV-1-Infected Cells Is Efficiently Triggered by IgA Targeting HIV-1 Envelope Subunit gp41. Front. Immunol. 2020, 11, 1141. [Google Scholar] [CrossRef] [PubMed]
- Gunn, B.; Schneider, J.; Shansab, M.; Bastian, A.R.; Fahrbach, K.; Smith, A.t.; Mahan, A.; Karim, M.; Licht, A.; Zvonar, I.; et al. Enhanced binding of antibodies generated during chronic HIV infection to mucus component MUC16. Mucosal Immunol. 2016, 9, 1549–1558. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, X.; Han, Y.; Qiu, Z.; Song, X.; Li, B.; Zhang, H.; Wang, H.; Feng, K.; Liu, L.; et al. High APRIL Levels Are Associated With Slow Disease Progression and Low Immune Activation in Chronic HIV-1-Infected Patients. Front. Med. 2020, 7, 299. [Google Scholar] [CrossRef]
- Huynh, J.P.; Lin, C.C.; Kimmey, J.M.; Jarjour, N.N.; Schwarzkopf, E.A.; Bradstreet, T.R.; Shchukina, I.; Shpynov, O.; Weaver, C.T.; Taneja, R.; et al. Bhlhe40 is an essential repressor of IL-10 during Mycobacterium tuberculosis infection. J. Exp. Med. 2018, 215, 1823–1838. [Google Scholar] [CrossRef]
- Zhu, J.; Luo, L.; Tian, L.; Yin, S.; Ma, X.; Cheng, S.; Tang, W.; Yu, J.; Ma, W.; Zhou, X.; et al. Aryl Hydrocarbon Receptor Promotes IL-10 Expression in Inflammatory Macrophages Through Src-STAT3 Signaling Pathway. Front. Immunol. 2018, 9, 2033. [Google Scholar] [CrossRef] [PubMed]
- Bandukwala, H.S.; Rao, A. 'Nurr'ishing Treg cells: Nr4a transcription factors control Foxp3 expression. Nat. Immunol. 2013, 14, 201–203. [Google Scholar] [CrossRef]
- Fujimoto, Y.; Tedder, T.F. CD83: A regulatory molecule of the immune system with great potential for therapeutic application. J. Med. Dent. Sci. 2006, 53, 85–91. [Google Scholar]
- Duren, R.P.; Boudreaux, S.P.; Conneely, O.M. Genome Wide Mapping of NR4A Binding Reveals Cooperativity with ETS Factors to Promote Epigenetic Activation of Distal Enhancers in Acute Myeloid Leukemia Cells. PLoS ONE 2016, 11, e0150450. [Google Scholar] [CrossRef]
- Chen, L.; Fan, F.; Wu, L.; Zhao, Y. The nuclear receptor 4A family members: Mediators in human disease and autophagy. Cell. Mol. Biol. Lett. 2020, 25, 48. [Google Scholar] [CrossRef]
- Boudreaux, S.P.; Duren, R.P.; Call, S.G.; Nguyen, L.; Freire, P.R.; Narayanan, P.; Redell, M.S.; Conneely, O.M. Drug targeting of NR4A nuclear receptors for treatment of acute myeloid leukemia. Leukemia 2019, 33, 52–63. [Google Scholar] [CrossRef]
- Tel-Karthaus, N.; Kers-Rebel, E.D.; Looman, M.W.; Ichinose, H.; de Vries, C.J.; Ansems, M. Nuclear Receptor Nur77 Deficiency Alters Dendritic Cell Function. Front. Immunol. 2018, 9, 1797. [Google Scholar] [CrossRef]
- Safe, S.; Jin, U.H.; Morpurgo, B.; Abudayyeh, A.; Singh, M.; Tjalkens, R.B. Nuclear receptor 4A (NR4A) family-orphans no more. J. Steroid Biochem. Mol. Biol. 2016, 157, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Dobbs, K.R.; Crabtree, J.N.; Dent, A.E. Innate immunity to malaria-The role of monocytes. Immunol. Rev. 2020, 293, 8–24. [Google Scholar] [CrossRef]
- Kapellos, T.S.; Bonaguro, L.; Gemünd, I.; Reusch, N.; Saglam, A.; Hinkley, E.R.; Schultze, J.L. Human Monocyte Subsets and Phenotypes in Major Chronic Inflammatory Diseases. Front. Immunol. 2019, 10, 2035. [Google Scholar] [CrossRef]
- Carlin, L.M.; Stamatiades, E.G.; Auffray, C.; Hanna, R.N.; Glover, L.; Vizcay-Barrena, G.; Hedrick, C.C.; Cook, H.T.; Diebold, S.; Geissmann, F. Nr4a1-dependent Ly6C(low) monocytes monitor endothelial cells and orchestrate their disposal. Cell 2013, 153, 362–375. [Google Scholar] [CrossRef]
