Studying the Interactions of U24 from HHV-6 in Order to Further Elucidate Its Potential Role in MS
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Synthesis and Purification of 15-Residue Peptides Representing the Polyproline-Rich N-Terminal Region of U24 from HHV-6A and HHV-6B
2.3. Expression and Purification of hNedd4L-WW3*
2.4. Expression and Purification of 15N-Fyn-SH3
2.5. Subjects
2.6. Peripheral Blood Mononuclear Cell (PBMC) Culture
2.7. T Cell Protein Transfection
2.8. Lactate Dehydrogenase (LDH) Assay
2.9. Proliferation Analysis
2.10. Smad2 and TGF-β1 Analysis
2.11. Nedd4L
2.12. Stable Cell Lines
2.13. Granzyme B ELISPOT Assay
2.14. Calcein Acetoxymethyl Ester (CAM) Cytotoxicity Assay
2.15. Cytometric Analysis and CD107a Degranulation Assay
2.16. Isothermal Titration Calorimetry Analysis
2.17. 1H−15N HSQC NMR Titrations and Kd Calculations for the Fyn-SH3 Protein and U24 Peptide Interaction
2.18. ELISA Assay for U24, pU24 and MBP IgG Titration
2.19. Statistics
3. Results
3.1. Endocytic Recycling Model: Interactions with hNedd4L-WW3*
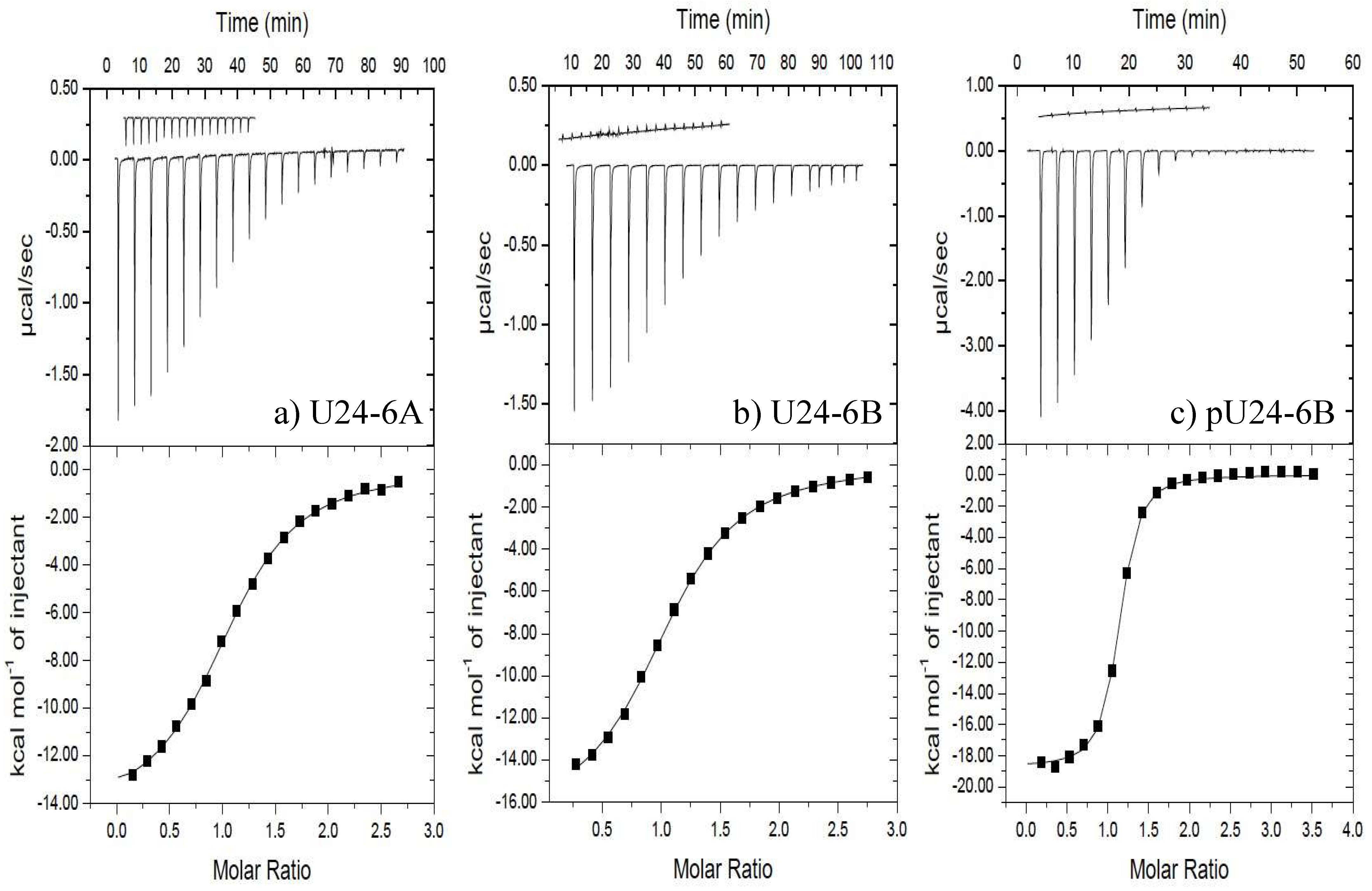
3.1.1. U24-6A and -6B Bind hNedd4L-WW3* with Micromolar Affinity
3.1.2. Phosphorylation at Thr6 of U24-6B Enhances the Affinity with hNedd4L-WW3*
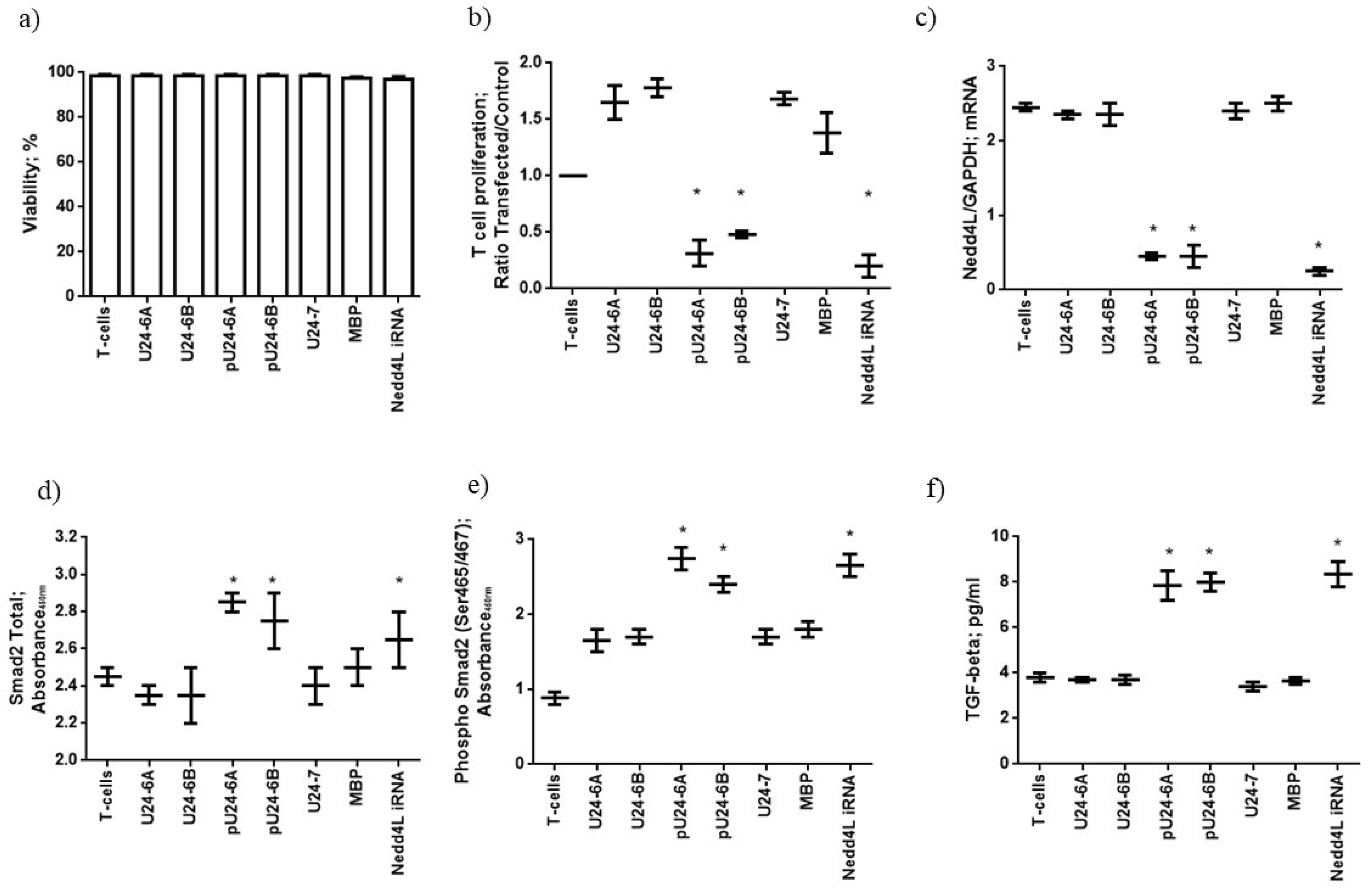
3.1.3. Phosphorylated U24-6A and -6B Affect T-Cell Proliferation via Nedd4L Knockdown
3.2. Mimicry Model: Interactions with Fyn-SH3
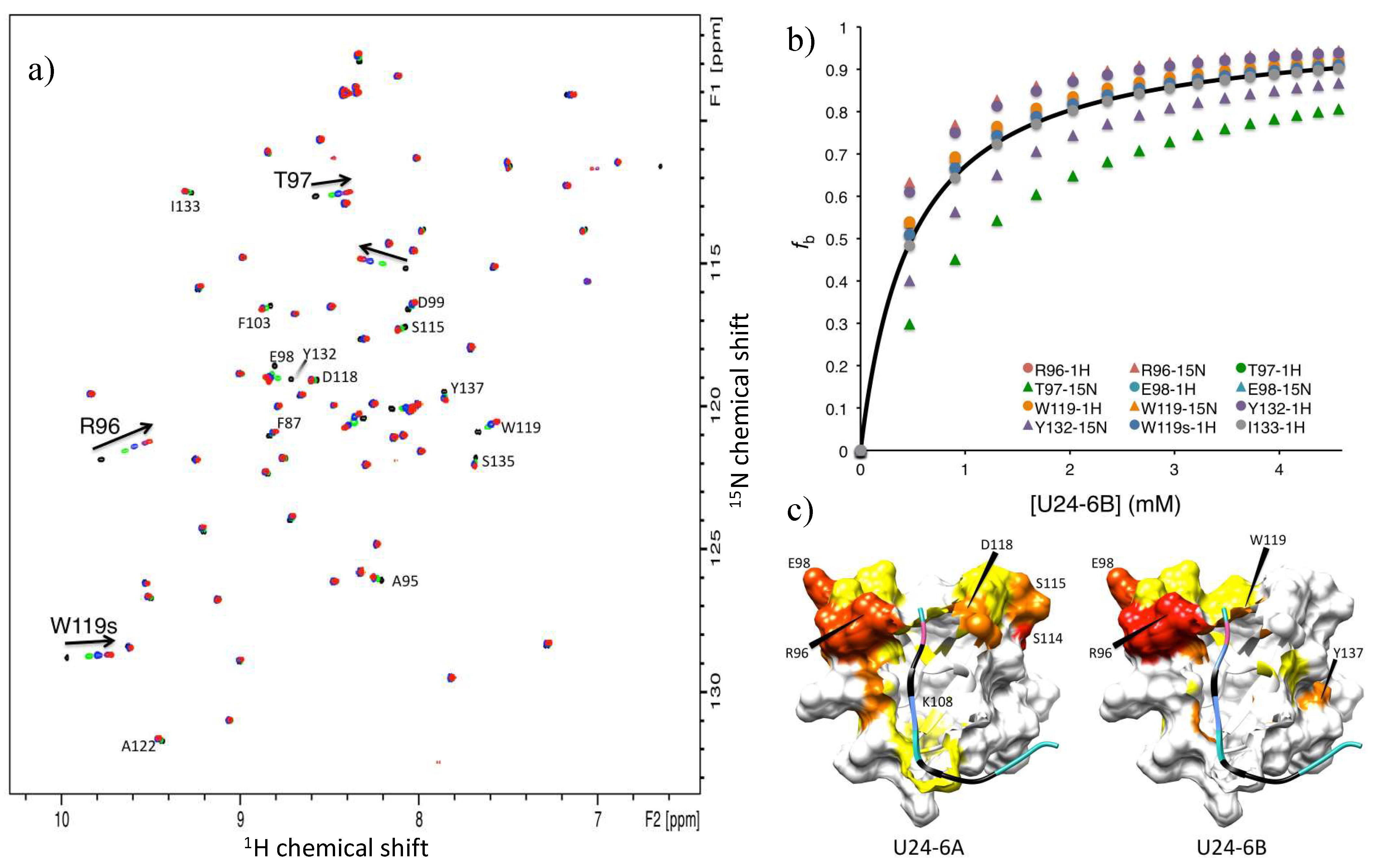
3.2.1. U24-6B Has a Higher Binding Affinity for Fyn-SH3 Than U24-6A
3.2.2. Phosphorylation of U24 Lowers Binding Affinity to Fyn-SH3
3.3. Immune Control Model: Interaction with KIR Receptors
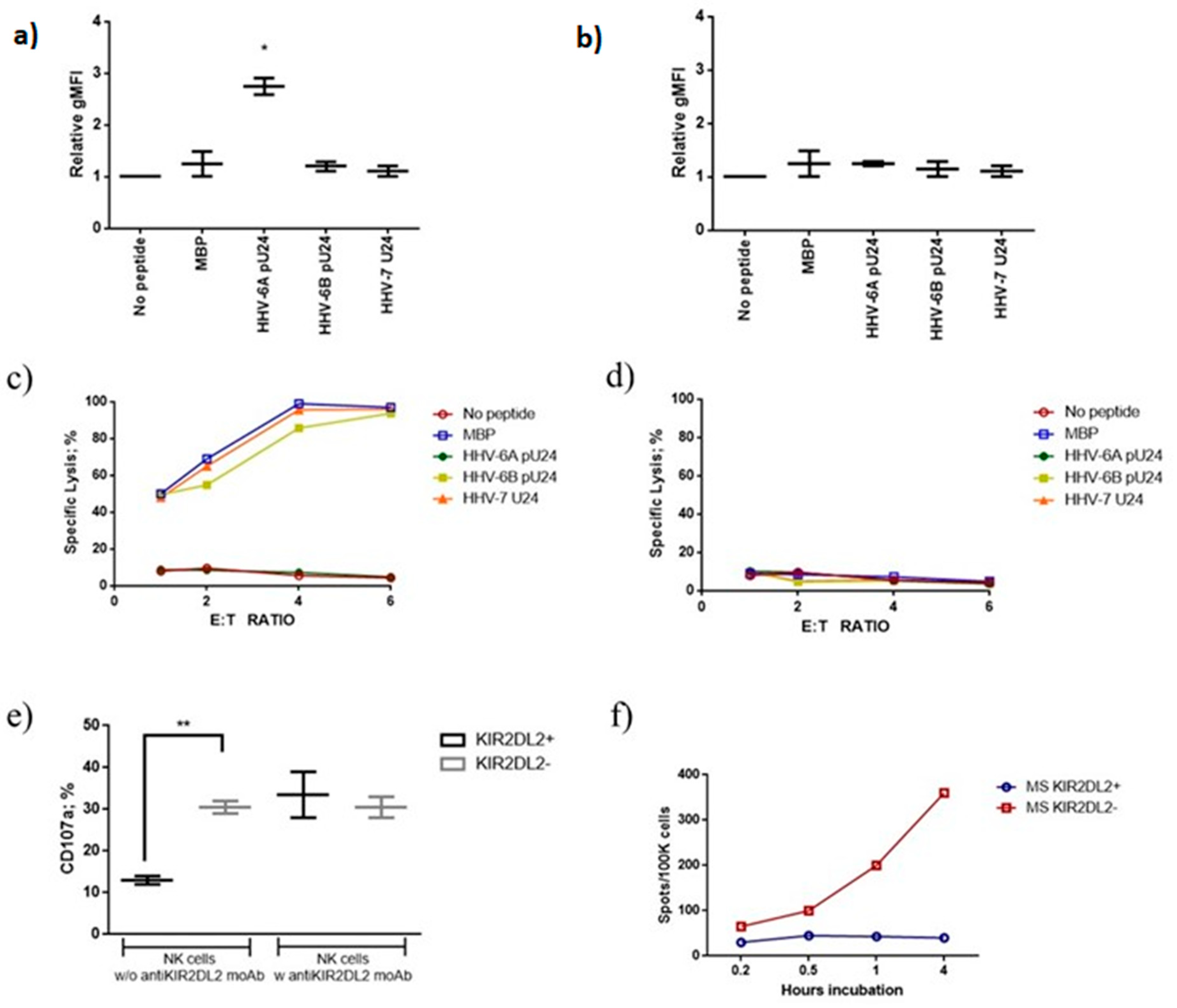
3.3.1. pU24-6A Controls NK Cell Activation
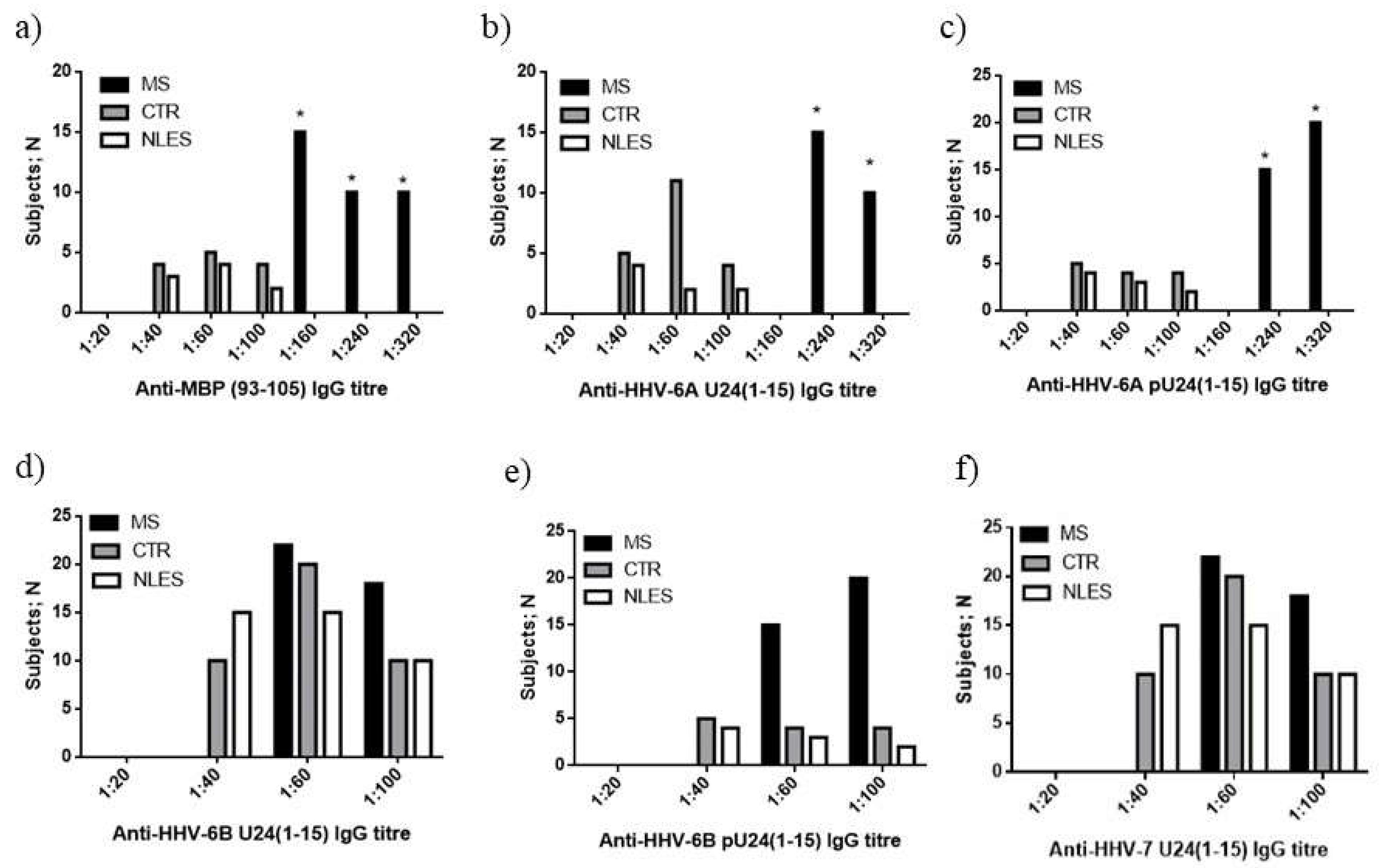
3.3.2. MS Patients Showed Increased Levels of U24 Derived Peptide IgG
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sola, P.; Merelli, E.; Marasca, R.; Poggi, M.; Luppi, M.; Montorsi, M.; Torelli, G. Human herpesvirus 6 and multiple sclerosis: Survey of anti-HHV-6 antibodies by immunofluorescence analysis and of viral sequences by polymerase chain reaction. J. Neurol. Neurosurg. Psychiatry 1993, 56, 917–919. [Google Scholar] [CrossRef] [PubMed]
- Salahuddin, S.Z.; Ablashi, D.V.; Markham, P.D.; Josephs, S.F.; Sturzenegger, S.; Kaplan, M.; Halligan, G.; Biberfeld, P.; Wong-Staal, F.; Kramarsky, B.; et al. Isolation of a new virus, HBLV, in patients with lymphoproliferative disorders. Science 1986, 234, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Leibovitch, E.C.; Jacobson, S. Human Herpesvirus 6 as a Viral Trigger in Mesial Temporal Lobe Epilepsy. J. Infect. Dis. 2015, 212, 1011–1013. [Google Scholar] [CrossRef]
- Theodore, W.H.; Leibovitch, E.; Billioux, B.J.; Inati, S.K.; Zaghloul, K.; Heiss, J.; Gaillard, W.D.; Jacobson, S. Human herpesvirus 6 and epilepsy. Epilepsia Open 2021, 6, 777–780. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, R.; Bortolotti, D.; Gentili, V.; Rotola, A.; Bolzani, S.; Caselli, E.; Tola, M.R.; Di Luca, D. KIR2DS2/KIR2DL2/HLA-C1 Haplotype Is Associated with Alzheimer’s Disease: Implication for the Role of Herpesvirus Infections. J. Alzheimer’s Dis. 2019, 67, 1379–1389. [Google Scholar] [CrossRef] [PubMed]
- Allnutt, M.A.; Johnson, K.; Bennett, D.A.; Connor, S.M.; Troncoso, J.C.; Pletnikova, O.; Albert, M.S.; Resnick, S.M.; Scholz, S.W.; De Jager, P.L.; et al. Human Herpesvirus 6 Detection in Alzheimer’s Disease Cases and Controls across Multiple Cohorts. Neuron 2020, 105, 1027–1035. [Google Scholar] [CrossRef] [PubMed]
