Characterization of a Broadly Neutralizing Monoclonal Antibody against SARS-CoV-2 Variants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Line and Virus
2.2. Production of mAbs
2.3. Immunofluorescence Assay (IFA)
2.4. Authentic Virus Microneutralization (VMN) Activity of mAbs against SARS-CoV-2
2.5. SARS-CoV-2 Surrogate Virus Neutralization Test (sVNT)
2.6. Mapping of Neutralizing Epitopes in the Spike RBD by Escape Mutant Analysis Using 9G8 mAb
2.7. Epitope Distribution Analysis
3. Results
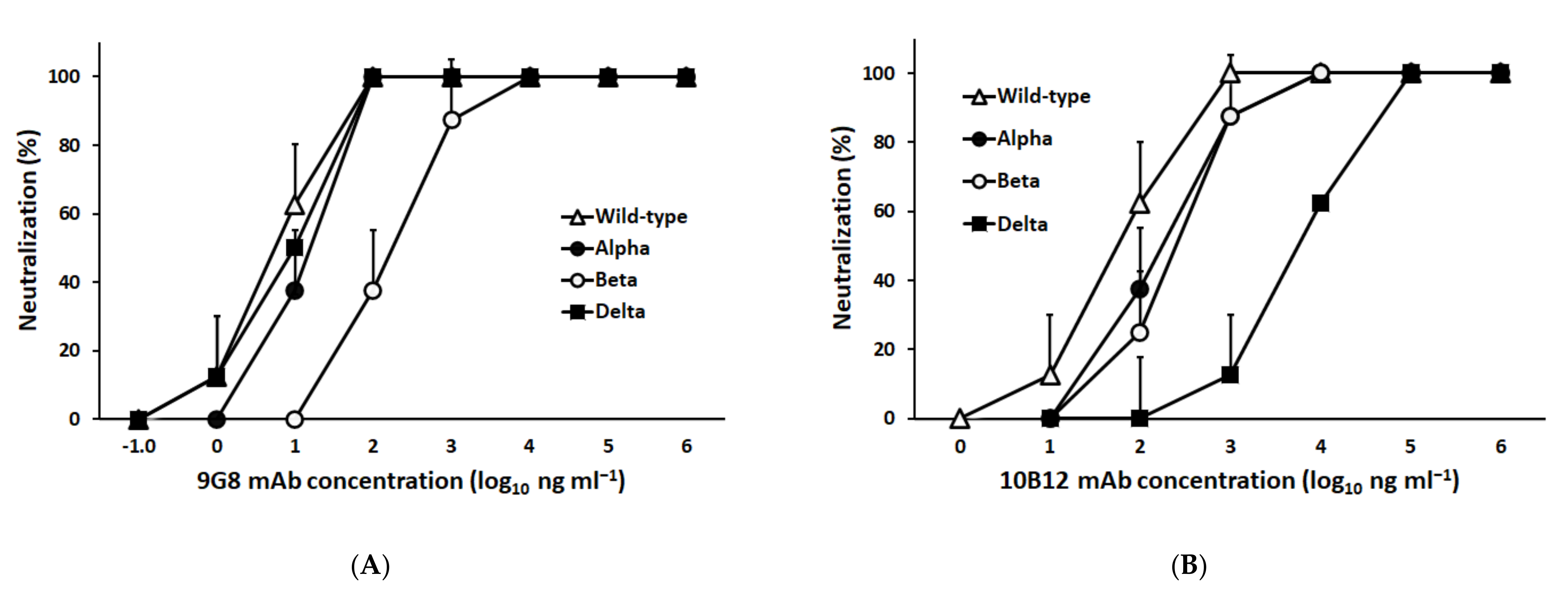
3.1. Characterization of a Broadly Neutralizing Monoclonal Antibody against Spike-RBD
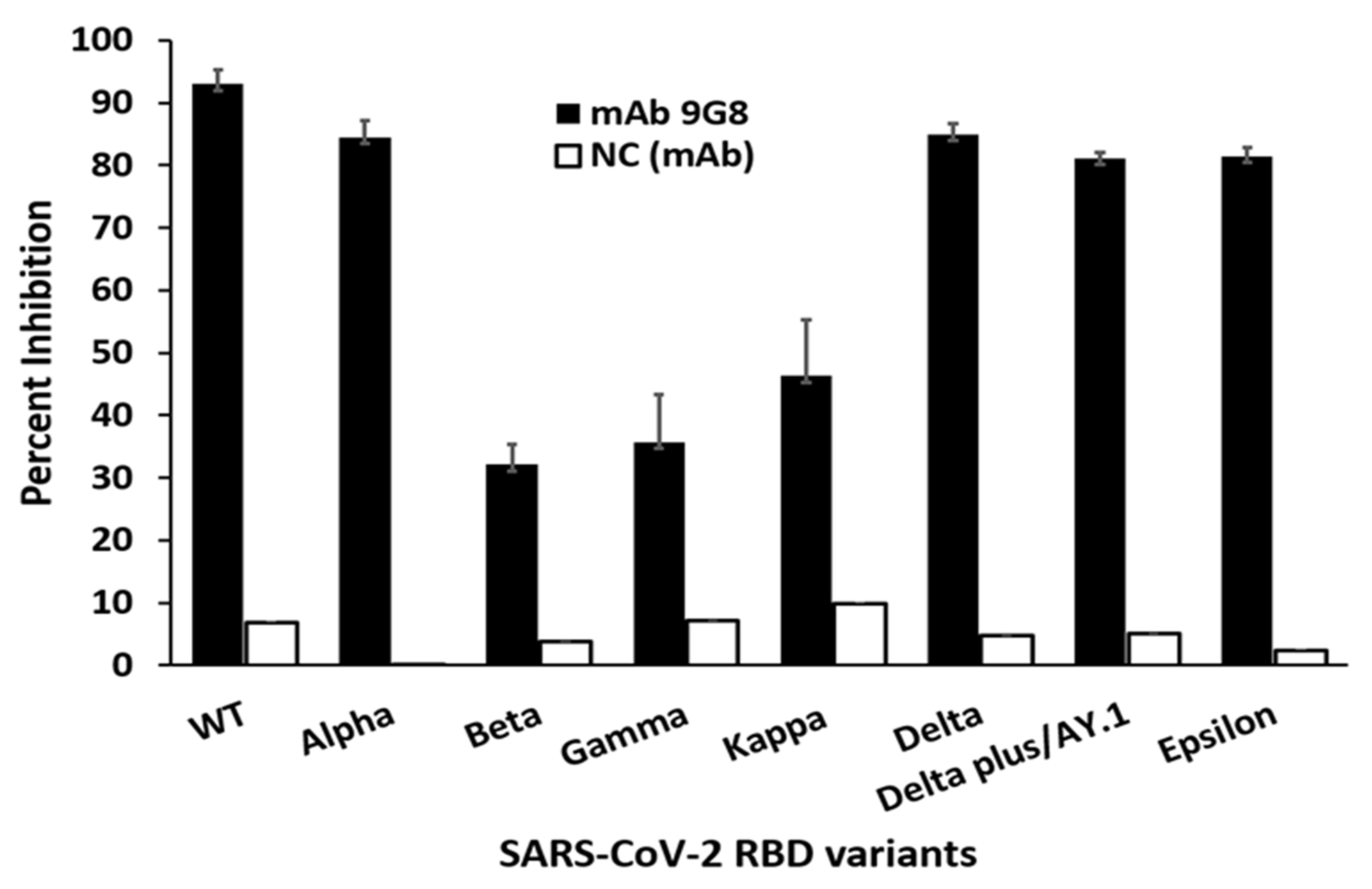
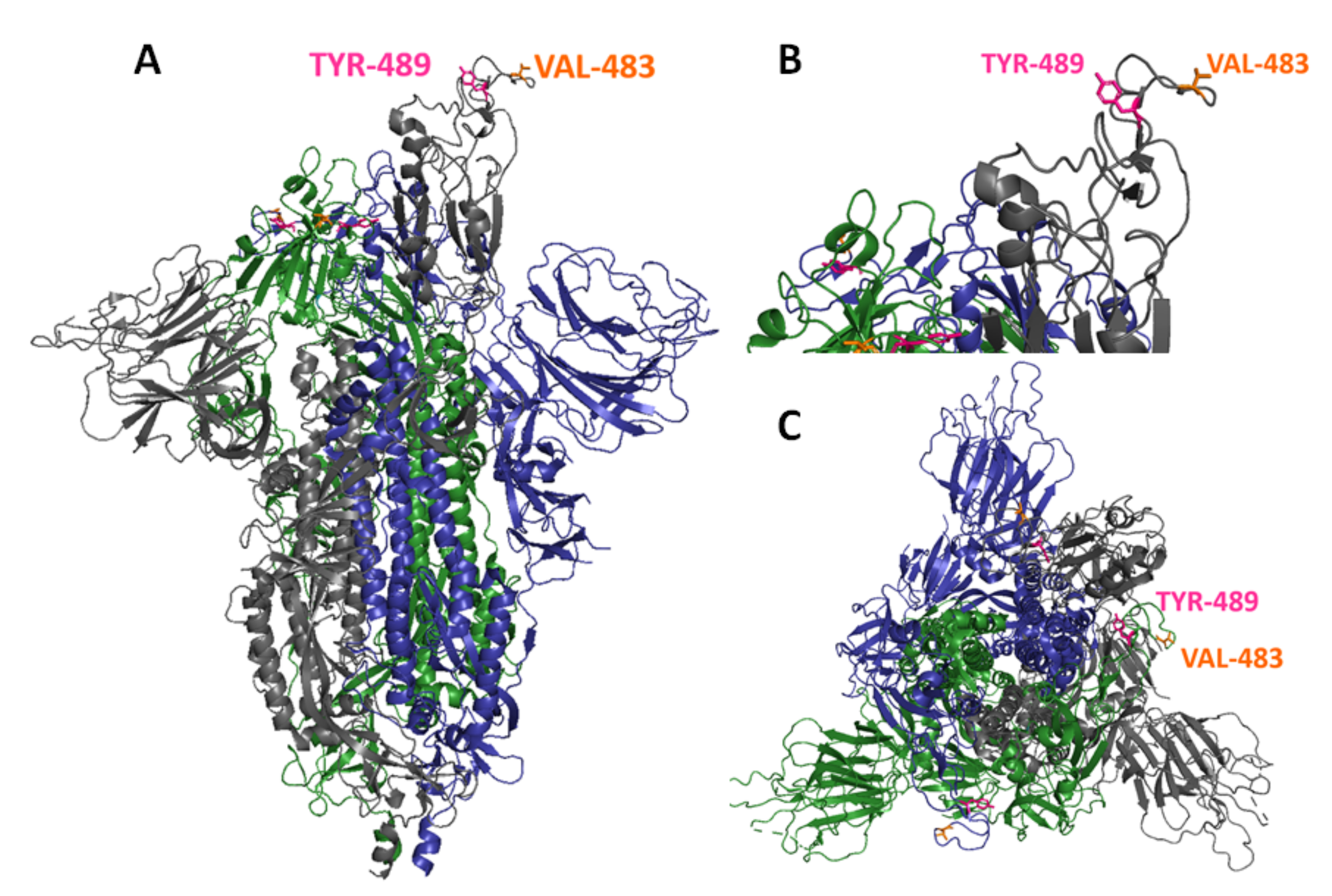
3.2. Generation of In Vitro Escape Mutants against mAb 9G8
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Hoffmann, M.; Arora, P.; Groß, R.; Seidel, A.; Hörnich, B.F.; Hahn, A.S.; Krüger, N.; Graichen, L.; Hofmann-Winkler, H.; Kempf, A.; et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell 2021, 184, 2384–2393. [Google Scholar] [CrossRef] [PubMed]
