The Parapoynx stagnalis Nucleopolyhedrovirus (PastNPV), a Divergent Member of the Alphabaculovirus Group I Clade, Encodes a Homolog of Ran GTPase
Abstract
1. Introduction
2. Materials and Methods
2.1. Virus Sample
2.2. DNA Isolation and Sequencing
2.3. Genome Annotation
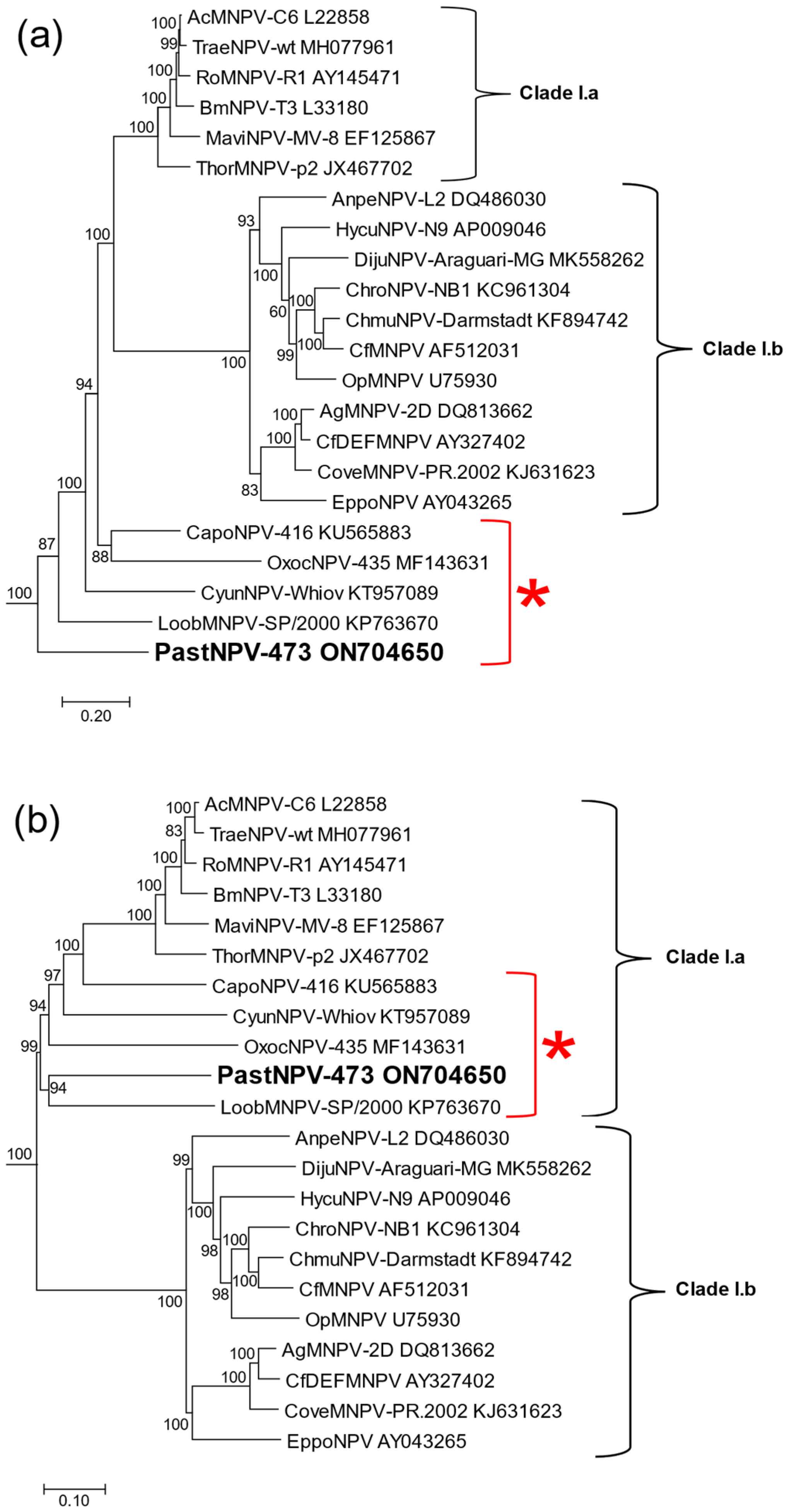
2.4. Phylogeny
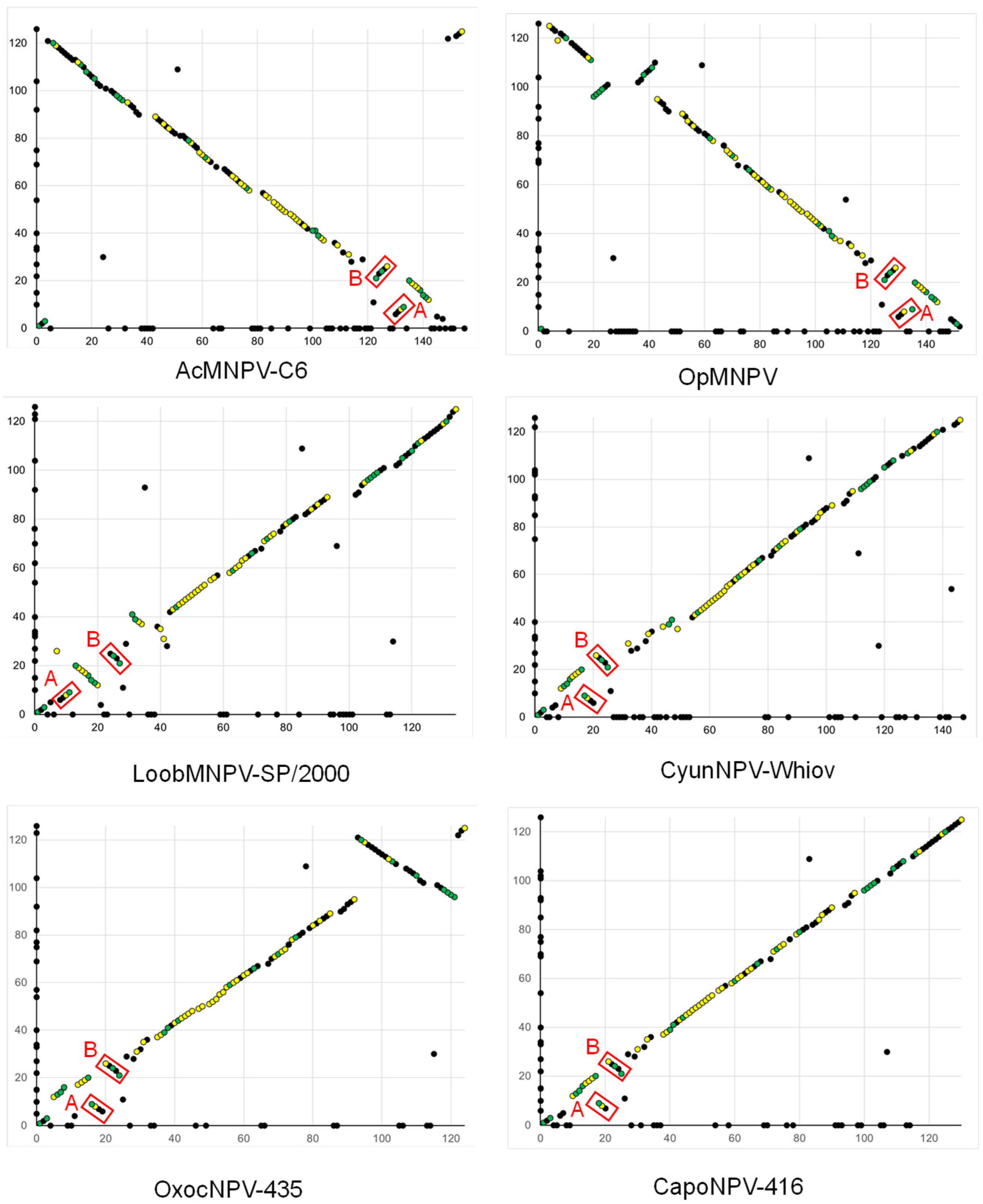
2.5. ORF Synteny and Pairwise Distance Estimation
3. Results
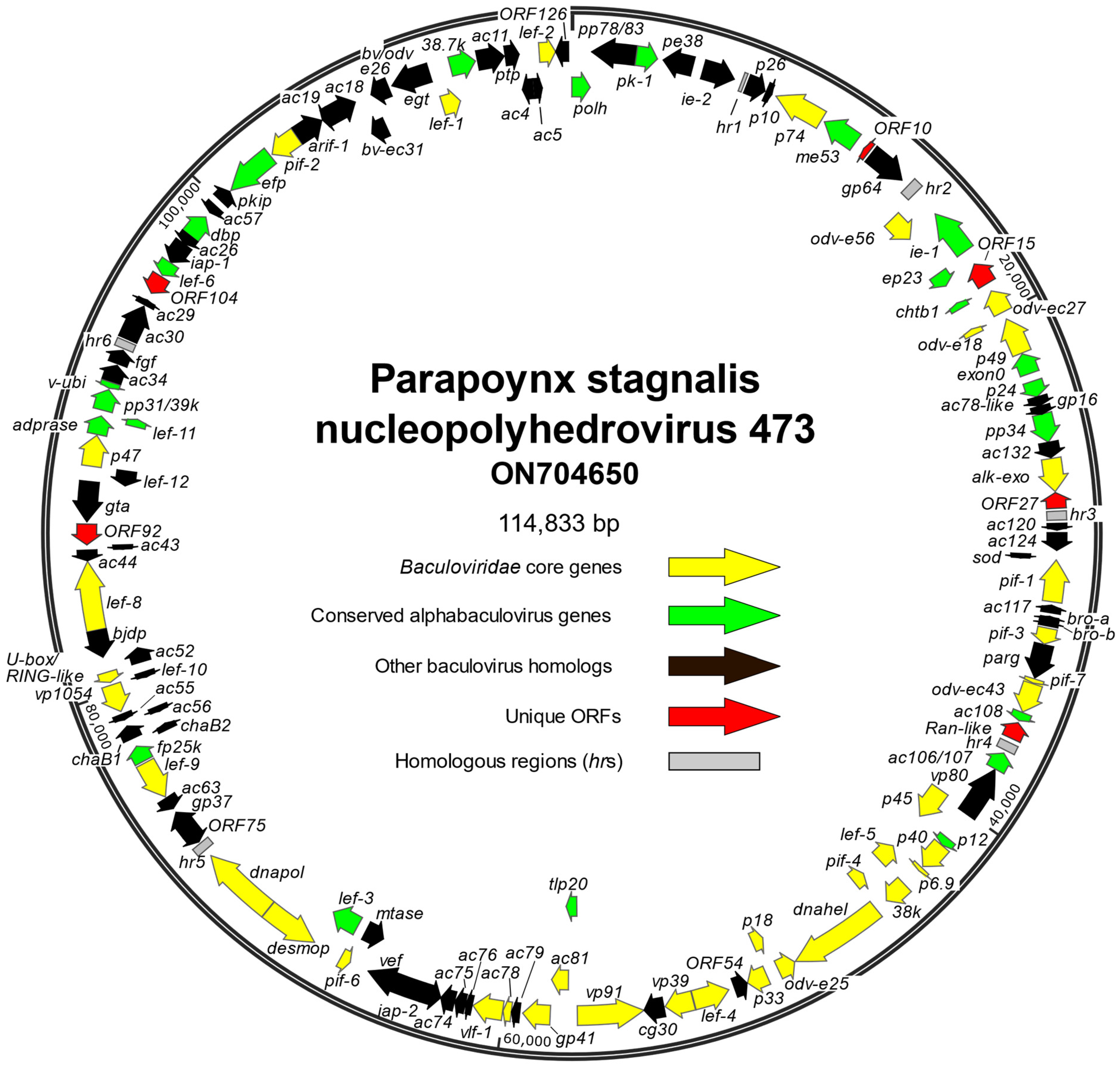
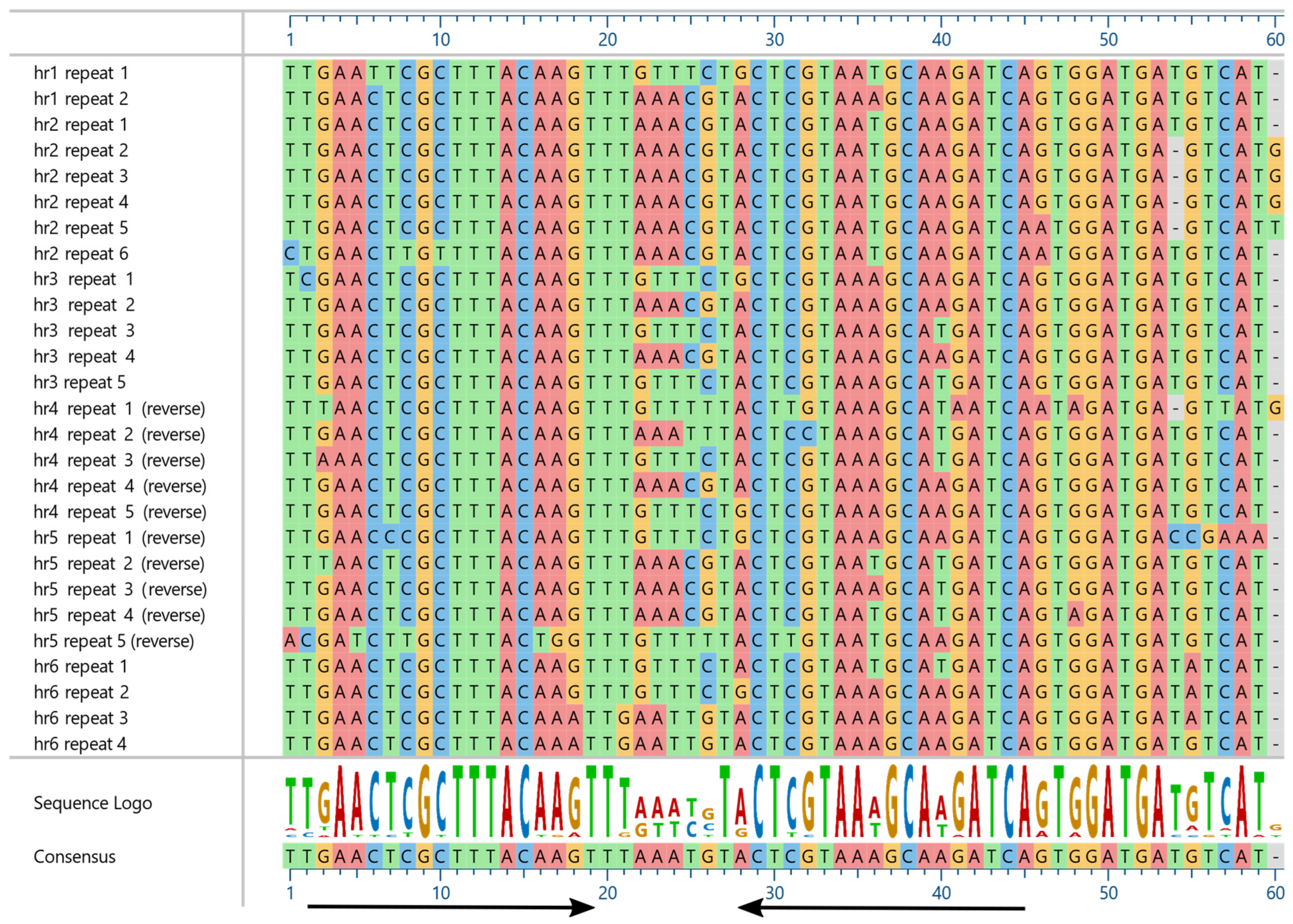
3.1. Characteristics of the PastNPV-473 Genome
3.2. Comparisons of PastNPV-473 with Other Viruses
3.3. ORF Content of PastNPV-473
3.3.1. ORFs with Homologs in Other Alphabaculoviruses
3.3.2. ORFs Not Found in Other Baculovirus Genomes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Rohrmann, G.F. Baculovirus Molecular Biology, 4th ed.; National Center for Biotechnology Information: Bethesda, MD, USA, 2019. [Google Scholar]
- Lacey, L.A.; Grzywacz, D.; Shapiro-Ilan, D.I.; Frutos, R.; Brownbridge, M.; Goettel, M.S. Insect pathogens as biological control agents: Back to the future. J. Invertebr. Pathol. 2015, 132, 1–41. [Google Scholar] [CrossRef] [PubMed]
- Harrison, R.L.; Herniou, E.A.; Jehle, J.A.; Theilmann, D.A.; Burand, J.P.; Becnel, J.J.; Krell, P.J.; van Oers, M.M.; Mowery, J.D.; Bauchan, G.R.; et al. ICTV virus taxonomy profile: Baculoviridae. J. Gen. Virol. 2018, 99, 1185–1186. [Google Scholar] [CrossRef] [PubMed]
- Knox, C.; Moore, S.D.; Luke, G.A.; Hill, M.P. Baculovirus-based strategies for the management of insect pests: A focus on development and application in South Africa. Biocontrol Sci. Technol. 2015, 25, 1–20. [Google Scholar] [CrossRef]
- Landwehr, A. Benefits of baculovirus use in IPM strategies for open field and protected vegetables. Front. Sustain. Food Syst. 2021, 4, 593796. [Google Scholar] [CrossRef]
- Jacob, A.; Pillai, K.S.; Ratan Asari, P.A. A nuclear polyhedrosis virus from rice caseworm Nymphula depunctalis Guen. (Lepidoptera: Pyralidae). Curr. Sci. India 1978, 47, 928–929. [Google Scholar]