- Caserta, M.T.; Hall, C.B.; Schnabel, K.; Mc Intyre, K.; Long, C.; Costanzo, M.; Dewhurst, S.; Insel, R.; Epstein, L.G. Neuroinvasion and persistence of human herpesvirus 6 inchildren. J. Infect. Dis. 1994, 170, 1586–1589. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, K.L.; Richardson, S.E.; Ward, K.N.; Donaldson, C.; MacGregor, D.; Banwell, B.; Mahant, S.; Bitnun, A. Delayed Primary HHV-7 Infection and Neurologic Disease. Pediatrics 2014, 133, e1541–e1547. [Google Scholar] [CrossRef]
- Wucherpfennig, K.W.; Strominger, J.L. Molecular mimicry in T cell-mediated autoimmunity: Viral peptides activate human T cell clones specific for myelin basic protein. Cell 1995, 80, 695–705. [Google Scholar] [CrossRef]
- Leibovitch, E.C.; Jacobson, S. Evidence linking HHV-6 with multiple sclerosis: An update. Curr. Opin. Virol. 2014, 9, 127–133. [Google Scholar] [CrossRef]
- Tarlinton, R.E.; Martynova, E.; Rizvanov, A.A.; Khaiboullina, S.; Verma, S. Role of viruses in the pathogenesis of multiple sclerosis. Viruses 2020, 12, 643. [Google Scholar] [CrossRef] [PubMed]
- Leibovitch, E.C.; Jacobson, S. Viruses in chronic progressive neurologic disease. Mult. Scler. 2018, 24, 48–52. [Google Scholar] [CrossRef] [PubMed]
- Pi, K.S.; Sang, Y.; Straus, S.K. Viral Proteins with PxxP and PY Motifs May Play a Role in Multiple Sclerosis. Viruses 2022, 14, 281. [Google Scholar] [CrossRef] [PubMed]
- Bjornevik, K.; Cortese, M.; Healy, B.C.; Kuhle, J.; Mina, M.J.; Leng, Y.; Elledge, S.J.; Niebuhr, D.W.; Scher, A.I.; Munger, K.L.; et al. Longitudinal analysis reveals high prevalence of Epstein-Barr virus associated with multiple sclerosis. Science 2022, 375, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Komaroff, A.L.; Pellett, P.E.; Jacobson, S. Human herpesviruses 6a and 6b in brain diseases: Association versus causation. Clin. Microbiol. Rev. 2021, 34, e00143-20. [Google Scholar] [CrossRef] [PubMed]
- Caselli, E.; D’Accolti, M.; Caccuri, F.; Soffritti, I.; Gentili, V.; Bortolotti, D.; Rotola, A.; Cassai, E.; Fiorentini, S.; Zani, A.; et al. The U94 Gene of Human Herpesvirus 6: A Narrative Review of Its Role and Potential Functions. Cells 2020, 9, 2608. [Google Scholar] [CrossRef] [PubMed]
- Kearns, P.K.A.; Casey, H.A.; Leach, J.P. Hypothesis: Multiple sclerosis is caused by three-hits, strictly in order, in genetically susceptible persons. Mult. Scler. Relat. Disord. 2018, 24, 157–174. [Google Scholar] [CrossRef]
- Longnecker, R.; Merchant, M.; Brown, M.E.; Fruehling, S.; Bickford, J.O.; Ikeda, M.; Harty, R.N. WW- and SH3-domain interactions with epstein-barr virus LMP2A. Exp. Cell Res. 2000, 257, 332–340. [Google Scholar] [CrossRef]
- Winberg, G.; Matskova, L.; Chen, F.; Plant, P.; Rotin, D.; Gish, G.; Ingham, R.; Ernberg, I.; Pawson, T. Latent Membrane Protein 2A of Epstein-Barr Virus Binds WW Domain E3 Protein-Ubiquitin Ligases That Ubiquitinate B-Cell Tyrosine Kinases. Mol. Cell. Biol. 2000, 20, 8526–8535. [Google Scholar] [CrossRef][Green Version]
- Panousis, C.G.; Rowe, D.T. Epstein-Barr virus latent membrane protein 2 associates with and is a substrate for mitogen-activated protein kinase. J. Virol. 1997, 71, 4752–4760. [Google Scholar] [CrossRef]
- Ben-Fredj, N.; Ben-Selma, W.; Rotola, A.; Nefzi, F.; Benedetti, S.; Frih-Ayed, M.; Di Luca, D.; Aouni, M.; Caselli, E. Prevalence of human herpesvirus U94/REP antibodies and DNA in Tunisian multiple sclerosis patients. J. Neurovirol. 2013, 19, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Koshizuka, T.; Kobayashi, T.; Ishioka, K.; Suzutani, T. Herpesviruses possess conserved proteins for interaction with Nedd4 family ubiquitin E3 ligases. Sci. Rep. 2018, 8, 4447. [Google Scholar] [CrossRef] [PubMed]
- Nobre, L.; Nightingale, K.; Ravenhill, B.J.; Antrobus, R.; Soday, L.; Nichols, J.; Davies, J.; Seirafian, S.; Wang, E.C.Y.; Davison, A.J.; et al. Human cytomegalovirus interactome analysis identifies degradation hubs, domain associations and viral protein functions. Elife 2019, 8, e49894. [Google Scholar] [CrossRef] [PubMed]