- Wibmer, C.K.; Ayres, F.; Hermanus, T.; Madzivhandila, M.; Kgagudi, P.; Oosthuysen, B.; Lambson, B.E.; de Oliveira, T.; Vermeulen, M.; van der Berg, K.; et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021, 27, 622–625. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Nair, M.S.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D.; et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 2021, 593, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.E.; Zhang, X.; Case, J.B.; Winkler, E.S.; Liu, Y.; VanBlargan, L.A.; Liu, J.; Errico, J.M.; Xie, X.; Suryadevara, N.; et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021, 27, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Planas, D.; Veyer, D.; Baidaliuk, A.; Staropoli, I.; Guivel-Benhassine, F.; Rajah, M.M.; Planchais, C.; Porrot, F.; Robillard, N.; Puech, J.; et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 2021, 596, 276–280. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Wang, H.; Chen, L.; Fang, Q.; Zhang, B.; Jiang, L.; Wu, Z.; Yang, Y.; Zhou, Y.; Chen, B.; et al. Non-RBM Mutations Impaired SARS-CoV-2 Spike Protein Regulated to the ACE2 Receptor Based on Molecular Dynamic Simulation. Front. Mol. Biosci. 2021, 8, 614443. [Google Scholar] [CrossRef] [PubMed]
- Plante, J.A.; Mitchell, B.M.; Plante, K.S.; Debbink, K.; Weaver, S.C.; Menachery, V.D. The variant gambit: COVID-19’s next move. Cell Host Microbe 2021, 29, 508–515. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, W.M.; Christensen, M.; Santos, G.D.; Miller, D. Production of monoclonal antibodies. Curr. Protoc. Immunol. 2013, 102, 2.5.1–2.5.29. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.W.; Chia, W.N.; Young, B.E.; Zhu, F.; Lim, B.L.; Sia, W.R.; Thein, T.L.; Chen, M.I.; Leo, Y.S.; Lye, D.C.; et al. Pan-Sarbecovirus Neutralizing Antibodies in BNT162b2-Immunized SARS-CoV-1 Survivors. N. Engl. J. Med. 2021, 385, 1401–1406. [Google Scholar] [CrossRef] [PubMed]
- Kaverin, N.V.; Rudneva, I.A.; Ilyushina, N.A.; Varich, N.L.; Lipatov, A.S.; Smirnov, Y.A.; Govorkova, E.A.; Gitelman, A.K.; Lvov, D.K.; Webster, R.G. Structure of antigenic sites on the haemagglutinin molecule of H5 avian influenza virus and phenotypic variation of escape mutants. J. Gen. Virol. 2002, 83, 2497–2505. [Google Scholar] [CrossRef] [PubMed]
- Callaway, E. Heavily mutated Omicron variant puts scientists on alert. Nature 2021, 600, 10. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, A.; Kerr, S.; Woolhouse, M.; McMenamin, J.; Robertson, C. Severity of Omicron Variant of Concern and Vaccine Effectiveness against Symptomatic Disease: National Cohort with Nested Test Negative Design Study in Scotland. Edinb. Res. Explor. 2021. Available online: https://www.pure.ed.ac.uk/ws/files/245818096/Severity_of_Omicron_variant_of_concern_and_vaccine_effectiveness_against_symptomatic_disease.pdf (accessed on 28 November 2021).