- Pathak, M.D.; Kahn, Z.R. Insect Pests of Rice; International Rice Research Institute: Los Banos, Philippines, 1994. [Google Scholar]
- Heinrichs, E.A.; Barrion, A.T. Rice-Feeding Insects and Selected Natural Enemies in West Africa: Biology, Ecology, Identification; International Rice Research Institute: Los Banos, Philippines, 2004. [Google Scholar]
- Devanesan, S.; Jacob, A. Studies on a nuclear polyhedrosis of rice caseworm Nymphula-depunctalis Guen (Pyraustidae, Lepidoptera). Entomon 1980, 5, 277–284. [Google Scholar]
- Harrison, R.L.; Rowley, D.L. Complete genome sequence of an Alphabaculovirus from the southern armyworm. Spodoptera eridania. Microbiol. Resour. Announc. 2019, 8, e01277-18. [Google Scholar] [CrossRef]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids. Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef]
- Zimmermann, L.; Stephens, A.; Nam, S.Z.; Rau, D.; Kubler, J.; Lozajic, M.; Gabler, F.; Soding, J.; Lupas, A.N.; Alva, V. A completely reimplemented MPI bioinformatics yoolkit with a new HHpred server at its core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef]
- van Oers, M.M.; Vlak, J.M. Baculovirus genomics. Curr. Drug Targets 2007, 8, 1051–1068. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids. Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids. Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Edler, D.; Klein, J.; Antonelli, A.; Silvestro, D. raxmlGUI 2.0: A graphical interface and toolkit for phylogenetic analyses using RAxML. Methods Ecol. Evol. 2021, 12, 373–377. [Google Scholar] [CrossRef]
- Le, S.Q.; Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11 Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Hu, Z.H.; Arif, B.M.; Jin, F.; Martens, J.W.; Chen, X.W.; Sun, J.S.; Zuidema, D.; Goldbach, R.W.; Vlak, J.M. Distinct gene arrangement in the Buzura suppressaria single-nucleocapsid nucleopolyhedrovirus genome. J. Gen. Virol. 1998, 79 Pt 11, 2841–2851. [Google Scholar] [CrossRef]