- Caselli, E.; Di Luca, D. Molecular biology and clinical associations of Roseoloviruses human herpesvirus 6 and human herpesvirus 7. New Microbiol. 2007, 30, 173–187. [Google Scholar]
- Yao, K.; Mandel, M.; Akyani, N.; Maynard, K.; Sengamalay, N.; Fotheringham, J.; Ghedin, E.; Kashanchi, F.; Jacobson, S. Differential HHV-6A Gene Expression in T Cells and Primary Human Astrocytes Based on Multi-Virus Array Analysis. Glia 2006, 53, 789–798. [Google Scholar] [CrossRef]
- Tejada-Simon, M.V.; Zang, Y.C.Q.; Hong, J.; Rivera, V.M.; Zhang, J.Z. Cross-reactivity with myelin basic protein and human herpesvirus-6 in multiple sclerosis. Ann. Neurol. 2003, 53, 189–197. [Google Scholar] [CrossRef]
- Leibovitch, E.; Wohler, J.E.; Cummings Macri, S.M.; Motanic, K.; Harberts, E.; Gaitán, M.I.; Maggi, P.; Ellis, M.; Westmoreland, S.; Silva, A.; et al. Novel Marmoset (Callithrix jacchus) Model of Human Herpesvirus 6A and 6B Infections: Immunologic, Virologic and Radiologic Characterization. PLoS Pathog. 2013, 9, e1003138. [Google Scholar] [CrossRef]
- Tait, A.R.; Straus, S.K. Phosphorylation of U24 from Human Herpes Virus type 6 (HHV-6) and its potential role in mimicking myelin basic protein (MBP) in multiple sclerosis. FEBS Lett. 2008, 582, 2685–2688. [Google Scholar] [CrossRef][Green Version]
- Sang, Y.; Tait, A.R.; Scott, W.R.P.; Creagh, A.L.; Kumar, P.; Haynes, C.A.; Straus, S.K. Probing the interaction between U24 and the SH3 domain of Fyn tyrosine kinase. Biochemistry 2014, 53, 6092–6102. [Google Scholar] [CrossRef]
- Sang, Y.; Zhang, R.; Scott, W.R.P.; Creagh, A.L.; Haynes, C.A.; Straus, S.K. U24 from Roseolovirus interacts strongly with Nedd4 WW Domains. Sci. Rep. 2017, 7, 39776. [Google Scholar] [CrossRef]
- Sang, Y.; Zhang, R.; Creagh, A.L.; Haynes, C.A.; Straus, S.K. Interactions of U24 from Roseolovirus with WW domains: Canonical vs. noncanonical. Biochem. Cell Biol. 2017, 95, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Compston, A.; Coles, A. Multiple sclerosis. Lancet 2008, 372, 1502–1517. [Google Scholar] [CrossRef]
- Vassall, K.A.; Bamm, V.V.; Harauz, G. MyelStones: The executive roles of myelin basic protein in myelin assembly and destabilization in multiple sclerosis. Biochem. J. 2015, 472, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Vassall, K.A.; Bamm, V.V.; Jenkins, A.D.; Velte, C.J.; Kattnig, D.R.; Boggs, J.M.; Hinderberger, D.; Harauz, G. Substitutions mimicking deimination and phosphorylation of 18.5-kDa myelin basic protein exert local structural effects that subtly influence its global folding. Biochim. Biophys. Acta Biomembr. 2016, 1858, 1262–1277. [Google Scholar] [CrossRef]
- Harauz, G.; Musse, A.A. A tale of two citrullines-Structural and functional aspects of myelin basic protein deimination in health and disease. Neurochem. Res. 2007, 32, 137–158. [Google Scholar] [CrossRef]
- Boggs, J.M.; Rangaraj, G.; Gao, W.; Heng, Y.M. Effect of phosphorylation of myelin basic protein by MAPK on its interactions with actin and actin binding to a lipid membrane in vitro. Biochemistry 2006, 45, 391–401. [Google Scholar] [CrossRef]
- Smith, G.S.T.; De Avila, M.; Paez, P.M.; Spreuer, V.; Wills, M.K.B.; Jones, N.; Boggs, J.M.; Harauz, G. Proline substitutions and threonine pseudophosphorylation of the SH3 ligand of 18.5-kDa myelin basic protein decrease its affinity for the Fyn-SH3 domain and alter process development and protein localization in oligodendrocytes. J. Neurosci. Res. 2012, 90, 28–47. [Google Scholar] [CrossRef]
- Atkins, C.M.; Yon, M.; Groome, N.P.; Sweatt, J.D. Regulation of myelin basic protein phosphorylation by mitogen-activated protein kinase during increased action potential firing in the hippocampus. J. Neurochem. 1999, 73, 1090–1097. [Google Scholar] [CrossRef]