- Thomas, G.; Tacke, R.; Hedrick, C.C.; Hanna, R.N. Nonclassical patrolling monocyte function in the vasculature. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 1306–1316. [Google Scholar] [CrossRef]
- Lajoie, J.; Massinga Loembe, M.; Poudrier, J.; Guédou, F.; Pépin, J.; Labbé, A.C.; Alary, M.; Roger, M. Blood soluble human leukocyte antigen G levels are associated with human immunodeficiency virus type 1 infection in Beninese commercial sex workers. Hum. Immunol. 2010, 71, 182–185. [Google Scholar] [CrossRef] [PubMed]
- Boyette, L.B.; Macedo, C.; Hadi, K.; Elinoff, B.D.; Walters, J.T.; Ramaswami, B.; Chalasani, G.; Taboas, J.M.; Lakkis, F.G.; Metes, D.M. Phenotype, function, and differentiation potential of human monocyte subsets. PLoS ONE 2017, 12, e0176460. [Google Scholar] [CrossRef]
- Wacleche, V.S.; Cattin, A.; Goulet, J.P.; Gauchat, D.; Gosselin, A.; Cleret-Buhot, A.; Zhang, Y.; Tremblay, C.L.; Routy, J.P.; Ancuta, P. CD16(+) monocytes give rise to CD103(+)RALDH2(+)TCF4(+) dendritic cells with unique transcriptional and immunological features. Blood Adv. 2018, 2, 2862–2878. [Google Scholar] [CrossRef] [PubMed]
- Randolph, G.J.; Sanchez-Schmitz, G.; Liebman, R.M.; Schäkel, K. The CD16(+) (FcgammaRIII(+)) subset of human monocytes preferentially becomes migratory dendritic cells in a model tissue setting. J. Exp. Med. 2002, 196, 517–527. [Google Scholar] [CrossRef]


| Non CSWs HIV- | CSWs HIV- | HESN | CSWs HIV+ | ||
|---|---|---|---|---|---|
| N = 10 | N = 11 | N = 10 | N = 12 | * p-value | |
| Age, mean (SD), years | 34 (6.5) | 34 (7.6) | 44 (7.9) | 45 (9) | NS |
| Duration of sex work (SD), years | N/A | 4 (0.9) | 12 (3.7) | 9 (4.5) | 0.001337 |
| Number of clients past week, mean (SD) | N/A | 15 (14.5) | 12 (14.4) | 18 (22.5) | NS |
| Condom always used with clients past week | N/A | 7 | 8 | 9 | NS |
| Vaginal douching | 10 | 11 | 10 | 12 | NS |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blondin-Ladrie, L.; Fourcade, L.; Modica, A.; Aranguren, M.; de Montigny, N.; Labbé, A.-C.; Alary, M.; Guédou, F.; Poudrier, J.; Roger, M. Monocyte Gene and Molecular Expression Profiles Suggest Distinct Effector and Regulatory Functions in Beninese HIV Highly Exposed Seronegative Female Commercial Sex Workers. Viruses 2022, 14, 361. https://doi.org/10.3390/v14020361
Blondin-Ladrie L, Fourcade L, Modica A, Aranguren M, de Montigny N, Labbé A-C, Alary M, Guédou F, Poudrier J, Roger M. Monocyte Gene and Molecular Expression Profiles Suggest Distinct Effector and Regulatory Functions in Beninese HIV Highly Exposed Seronegative Female Commercial Sex Workers. Viruses. 2022; 14(2):361. https://doi.org/10.3390/v14020361
Chicago/Turabian StyleBlondin-Ladrie, Laurence, Lyvia Fourcade, Alessandro Modica, Matheus Aranguren, Nicolas de Montigny, Annie-Claude Labbé, Michel Alary, Fernand Guédou, Johanne Poudrier, and Michel Roger. 2022. "Monocyte Gene and Molecular Expression Profiles Suggest Distinct Effector and Regulatory Functions in Beninese HIV Highly Exposed Seronegative Female Commercial Sex Workers" Viruses 14, no. 2: 361. https://doi.org/10.3390/v14020361
APA StyleBlondin-Ladrie, L., Fourcade, L., Modica, A., Aranguren, M., de Montigny, N., Labbé, A.-C., Alary, M., Guédou, F., Poudrier, J., & Roger, M. (2022). Monocyte Gene and Molecular Expression Profiles Suggest Distinct Effector and Regulatory Functions in Beninese HIV Highly Exposed Seronegative Female Commercial Sex Workers. Viruses, 14(2), 361. https://doi.org/10.3390/v14020361