- Jehle, J.A.; Lange, M.; Wang, H.; Hu, Z.; Wang, Y.; Hauschild, R. Molecular identification and phylogenetic analysis of baculoviruses from Lepidoptera. Virology 2006, 346, 180–193. [Google Scholar] [CrossRef]
- Garavaglia, M.J.; Miele, S.A.; Iserte, J.A.; Belaich, M.N.; Ghiringhelli, P.D. The ac53, ac78, ac101, and ac103 genes are newly discovered core genes in the family Baculoviridae. J. Virol. 2012, 86, 12069–12079. [Google Scholar] [CrossRef] [PubMed]
- Javed, M.A.; Biswas, S.; Willis, L.G.; Harris, S.; Pritchard, C.; van Oers, M.M.; Donly, B.C.; Erlandson, M.A.; Hegedus, D.D.; Theilmann, D.A. Autographa californica multiple nucleopolyhedrovirus AC83 is a per os infectivity factor (PIF) protein required for occlusion-derived virus (ODV) and budded virus nucleocapsid assembly as well as assembly of the PIF complex in ODV envelopes. J. Virol. 2017, 91, e02115-16. [Google Scholar] [CrossRef] [PubMed]
- Schneider, T.D.; Stephens, R.M. Sequence logos—A new way to display consensus sequences. Nucleic Acids Res. 1990, 18, 6097–6100. [Google Scholar] [CrossRef]
- Aragao-Silva, C.W.; Andrade, M.S.; Ardisson-Araujo, D.M.; Fernandes, J.E.; Morgado, F.S.; Bao, S.N.; Moraes, R.H.; Wolff, J.L.; Melo, F.L.; Ribeiro, B.M. The complete genome of a baculovirus isolated from an insect of medical interest: Lonomia obliqua (Lepidoptera: Saturniidae). Sci. Rep. 2016, 6, 23127. [Google Scholar] [CrossRef]
- Gomi, S.; Majima, K.; Maeda, S. Sequence analysis of the genome of Bombyx mori nucleopolyhedrovirus. J. Gen. Virol. 1999, 80, 1323–1337. [Google Scholar] [CrossRef] [PubMed]
- Harrison, R.L.; Bonning, B.C. Comparative analysis of the genomes of Rachiplusia ou and Autographa californica multiple nucleopolyhedroviruses. J. Gen. Virol. 2003, 84, 1827–1842. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, Z.; Zhang, L.; Hou, D.; Wang, M.; Arif, B.; Kou, Z.; Wang, H.; Deng, F.; Hu, Z. Genome sequencing and analysis of Catopsilia pomona nucleopolyhedrovirus: A distinct species in group I Alphabaculovirus. PLoS ONE 2016, 11, e0155134. [Google Scholar] [CrossRef]