- De Avila, M.; Vassall, K.A.; Smith, G.S.T.; Bamm, V.V.; Harauz, G. The proline-rich region of 18.5 kDa myelin basic protein binds to the SH3-domain of Fyn tyrosine kinase with the aid of an upstream segment to form a dynamic complex in vitro. Biosci. Rep. 2014, 34, 775–788. [Google Scholar] [CrossRef]
- Sullivan, B.M.; Coscoy, L. Downregulation of the T-cell receptor complex and impairment of T-cell activation by human herpesvirus 6 u24 protein. J. Virol. 2008, 82, 602–608. [Google Scholar] [CrossRef]
- Sullivan, B.M.; Coscoy, L. The U24 protein from human herpesvirus 6 and 7 affects endocytic recycling. J. Virol. 2010, 84, 1265–1275. [Google Scholar] [CrossRef] [PubMed]
- Kawabe, H.; Neeb, A.; Dimova, K.; Young, S.M.; Takeda, M.; Katsurabayashi, S.; Mitkovski, M.; Malakhova, O.A.; Zhang, D.E.; Umikawa, M.; et al. Regulation of Rap2A by the Ubiquitin Ligase Nedd4-1 Controls Neurite Development. Neuron 2010, 65, 358–372. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Lu, Y.; Lee, J.K.; Samara, R.; Willenberg, R.; Sears-Kraxberger, I.; Tedeschi, A.; Park, K.K.; Jin, D.; Cai, B.; et al. PTEN deletion enhances the regenerative ability of adult corticospinal neurons. Nat. Neurosci. 2010, 13, 1075–1081. [Google Scholar] [CrossRef] [PubMed]
- Georgieva, M.V.; de Pablo, Y.; Sanchis, D.; Comella, J.X.; Llovera, M. Ubiquitination of TrkA by Nedd4-2 regulates receptor lysosomal targeting and mediates receptor signaling. J. Neurochem. 2011, 117, 479–493. [Google Scholar] [CrossRef] [PubMed]
- Arévalo, J.C.; Waite, J.; Rajagopal, R.; Beyna, M.; Chen, Z.Y.; Lee, F.S.; Chao, M.V. Cell Survival through Trk Neurotrophin Receptors Is Differentially Regulated by Ubiquitination. Neuron 2006, 50, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Ben Fredj, N.; Rizzo, R.; Bortolotti, D.; Nefzi, F.; Chebel, S.; Rotola, A.; Frih-Ayed, M.; Di Luca, D.; Aouni, M. Evaluation of the implication of KIR2DL2 receptor in multiple sclerosis and herpesvirus susceptibility. J. Neuroimmunol. 2014, 271, 30–35. [Google Scholar] [CrossRef]
- Rizzo, R.; Gentili, V.; Casetta, I.; Caselli, E.; De Gennaro, R.; Granieri, E.; Cassai, E.; Di Luca, D.; Rotola, A. Altered natural killer cells’ response to herpes virus infection in multiple sclerosis involves KIR2DL2 expression. J. Neuroimmunol. 2012, 251, 55–64. [Google Scholar] [CrossRef]
- Rizzo, R.; Bortolotti, D.; Fainardi, E.; Gentili, V.; Bolzani, S.; Baldi, E.; Casetta, I.; Granieri, E.; Rotola, A.; Furlan, R.; et al. KIR2DL2 inhibitory pathway enhances Th17 cytokine secretion by NK cells in response to herpesvirus infection in multiple sclerosis patients. J. Neuroimmunol. 2016, 294, 1–5. [Google Scholar] [CrossRef]
- Velarde-de la Cruz, E.E.; Sánchez-Hernández, P.E.; Muñoz-Valle, J.F.; Palafox-Sánchez, C.A.; Ramírez-de los Santos, S.; Graciano-Machuca, O.; García-Iglesias, T.; Montoya-Buelna, M.; Ramírez-Dueñas, M.G. KIR2DL2 and KIR2DS2 as genetic markers to the methotrexate response in rheumatoid arthritis patients. Immunopharmacol. Immunotoxicol. 2016, 38, 303–309. [Google Scholar] [CrossRef]
- Littera, R.; Chessa, L.; Deidda, S.; Angioni, G.; Campagna, M.; Lai, S.; Melis, M.; Cipri, S.; Firinu, D.; Santus, S.; et al. Natural killer-cell immunoglobulin-like receptors trigger differences in immune response to SARS-CoV-2 infection. PLoS ONE 2021, 16, e0255608. [Google Scholar] [CrossRef]
- Gianchecchi, E.; Delfino, D.V.; Fierabracci, A. Natural Killer Cells: Potential Biomarkers and Therapeutic Target in Autoimmune Diseases? Front. Immunol. 2021, 12, 616853. [Google Scholar] [PubMed]
- Dizaji Asl, K.; Velaei, K.; Rafat, A.; Tayefi Nasrabadi, H.; Movassaghpour, A.A.; Mahdavi, M.; Nozad Charoudeh, H. The role of KIR positive NK cells in diseases and its importance in clinical intervention. Int. Immunopharmacol. 2021, 92, 107361. [Google Scholar] [PubMed]
