Recent Emergence of Bovine Coronavirus Variants with Mutations in the Hemagglutinin-Esterase Receptor Binding Domain in U.S. Cattle
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples and Ethics Statement
2.2. Sample Preparation and Illumina Sequencing
2.3. Phylogenetic Analysis
2.4. Recombination Detection in HE Gene Sequences
2.5. Expression of Wild Type and Variant Hemagglutinin Esterases
2.6. Hemagglutinin Esterase Lectin Activity
2.7. Hemagglutinin Esterase Structure Modeling
3. Results
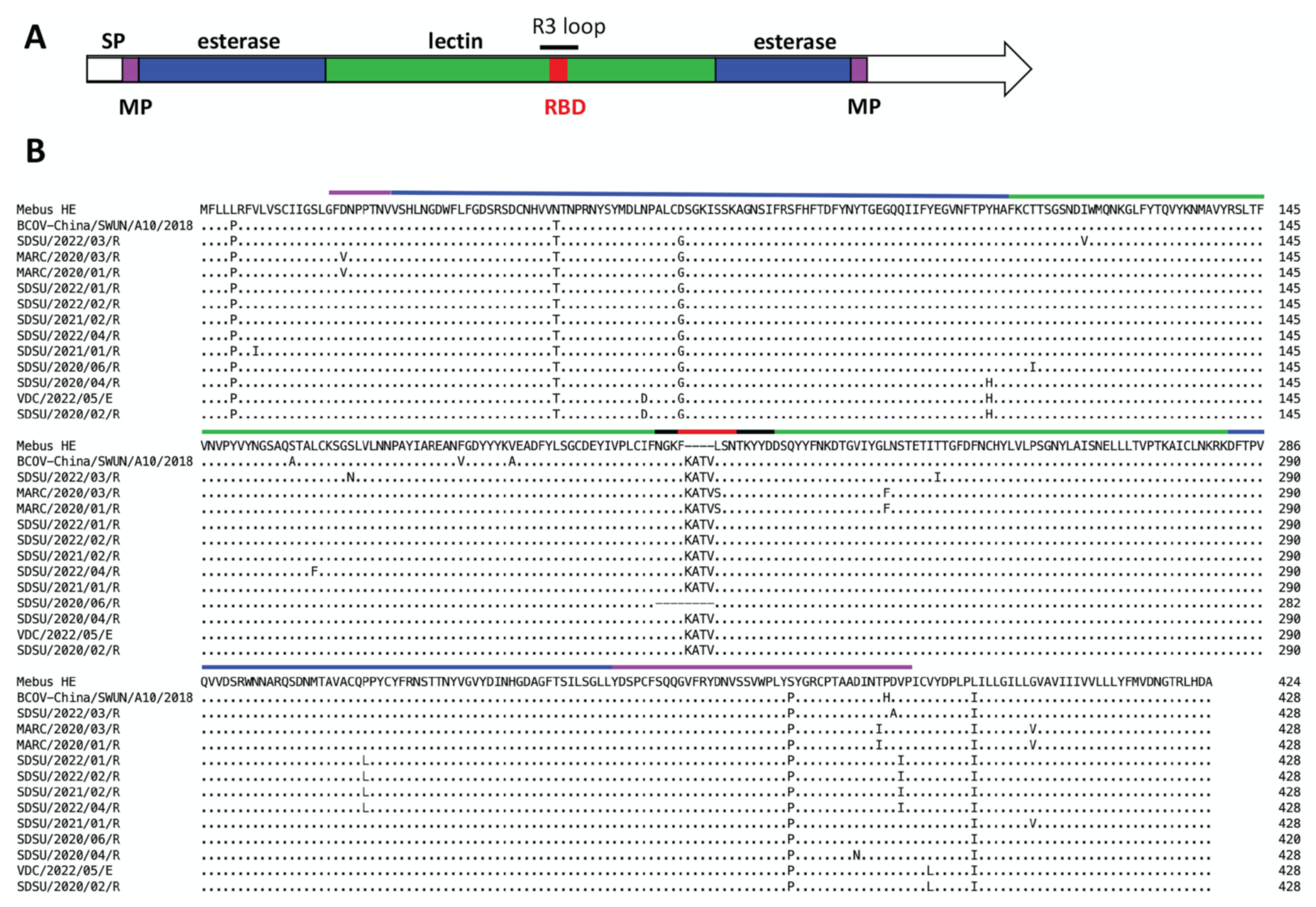
3.1. Whole Genome Sequence Analysis
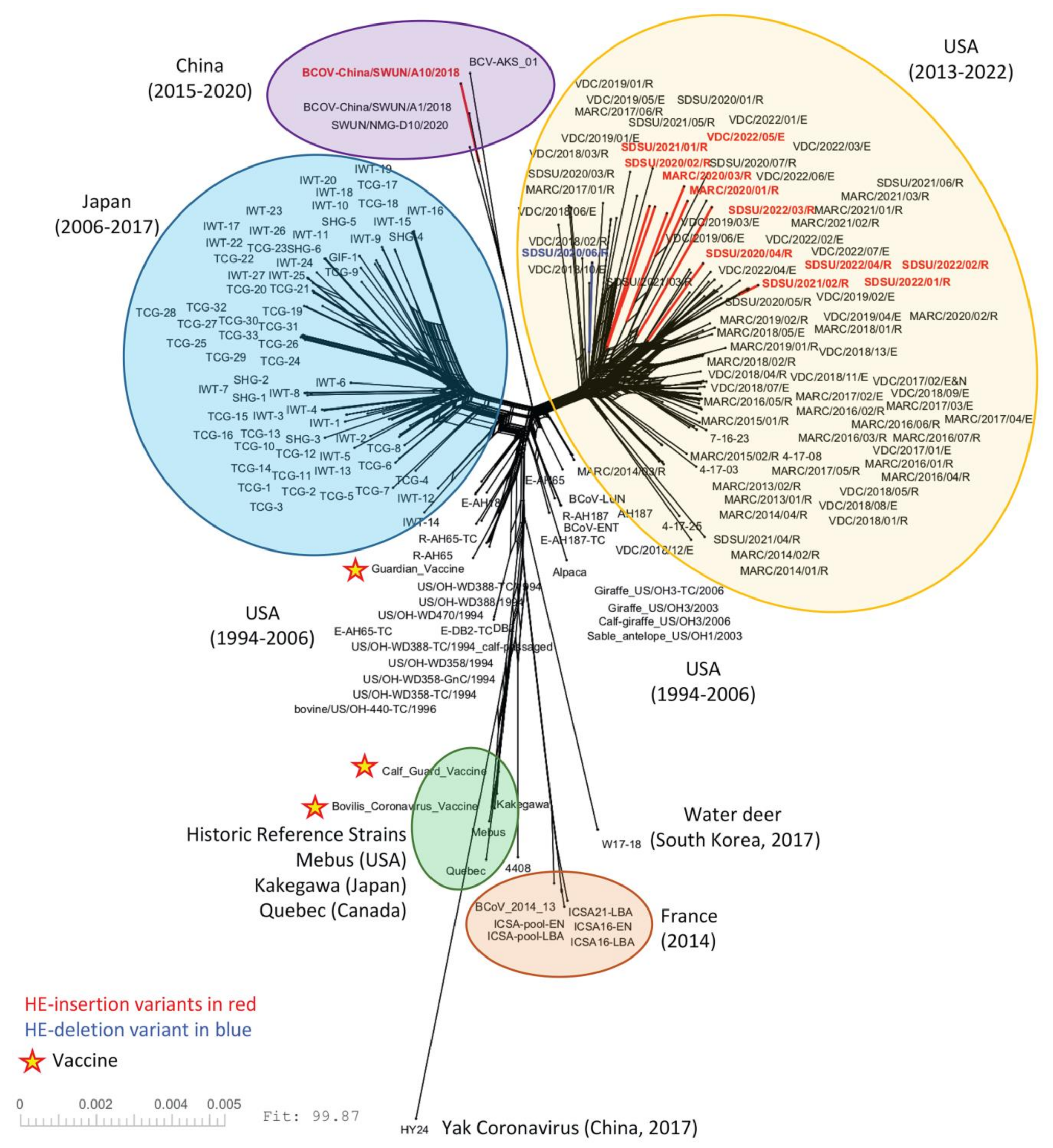
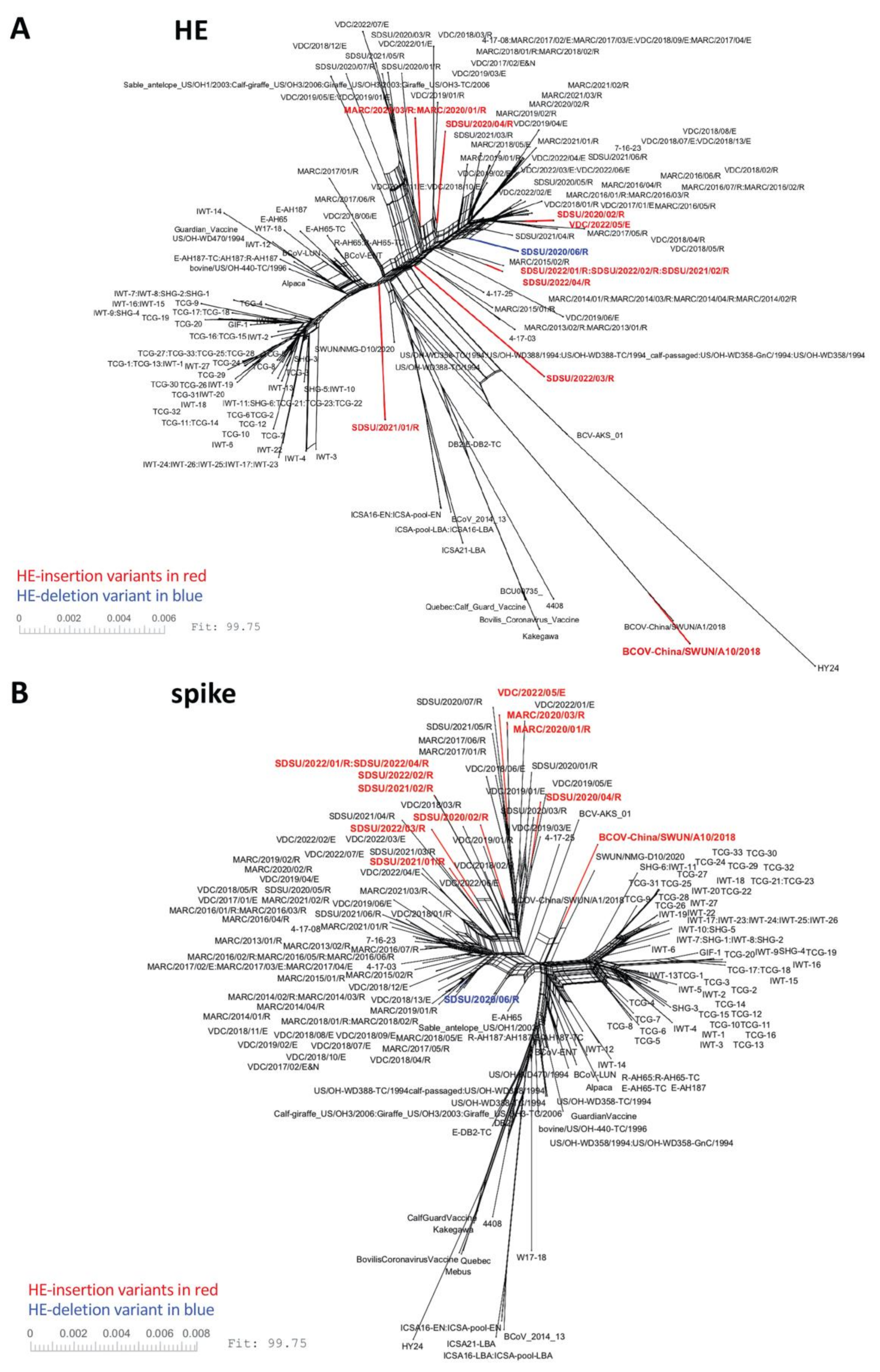
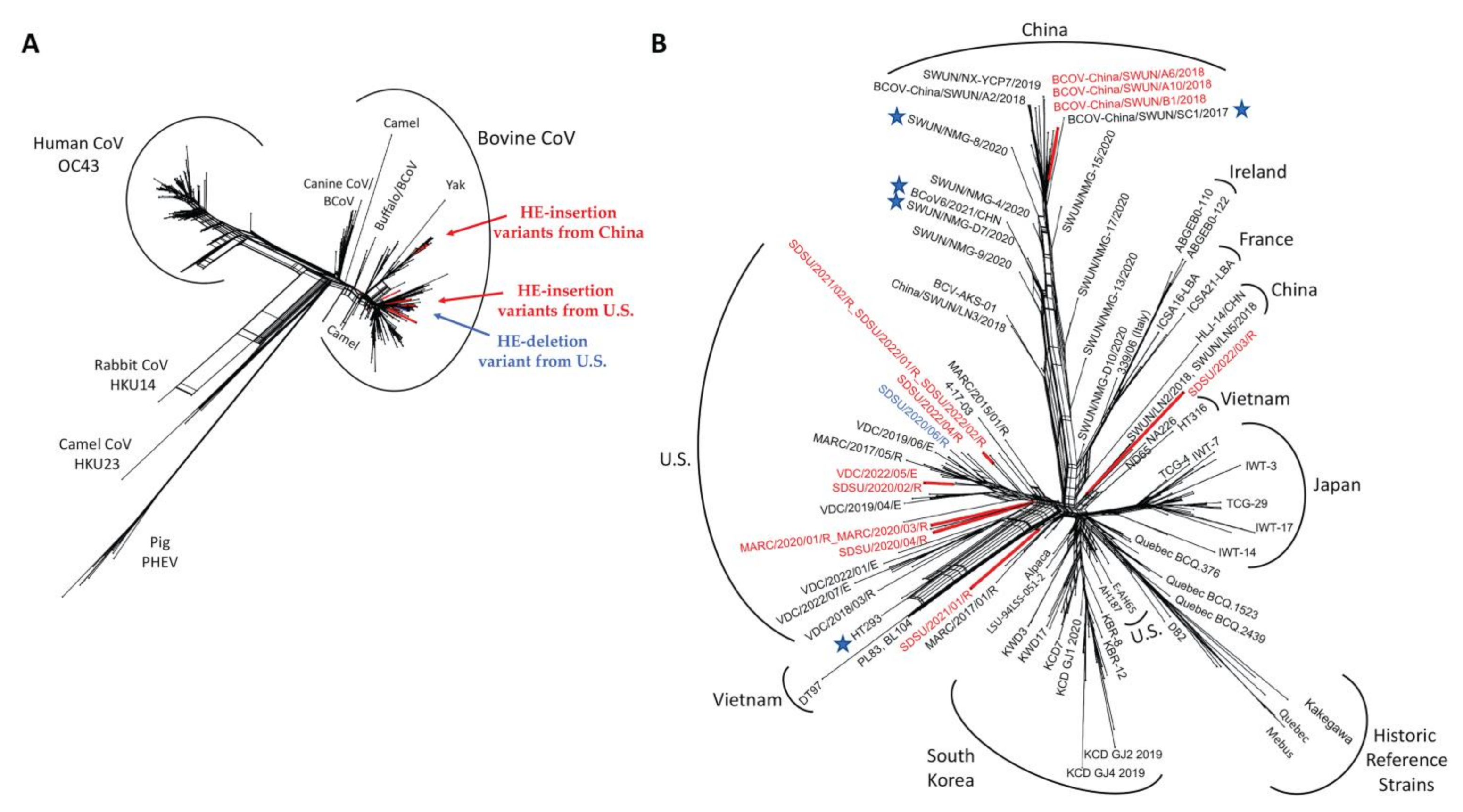
3.2. Phylogenetic Analysis
3.3. Recombination Detection
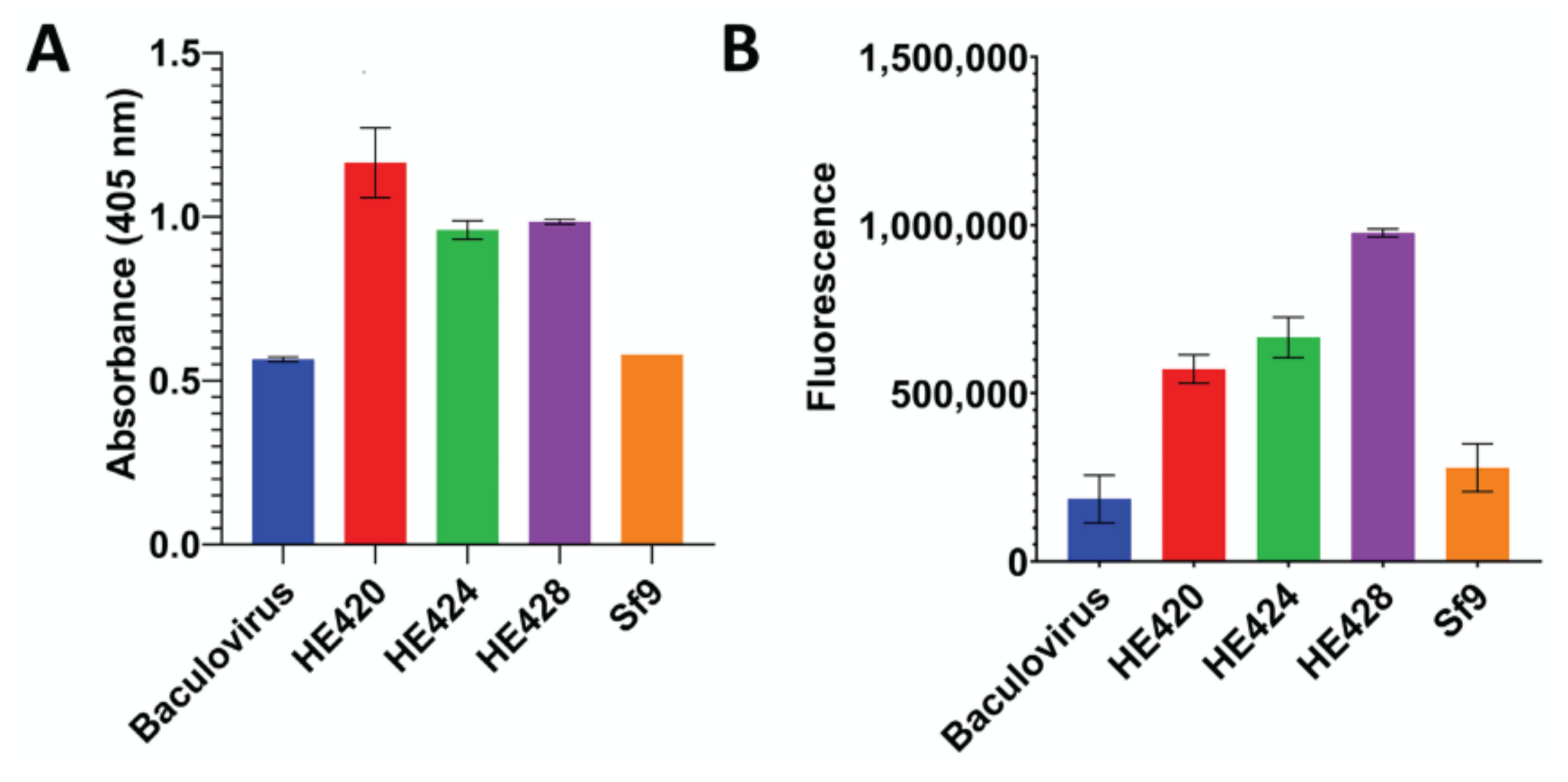