- Koehler, M.; Ray, A.; Moreira, R.A.; Juniku, B.; Poma, A.B.; Alsteens, D. Molecular insights into receptor binding energetics and neutralization of SARS-CoV-2 variants. Nat. Commun. 2021, 12, 6977. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.; Woo, H.G. Omicron: A heavily mutated SARS-CoV-2 variant exhibits stronger binding to ACE2 and potently escape approved COVID-19 therapeutic antibodies. bioRxiv 2021. [Google Scholar] [CrossRef]
- Ashwaq, O.; Manickavasagam, P.; Haque, S.M. V483A: An emerging mutation hotspot of SARS-CoV-2. Future Virol. 2021, 16, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.; Pathirana, P.N.; Nguyen, T.; Nguyen, Q.V.H.; Bhatti, A.; Nguyen, D.C.; Nguyen, D.T.; Nguyen, N.D.; Creighton, D.; Abdelrazek, M. Genomic mutations and changes in protein secondary structure and solvent accessibility of SARS-CoV-2 (COVID-19 virus). Sci. Rep. 2021, 11, 3487. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wu, J.; Nie, J.; Zhang, L.; Hao, H.; Liu, S.; Zhao, C.; Zhang, Q.; Liu, H.; Nie, L.; et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell 2020, 182, 1284–1294. [Google Scholar] [CrossRef] [PubMed]
- Adams, L.E.; Dinnon, K.H., 3rd; Hou, Y.J.; Sheahan, T.P.; Heise, M.T.; Baric, R.S. Critical ACE2 Determinants of SARS-CoV-2 and Group 2B Coronavirus Infection and Replication. mBio 2021, 12, e03149-20. [Google Scholar] [CrossRef] [PubMed]
- Tsai, K.C.; Lee, Y.C.; Tseng, T.S. Comprehensive Deep Mutational Scanning Reveals the Immune-Escaping Hotspots of SARS-CoV-2 Receptor-Binding Domain Targeting Neutralizing Antibodies. Front. Microbiol. 2021, 12, 698365. [Google Scholar] [CrossRef] [PubMed]
| SARS-CoV-2 Variant | Lineage | RBD Mutations |
|---|---|---|
| L | Wuhan-Hu-1 | N/A |
| Alpha | B.1.1.7 | N501Y |
| Beta | B.1.351 | K417N, E484K, N501Y |
| Gamma | P.1 | K417T, E484K, N501Y |
| Kappa | B.1.617.1 | L452R, E484Q |
| Delta | B.1.617.2 | L452R, T478K |
| Delta Plus/AY.1 | B.1.617.2.1 | K417T, L452R, T478K |
| Epsilon | B.1.429 | L452R |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zakir, T.S.; Meng, T.; Carmen, L.C.P.; Chu, J.J.H.; Lin, R.T.P.; Prabakaran, M. Characterization of a Broadly Neutralizing Monoclonal Antibody against SARS-CoV-2 Variants. Viruses 2022, 14, 230. https://doi.org/10.3390/v14020230
Zakir TS, Meng T, Carmen LCP, Chu JJH, Lin RTP, Prabakaran M. Characterization of a Broadly Neutralizing Monoclonal Antibody against SARS-CoV-2 Variants. Viruses. 2022; 14(2):230. https://doi.org/10.3390/v14020230
Chicago/Turabian StyleZakir, Tasnim Saifudin, Tao Meng, Lee Ching Pei Carmen, Justin Jang Hann Chu, Raymond Tzer Pin Lin, and Mookkan Prabakaran. 2022. "Characterization of a Broadly Neutralizing Monoclonal Antibody against SARS-CoV-2 Variants" Viruses 14, no. 2: 230. https://doi.org/10.3390/v14020230
APA StyleZakir, T. S., Meng, T., Carmen, L. C. P., Chu, J. J. H., Lin, R. T. P., & Prabakaran, M. (2022). Characterization of a Broadly Neutralizing Monoclonal Antibody against SARS-CoV-2 Variants. Viruses, 14(2), 230. https://doi.org/10.3390/v14020230