- Alenda, R.; Álvarez-Lafuente, R.; Costa-Frossard, L.; Arroyo, R.; Mirete, S.; Álvarez-Cermeño, J.C.; Villar, L.M. Identification of the major HHV-6 antigen recognized by cerebrospinal fluid IgG in multiple sclerosis. Eur. J. Neurol. 2014, 21, 1096–1101. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Mozo, M.I.; Nieto-Guerrero, A.; Pérez-Pérez, S.; García-Martínez, M.A.; Arroyo, R.; Alvarez-Lafuente, R. MicroRNAs of Human Herpesvirus 6A and 6B in Serum and Cerebrospinal Fluid of Multiple Sclerosis Patients. Front. Immunol. 2020, 11, 2142. [Google Scholar] [CrossRef] [PubMed]
- Engdahl, E.; Gustafsson, R.; Huang, J.; Biström, M.; Lima Bomfim, I.; Stridh, P.; Khademi, M.; Brenner, N.; Butt, J.; Michel, A.; et al. Increased Serological Response Against Human Herpesvirus 6A Is Associated with Risk for Multiple Sclerosis. Front. Immunol. 2019, 10, 2715. [Google Scholar] [CrossRef]
- Renzoni, D.A.; Pugh, D.J.R.; Siligardi, G.; Das, P.; Morton, C.J.; Rossi, C.; Waterfield, M.D.; Campbell, I.D.; Ladbury, J.E. Structural and thermodynamic characterization of the interaction of the SH3 domain from Fyn with the proline-rich binding site on the p85 subunit of PI3-kinase. Biochemistry 1996, 35, 15646–15653. [Google Scholar] [CrossRef]
- Morton, C.J.; Pugh, D.J.R.; Brown, E.L.J.; Kahmann, J.D.; Renzoni, D.A.C.; Campbell, I.D. Solution structure and peptide binding of the SH3 domain from human Fyn. Structure 1996, 4, 705–714. [Google Scholar] [CrossRef]
- Demers, J.P.; Mittermaier, A. Binding mechanism of an SH3 domain studied by NMR and ITC. J. Am. Chem. Soc. 2009, 131, 4355–4367. [Google Scholar] [CrossRef]
- Meneses, E.; Mittermaier, A. Electrostatic interactions in the binding pathway of a transient protein complex studied by NMR and isothermal titration calorimetry. J. Biol. Chem. 2014, 289, 27911–27923. [Google Scholar] [CrossRef]
- Jia, C.Y.H.; Nie, J.; Wu, C.; Li, C.; Li, S.S.C. Novel Src homology 3 domain-binding motifs identified from proteomic screen of a pro-rich region. Mol. Cell. Proteomics 2005, 4, 1155–1166. [Google Scholar] [CrossRef]
- McDonald, W.I.; Compston, A.; Edan, G.; Goodkin, D.; Hartung, H.P.; Lublin, F.D.; McFarland, H.F.; Paty, D.W.; Polman, C.H.; Reingold, S.C.; et al. Recommended diagnostic criteria for multiple sclerosis: Guidelines from the International Panel on the Diagnosis of Multiple Sclerosis. Ann. Neurol. 2001, 50, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Kurtzke, J.F. Rating neurologic impairment in multiple sclerosis: An expanded disability status scale (EDSS). Neurology 1983, 33, 1444–1452. [Google Scholar] [CrossRef] [PubMed]
- Lublin, F.D.; Reingold, S.C. Defining the clinical course of multiple sclerosis: Results of an international survey. Neurology 1996, 46, 907–911. [Google Scholar] [CrossRef]
- Hochberg, M.C. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997, 40, 1725. [Google Scholar] [CrossRef] [PubMed]
- Alunno, A.; Najm, A.; Machado, P.M.; Bertheussen, H.; Burmester, G.R.R.; Carubbi, F.; De Marco, G.; Giacomelli, R.; Hermine, O.; Isaacs, J.D.; et al. 2021 update of the EULAR points to consider on the use of immunomodulatory therapies in COVID-19. Ann. Rheum. Dis. 2022, 81, 34–40. [Google Scholar] [CrossRef]
- Bortoluzzi, A.; Fanouriakis, A.; Appenzeller, S.; Costallat, L.; Scirè, C.A.; Murphy, E.; Bertsias, G.; Hanly, J.; Govoni, M. Validity of the Italian algorithm for the attribution of neuropsychiatric events in systemic lupus erythematosus: A retrospective multicentre international diagnostic cohort study. BMJ Open 2017, 7, 15546. [Google Scholar] [CrossRef]
- Bortolotti, D.; Gentili, V.; Rotola, A.; Cultrera, R.; Marci, R.; Di Luca, D.; Rizzo, R. HHV-6A infection of endometrial epithelial cells affects immune profile and trophoblast invasion. Am. J. Reprod. Immunol. 2019, 82, e13174. [Google Scholar] [CrossRef]