3.4. Baculovirus-Mediated Expression of Hemagglutinin Esterase Variants in Sf9 Cells
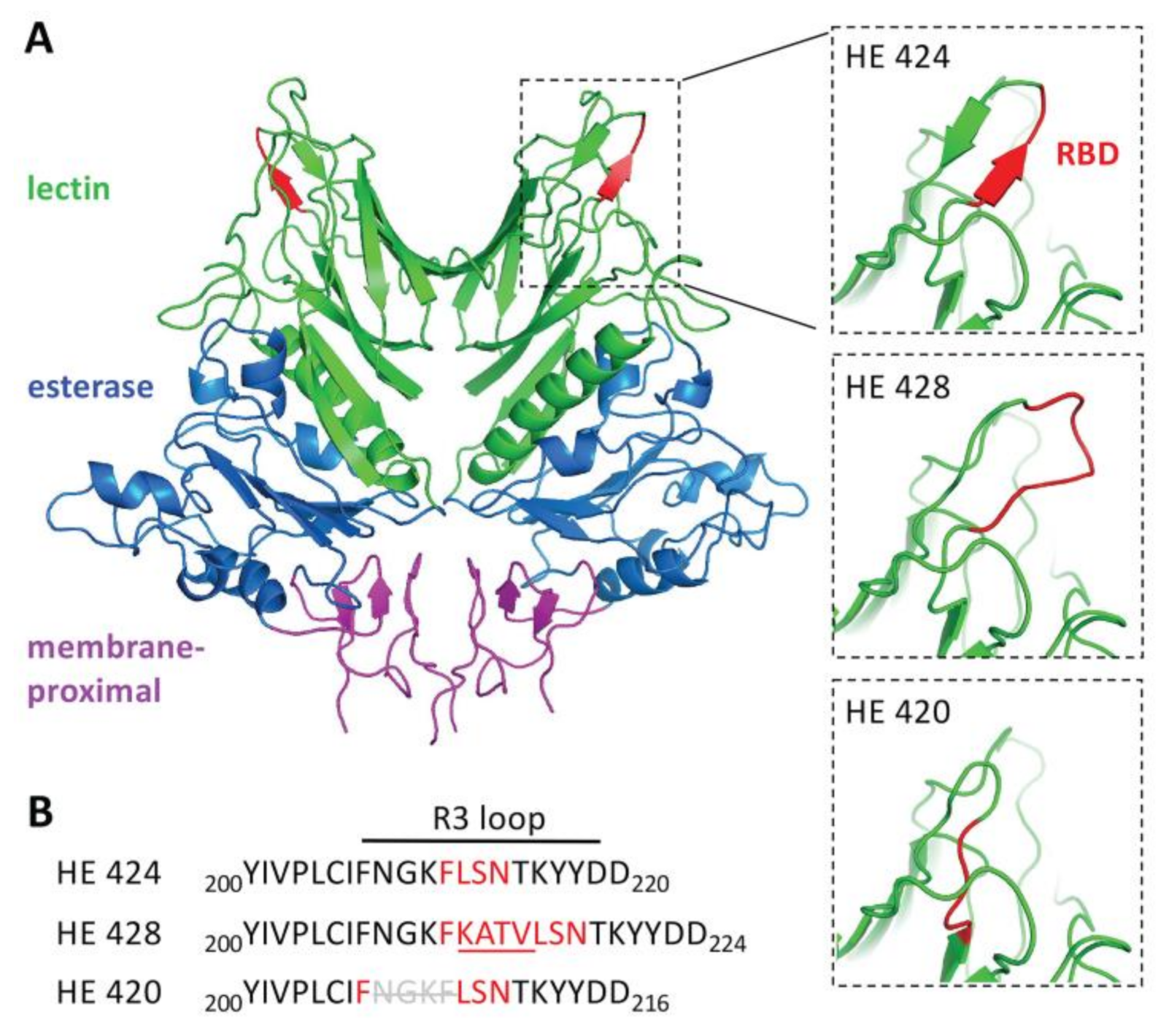
3.5. Hemagglutinin Esterase Structural Modeling
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hasoksuz, M.; Sreevatsan, S.; Cho, K.O.; Hoet, A.E.; Saif, L.J. Molecular analysis of the S1 subunit of the spike glycoprotein of respiratory and enteric bovine coronavirus isolates. Virus Res. 2002, 84, 101–109. [Google Scholar] [CrossRef]
- Saif, L.J. Bovine respiratory coronavirus. Vet. Clin. N. Am. Food Anim. Pract. 2010, 26, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Griffin, D.; Chengappa, M.M.; Kuszak, J.; McVey, D.S. Bacterial pathogens of the bovine respiratory disease complex. Vet. Clin. N. Am. Food Anim. Pract. 2010, 26, 381–394. [Google Scholar] [CrossRef]