- Wang, Y.S.; Huang, G.H.; Cheng, X.H.; Wang, X.; Garretson, T.A.; Dai, L.Y.; Zhang, C.X.; Cheng, X.W. Genome of Thysanoplusia orichalcea multiple nucleopolyhedrovirus lacks the superoxide dismutase gene. J. Virol. 2012, 86, 11948–11949. [Google Scholar] [CrossRef][Green Version]
- Ayres, M.D.; Howard, S.C.; Kuzio, J.; Lopez-Ferber, M.; Possee, R.D. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 1994, 202, 586–605. [Google Scholar] [CrossRef]
- Chen, Y.R.; Wu, C.Y.; Lee, S.T.; Wu, Y.J.; Lo, C.F.; Tsai, M.F.; Wang, C.H. Genomic and host range studies of Maruca vitrata nucleopolyhedrovirus. J. Gen. Virol. 2008, 89, 2315–2330. [Google Scholar] [CrossRef]
- Harrison, R.L.; Lynn, D.E. Genomic sequence analysis of a nucleopolyhedrovirus isolated from the diamondback moth, Plutella xylostella. Virus Genes 2007, 35, 857–873. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.F.; Chen, T.H.; Chang, Z.T.; Wang, T.C.; Lee, S.J.; Kim, J.C.; Kim, J.S.; Chiu, K.P.; Nai, Y.S. Genomic sequencing of Troides aeacus nucleopolyhedrovirus (TraeNPV) from golden birdwing larvae (Troides aeacus formosanus) to reveal defective Autographa californica NPV genomic features. Bmc Genom. 2019, 20, 419. [Google Scholar] [CrossRef] [PubMed]
- Pearson, M.N.; Rohrmann, G.F. Transfer, incorporation, and substitution of envelope fusion proteins among members of the Baculoviridae, Orthomyxoviridae, and Metaviridae (insect retrovirus) families. J. Virol. 2002, 76, 5301–5304. [Google Scholar] [CrossRef]
- Theze, J.; Lopez-Vaamonde, C.; Cory, J.S.; Herniou, E.A. Biodiversity, evolution and ecological specialization of baculoviruses: A treasure trove for future applied research. Viruses 2018, 10, 366. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Wang, J.; Wang, Q.; Yin, F.; Liu, X.; Hou, D.; Zhang, L.; Liu, H.; Li, J.; Arif, B.M.; et al. Genome characteristics of the Cyclophragma undans nucleopolyhedrovirus: A distinct species in group I of Alphabaculovirus. Virol. Sin. 2018, 33, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hou, D.; Wang, Q.; Kuang, W.; Zhang, L.; Li, J.; Shen, S.; Deng, F.; Wang, H.; Hu, Z.; et al. Genome analysis of a novel Group I Alphabaculovirus obtained from Oxyplax ochracea. PLoS ONE 2018, 13, e0192279. [Google Scholar] [CrossRef]
- Wennmann, J.T.; Keilwagen, J.; Jehle, J.A. Baculovirus Kimura two-parameter species demarcation criterion is confirmed by the distances of 38 core gene nucleotide sequences. J. Gen. Virol. 2018, 99, 1307–1320. [Google Scholar] [CrossRef]
- Guarino, L.A.; Gonzalez, M.A.; Summers, M.D. Complete sequence and enhancer function of the homologous DNA regions of Autographa californica nuclear polyhedrosis virus. J. Virol. 1986, 60, 224–229. [Google Scholar] [CrossRef]
- Harrison, R.L.; Mowery, J.D.; Rowley, D.L.; Bauchan, G.R.; Theilmann, D.A.; Rohrmann, G.F.; Erlandson, M.A. The complete genome sequence of a third distinct baculovirus isolated from the true armyworm, Mythimna unipuncta, contains two copies of the lef-7 gene. Virus Genes 2018, 54, 297–310. [Google Scholar] [CrossRef]
- Ishimwe, E.; Hodgson, J.J.; Clem, R.J.; Passarelli, A.L. Reaching the melting point: Degradative enzymes and protease inhibitors involved in baculovirus infection and dissemination. Virology 2015, 479–480, 637–649. [Google Scholar] [CrossRef]
- Snider, J.; Thibault, G.; Houry, W.A. The AAA plus superfamily of functionally diverse proteins. Genome Biol. 2008, 9, 216. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Qi, Y. Genome sequence of Leucania seperata nucleopolyhedrovirus. Virus Genes 2007, 35, 845–856. [Google Scholar] [CrossRef] [PubMed]
- Iyer, L.M.; Leipe, D.D.; Koonin, E.V.; Aravind, L. Evolutionary history and higher order classification of AAA plus ATPases. J. Struct. Biol. 2004, 146, 11–31. [Google Scholar] [CrossRef] [PubMed]