- Ogawa, K.; Utsunomiya, T.; Mimori, K.; Tanaka, F.; Inoue, H.; Nagahara, H.; Murayama, S.; Mori, M. Clinical significance of human kallikrein gene 6 messenger RNA expression in colorectal cancer. Clin. Cancer Res. 2005, 11, 2889–2893. [Google Scholar] [CrossRef]
- Gatfield, J.; Lammert, E.; Nickolaus, P.; Münz, C.; Rothenfusser, S.; Fisch, P.; Stevanović, S.; Schild, H.; Rammensee, H.G.; Arnold, D. Cell lines transfected with the TAP inhibitor ICP47 allow testing peptide binding to a variety of HLA class I molecules. Int. Immunol. 1998, 10, 1665–1672. [Google Scholar] [CrossRef][Green Version]
- Mal, T.K.; Matthews, S.J.; Kovacs, H.; Campbell, I.D.; Boyd, J. Some NMR experiments and a structure determination employing a {15N,2H} enriched protein. J. Biomol. NMR 1998, 12, 259–276. [Google Scholar] [CrossRef]
- Williamson, M.P. Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 2013, 73, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.A.; Li, M.O. TGF-β: Guardian of T Cell Function. J. Immunol. 2013, 191, 3973–3979. [Google Scholar] [CrossRef] [PubMed]
- Boggs, J.M.; Homchaudhuri, L.; Ranagaraj, G.; Liu, Y.; Smith, G.S.; Harauz, G. Interaction of myelin basic protein with cytoskeletal and signaling proteins in cultured primary oligodendrocytes and N19 oligodendroglial cells. BMC Res. Notes 2014, 7, 387. [Google Scholar] [CrossRef]
- Kim, J.K.; Mastronardi, F.G.; Wood, D.D.; Lubman, D.M.; Zand, R.; Moscarello, M.A. Multiple sclerosis: An important role for post-translational modifications of myelin basic protein in pathogenesis. Mol. Cell. Proteomics 2003, 2, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Sang, Y. Protein Interactions of Membrane Protein U24 from Roseolovirus and Implications for Its Function. Ph.D. Thesis, University of British Columbia, Vancouver, BC, Canada, 2016. [Google Scholar]
- Zarrine-Afsar, A.; Mittermaier, A.; Kay, L.E.; Davidson, A.R. Protein stabilization by specific binding of guanidinium to a functional arginine-binding surface on an SH3 domain. Protein Sci. 2006, 15, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera-A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Bortolotti, D.; Gentili, V.; Bortoluzzi, A.; Govoni, M.; Schiuma, G.; Beltrami, S.; Rizzo, S.; Baldi, E.; Caselli, E.; Pugliatti, M.; et al. Herpesvirus Infections in KIR2DL2-Positive Multiple Sclerosis Patients: Mechanisms Triggering Autoimmunity. Microorganisms 2022, 10, 494. [Google Scholar] [CrossRef]
- Schafer, J.L.; Ries, M.; Guha, N.; Connole, M.; Colantonio, A.D.; Wiertz, E.J.; Wilson, N.A.; Kaur, A.; Evans, D.T. Suppression of a Natural Killer Cell Response by Simian Immunodeficiency Virus Peptides. PLoS Pathog. 2015, 11, e1005145. [Google Scholar] [CrossRef]
- Tai, A.K.; Luka, J.; Ablashi, D.; Huber, B.T. HHV-6A infection induces expression of HERV-K18-encoded superantigen. J. Clin. Virol. 2009, 46, 47–48. [Google Scholar] [CrossRef]
- Grut, V.; Biström, M.; Salzer, J.; Stridh, P.; Jons, D.; Gustafsson, R.; Fogdell-Hahn, A.; Huang, J.; Brenner, N.; Butt, J.; et al. Cytomegalovirus seropositivity is associated with reduced risk of multiple sclerosis—A presymptomatic case–control study. Eur. J. Neurol. 2021, 28, 3072–3079. [Google Scholar] [CrossRef]
- Fleming, J.; Hernandez, G.; Hartman, L.; Maksimovic, J.; Nace, S.; Lawler, B.; Risa, T.; Cook, T.; Agni, R.; Reichelderfer, M.; et al. Safety and efficacy of helminth treatment in relapsing-remitting multiple sclerosis: Results of the HINT 2 clinical trial. Mult. Scler. J. 2019, 25, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Zuchero, J.B.; Fu, M.M.; Sloan, S.A.; Ibrahim, A.; Olson, A.; Zaremba, A.; Dugas, J.C.; Wienbar, S.; Caprariello, A.V.; Kantor, C.; et al. CNS Myelin Wrapping Is Driven by Actin Disassembly. Dev. Cell 2015, 34, 152–167. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.J.; Redmond, S.A.; Chan, J.R. Remodeling myelination: Implications for mechanisms of neural plasticity. Nat. Neurosci. 2016, 19, 190–197. [Google Scholar] [CrossRef]