- Foster, D.M.; Smith, G.W. Pathophysiology of diarrhea in calves. Vet. Clin. N. Am. Food Anim. Pract. 2009, 25, 13–36, xi. [Google Scholar] [CrossRef]
- Yoo, D.; Pei, Y. Full-length genomic sequence of bovine coronavirus (31 kb). Completion of the open reading frame 1a/1b sequences. Adv. Exp. Med. Biol. 2001, 494, 73–76. [Google Scholar]
- Brierley, I.; Boursnell, M.E.; Binns, M.M.; Bilimoria, B.; Blok, V.C.; Brown, T.D.; Inglis, S.C. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 1987, 6, 3779–3785. [Google Scholar] [CrossRef] [PubMed]
- Senanayake, S.D.; Brian, D.A. Bovine coronavirus I protein synthesis follows ribosomal scanning on the bicistronic N mRNA. Virus Res. 1997, 48, 101–105. [Google Scholar] [CrossRef]
- Sanchez, C.M.; Gebauer, F.; Sune, C.; Mendez, A.; Dopazo, J.; Enjuanes, L. Genetic evolution and tropism of transmissible gastroenteritis coronaviruses. Virology 1992, 190, 92–105. [Google Scholar] [CrossRef]
- Woo, P.C.; Lau, S.K.; Huang, Y.; Yuen, K.Y. Coronavirus diversity, phylogeny and interspecies jumping. Exp. Biol. Med. (Maywood) 2009, 234, 1117–1127. [Google Scholar] [CrossRef] [PubMed]
- Alekseev, K.P.; Vlasova, A.N.; Jung, K.; Hasoksuz, M.; Zhang, X.; Halpin, R.; Wang, S.; Ghedin, E.; Spiro, D.; Saif, L.J. Bovine-like coronaviruses isolated from four species of captive wild ruminants are homologous to bovine coronaviruses, based on complete genomic sequences. J. Virol. 2008, 82, 12422–12431. [Google Scholar] [CrossRef]
- Cebra, C.K.; Mattson, D.E.; Baker, R.J.; Sonn, R.J.; Dearing, P.L. Potential pathogens in feces from unweaned llamas and alpacas with diarrhea. J. Am. Vet. Med. Assoc. 2003, 223, 1806–1808. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Guo, Z.; Zhang, B.; Yue, H.; Tang, C. First detection of bovine coronavirus in Yak (Bos grunniens) and a bovine coronavirus genome with a recombinant HE gene. J. Gen. Virol. 2019, 100, 793–803. [Google Scholar] [CrossRef] [PubMed]
- Tsunemitsu, H.; el-Kanawati, Z.R.; Smith, D.R.; Reed, H.H.; Saif, L.J. Isolation of coronaviruses antigenically indistinguishable from bovine coronavirus from wild ruminants with diarrhea. J. Clin. Microbiol. 1995, 33, 3264–3269. [Google Scholar] [CrossRef] [PubMed]
- Hasoksuz, M.; Alekseev, K.; Vlasova, A.; Zhang, X.; Spiro, D.; Halpin, R.; Wang, S.; Ghedin, E.; Saif, L.J. Biologic, antigenic, and full-length genomic characterization of a bovine-like coronavirus isolated from a giraffe. J. Virol. 2007, 81, 4981–4990. [Google Scholar] [CrossRef]
- Majhdi, F.; Minocha, H.C.; Kapil, S. Isolation and characterization of a coronavirus from elk calves with diarrhea. J. Clin. Microbiol. 1997, 35, 2937–2942. [Google Scholar] [CrossRef]
- Wunschmann, A.; Frank, R.; Pomeroy, K.; Kapil, S. Enteric coronavirus infection in a juvenile dromedary (Camelus dromedarius). J. Vet. Diagn. Investig. 2002, 14, 441–444. [Google Scholar] [CrossRef] [PubMed]
- Chasey, D.; Reynolds, D.J.; Bridger, J.C.; Debney, T.G.; Scott, A.C. Identification of coronaviruses in exotic species of Bovidae. Vet. Rec. 1984, 115, 602–603. [Google Scholar] [CrossRef]
- Decaro, N.; Lorusso, A. Novel human coronavirus (SARS-CoV-2): A lesson from animal coronaviruses. Vet. Microbiol. 2020, 244, 108693. [Google Scholar] [CrossRef]
- Mounir, S.; Talbot, P.J. Sequence analysis of the membrane protein gene of human coronavirus OC43 and evidence for O-glycosylation. J. Gen. Virol. 1992, 73 Pt 10, 2731–2736. [Google Scholar] [CrossRef] [PubMed]