- de Jong, J.G.; Lauzon, H.A.; Dominy, C.; Poloumienko, A.; Carstens, E.B.; Arif, B.M.; Krell, P.J. Analysis of the Choristoneura fumiferana nucleopolyhedrovirus genome. J. Gen. Virol. 2005, 86, 929–943. [Google Scholar] [CrossRef]
- Slavicek, J.M. Baculovirus enhancins and their role in viral pathogenicity. In Molecular Virology; Adoga, M., Ed.; IntechOpen: London, UK, 2012; pp. 147–168. [Google Scholar]
- Derksen, A.C.; Granados, R.R. Alteration of a lepidopteran peritrophic membrane by baculoviruses and enhancement of viral infectivity. Virology 1988, 167, 242–250. [Google Scholar] [CrossRef]
- Toprak, U.; Harris, S.; Baldwin, D.; Theilmann, D.; Gillott, C.; Hegedus, D.D.; Erlandson, M.A. Role of enhancin in Mamestra configurata nucleopolyhedrovirus virulence: Selective degradation of host peritrophic matrix proteins. J. Gen. Virol. 2012, 93, 744–753. [Google Scholar] [CrossRef]
- Hegedus, D.; Erlandson, M.; Gillott, C.; Toprak, U. New insights into peritrophic matrix synthesis, architecture, and function. Annu. Rev. Entomol. 2009, 54, 285–302. [Google Scholar] [CrossRef]
- Uchima, K.; Harvey, J.P.; Omi, E.M.; Tanada, Y. Binding-sites on the midgut cell-membrane for the synergistic factor of a granulosis-virus of the armyworm (Pseudaletia-unipuncta). Insect Biochem. 1988, 18, 645–650. [Google Scholar] [CrossRef]
- Hoover, K.; Humphries, M.A.; Gendron, A.R.; Slavicek, J.M. Impact of viral enhancin genes on potency of Lymantria dispar multiple nucleopolyhedrovirus in L. dispar following disruption of the peritrophic matrix. J. Invertebr. Pathol. 2010, 104, 150–152. [Google Scholar] [CrossRef]
- Thumbi, D.K.; Beliveau, C.; Cusson, M.; Lapointe, R.; Lucarotti, C.J. Comparative genome sequence analysis of Choristoneura occidentalis Freeman and C. rosaceana Harris (Lepidoptera: Tortricidae) alphabaculoviruses. PLoS ONE 2013, 8, e68968. [Google Scholar] [CrossRef][Green Version]
- Rawlings, N.D.; Barrett, A.J. Evolutionary families of metallopeptidases. Method Enzymol. 1995, 248, 183–228. [Google Scholar]