- Krämer-Albers, E.M.; White, R. From axon-glial signalling to myelination: The integrating role of oligodendroglial Fyn kinase. Cell. Mol. Life Sci. 2011, 68, 2003–2012. [Google Scholar] [CrossRef] [PubMed]
- Yon, M.; Ackerley, C.A.; Mastronardi, F.G.; Groome, N.; Moscarello, M.A. Identification of a mitogen-activated protein kinase site in human myelin basic protein in situ. J. Neuroimmunol. 1996, 65, 55–59. [Google Scholar] [CrossRef]
- White, R.; Krämer-Albers, E.M. Axon-glia interaction and membrane traffic in myelin formation. Front. Cell. Neurosci. 2014, 7, 284. [Google Scholar] [CrossRef]
- Ding, X.; Jo, J.; Wang, C.Y.; Cristobal, C.D.; Zuo, Z.; Ye, Q.; Wirianto, M.; Lindeke-Myers, A.; Choi, J.M.; Mohila, C.A.; et al. The Daam2-VHL-Nedd4 axis governs developmental and regenerative oligodendrocyte differentiation. Genes Dev. 2020, 34, 1177–1189. [Google Scholar] [CrossRef]
- Schmiedel, D.; Tai, J.; Levi-Schaffer, F.; Dovrat, S.; Mandelboim, O. Human Herpesvirus 6B Downregulates Expression of Activating Ligands during Lytic Infection To Escape Elimination by Natural Killer Cells. J. Virol. 2016, 90, 9608–9617. [Google Scholar] [CrossRef]
- Sharon, E.; Frenkel, N. Human Herpesvirus 6A Exhibits Restrictive Propagation with Limited Activation of the Protein Kinase R-eIF2α Stress Pathway. J. Virol. 2017, 91, 9. [Google Scholar] [CrossRef]
- Reynaud, J.M.; Jegou, J.-F.; Welsch, J.C.; Horvat, B. Human Herpesvirus 6A Infection in CD46 Transgenic Mice: Viral Persistence in the Brain and Increased Production of Proinflammatory Chemokines via Toll-Like Receptor 9. J. Virol. 2014, 88, 5421–5436. [Google Scholar] [CrossRef]
- Biström, M.; Jons, D.; Engdahl, E.; Gustafsson, R.; Huang, J.; Brenner, N.; Butt, J.; Alonso-Magdalena, L.; Gunnarsson, M.; Vrethem, M.; et al. Epstein–Barr virus infection after adolescence and human herpesvirus 6A as risk factors for multiple sclerosis. Eur. J. Neurol. 2021, 28, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, R.K.L.; Engdahl, E.E.; Hammarfjord, O.; Adikari, S.B.; Lourda, M.; Klingström, J.; Svensson, M.; Fogdell-Hahn, A. Human Herpesvirus 6A Partially Suppresses Functional Properties of DC without Viral Replication. PLoS ONE 2013, 8, e58122. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Engdahl, E.; Gustafsson, R.; Fogdell-Hahn, A.; Waterboer, T.; Hillert, J.; Olsson, T.; Alfredsson, L.; Hedström, A.K. High antibody levels against human herpesvirus-6A interact with lifestyle factors in multiple sclerosis development. Mult. Scler. J. 2021, 28, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Holliday, A.E.; Shi, H.; Zhu, F.; Ewing, M.A.; Russell, D.H.; Clemmer, D.E. Characterizing intermediates along the transition from polyproline i to polyproline II using ion mobility spectrometry-mass spectrometry. J. Am. Chem. Soc. 2014, 136, 12702–12711. [Google Scholar] [CrossRef]
- Polverini, E.; Rangaraj, G.; Libich, D.S.; Boggs, J.M.; Harauz, G. Binding of the proline-rich segment of myelin basic protein to SH3 domains: Spectroscopic, microarray, and modeling studies of ligand conformation and effects of posttranslational modifications. Biochemistry 2008, 47, 267–282. [Google Scholar] [CrossRef]
- Macdonald, A.; Mazaleyrat, S.; McCormick, C.; Street, A.; Burgoyne, N.J.; Jackson, R.M.; Cazeaux, V.; Shelton, H.; Saksela, K.; Harris, M. Further studies on hepatitis C virus NS5A-SH3 domain interactions: Identification of residues critical for binding and implications for viral RNA replication and modulation of cell signalling. J. Gen. Virol. 2005, 86, 1035–1044. [Google Scholar] [CrossRef]
- Akhyani, N.; Berti, R.; Brennan, M.B.; Soldan, S.S.; Eaton, J.M.; McFarland, H.F.; Jacobson, S. Tissue distribution and variant characterization of human herpesvirus (HHV)-6: Increased prevalence of HHV-6A in patients with multiple sclerosis. J. Infect. Dis. 2000, 182, 1321–1325. [Google Scholar] [CrossRef]
- Aloisi, F.; Serafini, B.; Magliozzi, R.; Howell, O.W.; Reynolds, R. Detection of Epstein-Barr virus and B-cell follicles in the multiple sclerosis brain: What you find depends on how and where you look. Brain 2010, 133, e157. [Google Scholar] [CrossRef]
- Veroni, C.; Aloisi, F. The CD8 T Cell-Epstein-Barr Virus-B Cell Trialogue: A Central Issue in Multiple Sclerosis Pathogenesis. Front. Immunol. 2021, 12, 665718. [Google Scholar] [CrossRef]
- Serafini, B.; Rosicarelli, B.; Veroni, C.; Mazzola, G.A.; Aloisi, F. Epstein-Barr Virus-Specific CD8 T Cells Selectively Infiltrate the Brain in Multiple Sclerosis and Interact Locally with Virus-Infected Cells: Clue for a Virus-Driven Immunopathological Mechanism. J. Virol. 2019, 93, 00980-19. [Google Scholar] [CrossRef]
- Kofler, D.M.; Severson, C.A.; Mousissian, N.; De Jager, P.L.; Hafler, D.A. The CD6 Multiple Sclerosis Susceptibility Allele Is Associated with Alterations in CD4+ T Cell Proliferation. J. Immunol. 2011, 187, 3286–3291. [Google Scholar] [CrossRef] [PubMed]
- Mimpen, M.; Smolders, J.; Hupperts, R.; Damoiseaux, J. Natural killer cells in multiple sclerosis: A review. Immunol. Lett. 2020, 222, 1–11. [Google Scholar] [CrossRef] [PubMed]

| Name | Sequence | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MBP93-107 | I | V | T | P | R | T | P | P | P | S | Q | G | K | G | R | |
| U24-6A1-15 | M | D | P | P | R | T | P | P | P | S | Y | S | E | V | L | |
| pU24-6A1-15 | M | D | P | P | R | pT | P | P | P | S | Y | S | E | V | L | |
| U24-6B1-15 | M | D | R | P | R | T | P | P | P | S | Y | S | E | V | L | |
| pU24-6B1-15 | M | D | R | P | R | pT | P | P | P | S | Y | S | E | V | L | |
| U24-71-15 | M | - | T | H | E | T | P | P | P | S | Y | N | D | V | M | L |
| Target: hNedd4L-WW3* | ||||
|---|---|---|---|---|
| Ligand | Kd (μM) | ΔH° (kJ/mol) | ΔS° (J/mol K) | ΔG° (kJ/mol) |
| U24-6A | 8.5 ± 0.7 | −59.5 ± 0.9 | −103 ± 2 | (−2.9 ± 0.1) × 101 |
| U24-6B | 8.9 ± 0.9 | −71 ± 4 | −141.6 ± 0.9 | (−2.9 ± 0.4) × 101 |
| pU24-6B | 0.88 ± 0.01 | −78.4 ± 0.6 | −147 ± 2 | (−3.46 ± 0.08) × 101 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pi, K.-S.; Bortolotti, D.; Sang, Y.; Schiuma, G.; Beltrami, S.; Rizzo, S.; Bortoluzzi, A.; Baldi, E.; Creagh, A.L.; Haynes, C.A.; et al. Studying the Interactions of U24 from HHV-6 in Order to Further Elucidate Its Potential Role in MS. Viruses 2022, 14, 2384. https://doi.org/10.3390/v14112384
Pi K-S, Bortolotti D, Sang Y, Schiuma G, Beltrami S, Rizzo S, Bortoluzzi A, Baldi E, Creagh AL, Haynes CA, et al. Studying the Interactions of U24 from HHV-6 in Order to Further Elucidate Its Potential Role in MS. Viruses. 2022; 14(11):2384. https://doi.org/10.3390/v14112384
Chicago/Turabian StylePi, Keng-Shuo, Daria Bortolotti, Yurou Sang, Giovanna Schiuma, Silvia Beltrami, Sabrina Rizzo, Alessandra Bortoluzzi, Eleonora Baldi, A. Louise Creagh, Charles A. Haynes, and et al. 2022. "Studying the Interactions of U24 from HHV-6 in Order to Further Elucidate Its Potential Role in MS" Viruses 14, no. 11: 2384. https://doi.org/10.3390/v14112384
APA StylePi, K.-S., Bortolotti, D., Sang, Y., Schiuma, G., Beltrami, S., Rizzo, S., Bortoluzzi, A., Baldi, E., Creagh, A. L., Haynes, C. A., Rizzo, R., & Straus, S. K. (2022). Studying the Interactions of U24 from HHV-6 in Order to Further Elucidate Its Potential Role in MS. Viruses, 14(11), 2384. https://doi.org/10.3390/v14112384