- Wennmann, J.T.; Gueli Alletti, G.; Jehle, J.A. The genome sequence of Agrotis segetum nucleopolyhedrovirus B (AgseNPV-B) reveals a new baculovirus species within the Agrotis baculovirus complex. Virus Genes 2015, 50, 260–276. [Google Scholar] [CrossRef] [PubMed]
- Abd-Alla, A.; Jousset, F.X.; Li, Y.; Fediere, G.; Cousserans, F.; Bergoin, M. NS-3 protein of the Junonia coenia densovirus is essential for viral DNA replication in an Ld 652 cell line and Spodoptera littoralis larvae. J. Virol. 2004, 78, 790–797. [Google Scholar] [CrossRef]
- Fan, Q.; Li, S.; Wang, L.; Zhang, B.; Ye, B.; Zhao, Z.; Cui, L. The genome sequence of the multinucleocapsid nucleopolyhedrovirus of the Chinese oak silkworm Antheraea pernyi. Virology 2007, 366, 304–315. [Google Scholar] [CrossRef]
- Oliveira, J.V.; Wolff, J.L.; Garcia-Maruniak, A.; Ribeiro, B.M.; de Castro, M.E.; de Souza, M.L.; Moscardi, F.; Maruniak, J.E.; Zanotto, P.M. Genome of the most widely used viral biopesticide: Anticarsia gemmatalis multiple nucleopolyhedrovirus. J. Gen. Virol. 2006, 87, 3233–3250. [Google Scholar] [CrossRef] [PubMed]
- Hyink, O.; Dellow, R.A.; Olsen, M.J.; Caradoc-Davies, K.M.; Drake, K.; Herniou, E.A.; Cory, J.S.; O’Reilly, D.R.; Ward, V.K. Whole genome analysis of the Epiphyas postvittana nucleopolyhedrovirus. J. Gen. Virol. 2002, 83, 957–971. [Google Scholar] [CrossRef] [PubMed]
- Boudhraa, Z.; Carmona, E.; Provencher, D.; Mes-Masson, A.M. Ran GTPase: A key player in tumor progression and metastasis. Front. Cell. Dev. Biol. 2020, 8, 345. [Google Scholar] [CrossRef] [PubMed]
- Joseph, J. Ran at a glance. J. Cell Sci. 2006, 119, 3481–3484. [Google Scholar] [CrossRef][Green Version]
- Stewart, M.; Kent, H.M.; McCoy, A.J. Structural basis for molecular recognition between nuclear transport factor 2 (NTF2) and the GDP-bound form of the Ras-family GTPase Ran. J. Mol. Biol. 1998, 277, 635–646. [Google Scholar] [CrossRef]
- Vetter, I.R.; Nowak, C.; Nishimoto, T.; Kuhlmann, J.; Wittinghofer, A. Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: Implications for nuclear transport. Nature 1999, 398, 39–46. [Google Scholar] [CrossRef]
- Ahrens, C.H.; Russell, R.L.; Funk, C.J.; Evans, J.T.; Harwood, S.H.; Rohrmann, G.F. The sequence of the Orgyia pseudotsugata multinucleocapsid nuclear polyhedrosis virus genome. Virology 1997, 229, 381–399. [Google Scholar] [CrossRef] [PubMed]
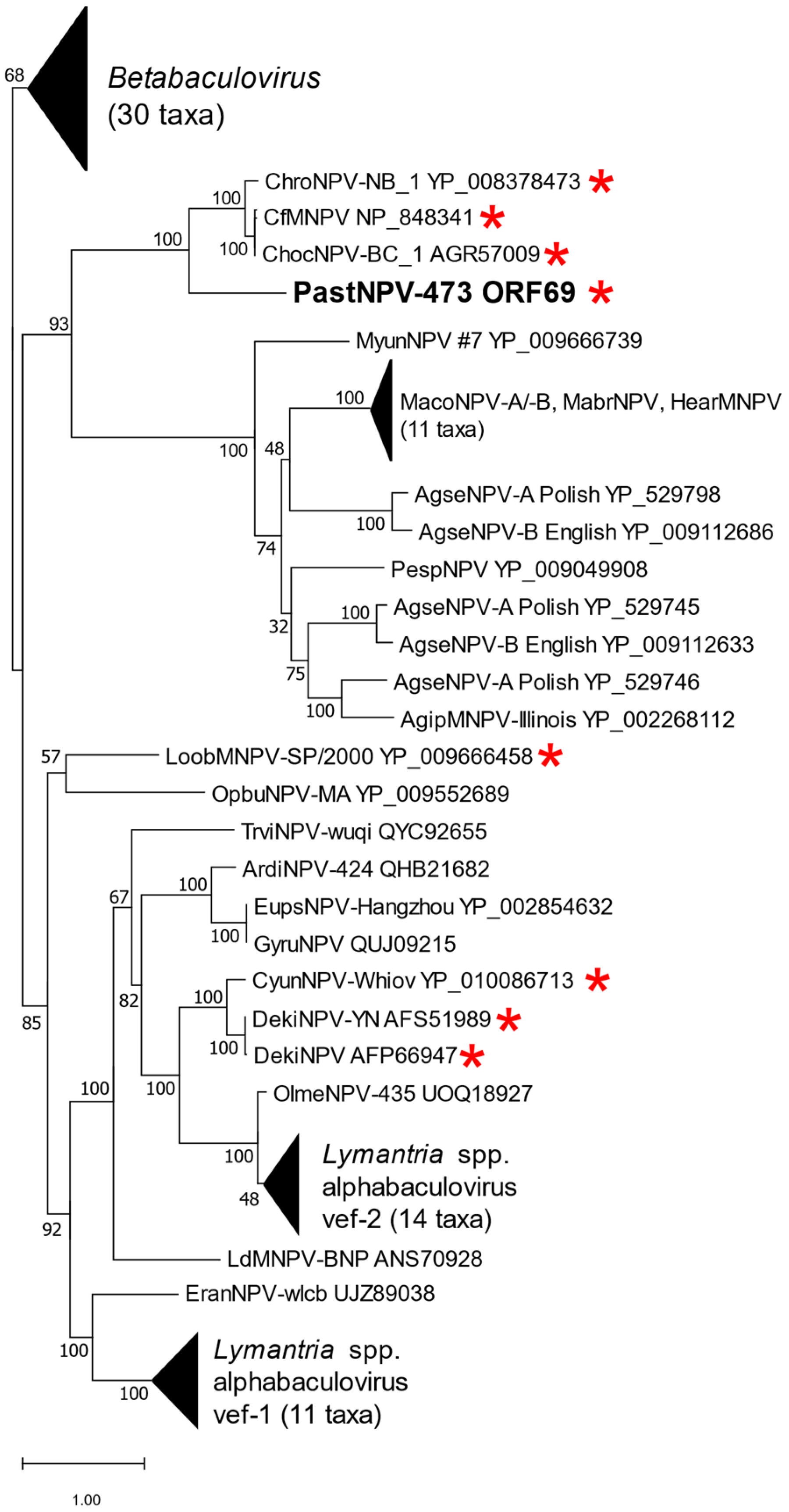
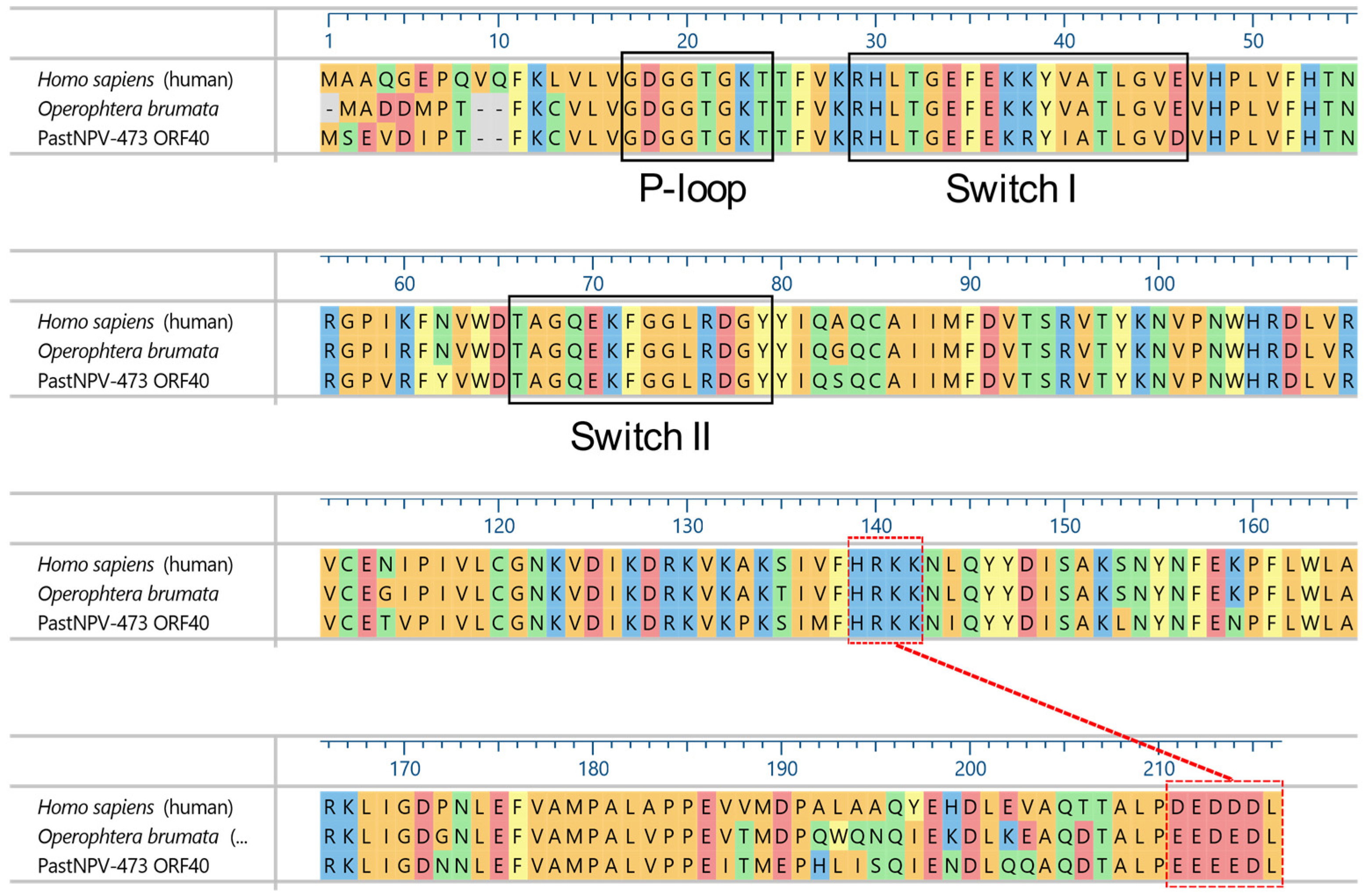
| ORF | Position (nt) | Amino Acids/kDa | Promoter Motifs 1 | BLASTx Match |
|---|---|---|---|---|
| 10 | 11799⟵12035 | 78/8.7 | C, L | hypothetical protein [Chitinophaga caeni] |
| 15 | 17875⟵18756 | 293/35.5 | C, L, T | - |
| 27 | 27159⟵27761 | 200/23.3 | C | - |
| 40 | 36077⟵36721 | 214/24.6 | C | PREDICTED: GTP-binding nuclear protein Ran [Amyelois transitella] |
| 92 | 85688⟵86476 | 262/31.6 | C, L, T | - |
| 104 | 95660⟵96394 | 244/29.2 | C, T | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harrison, R.L.; Rowley, D.L. The Parapoynx stagnalis Nucleopolyhedrovirus (PastNPV), a Divergent Member of the Alphabaculovirus Group I Clade, Encodes a Homolog of Ran GTPase. Viruses 2022, 14, 2289. https://doi.org/10.3390/v14102289
Harrison RL, Rowley DL. The Parapoynx stagnalis Nucleopolyhedrovirus (PastNPV), a Divergent Member of the Alphabaculovirus Group I Clade, Encodes a Homolog of Ran GTPase. Viruses. 2022; 14(10):2289. https://doi.org/10.3390/v14102289
Chicago/Turabian StyleHarrison, Robert L., and Daniel L. Rowley. 2022. "The Parapoynx stagnalis Nucleopolyhedrovirus (PastNPV), a Divergent Member of the Alphabaculovirus Group I Clade, Encodes a Homolog of Ran GTPase" Viruses 14, no. 10: 2289. https://doi.org/10.3390/v14102289
APA StyleHarrison, R. L., & Rowley, D. L. (2022). The Parapoynx stagnalis Nucleopolyhedrovirus (PastNPV), a Divergent Member of the Alphabaculovirus Group I Clade, Encodes a Homolog of Ran GTPase. Viruses, 14(10), 2289. https://doi.org/10.3390/v14102289