- Gelinas, A.M.; Sasseville, A.M.; Dea, S. Identification of specific variations within the HE, S1, and ORF4 genes of bovine coronaviruses associated with enteric and respiratory diseases in dairy cattle. Adv. Exp. Med. Biol. 2001, 494, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Vijgen, L.; Keyaerts, E.; Lemey, P.; Maes, P.; Van Reeth, K.; Nauwynck, H.; Pensaert, M.; Van Ranst, M. Evolutionary history of the closely related group 2 coronaviruses: Porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. J. Virol. 2006, 80, 7270–7274. [Google Scholar] [CrossRef]
- Saif, L.J.; Jung, K. Comparative Pathogenesis of Bovine and Porcine Respiratory Coronaviruses in the Animal Host Species and SARS-CoV-2 in Humans. J. Clin. Microbiol. 2020, 58, e01355-20. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Langereis, M.A.; van Vliet, A.L.; Huizinga, E.G.; de Groot, R.J. Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution. Proc. Natl. Acad. Sci. USA 2008, 105, 9065–9069. [Google Scholar] [CrossRef] [PubMed]
- Schultze, B.; Wahn, K.; Klenk, H.D.; Herrler, G. Isolated HE-protein from hemagglutinating encephalomyelitis virus and bovine coronavirus has receptor-destroying and receptor-binding activity. Virology 1991, 180, 221–228. [Google Scholar] [CrossRef]
- Lang, Y.; Li, W.; Li, Z.; Koerhuis, D.; van den Burg, A.C.S.; Rozemuller, E.; Bosch, B.J.; van Kuppeveld, F.J.M.; Boons, G.J.; Huizinga, E.G.; et al. Coronavirus hemagglutinin-esterase and spike proteins coevolve for functional balance and optimal virion avidity. Proc. Natl. Acad. Sci. USA 2020, 117, 25759–25770. [Google Scholar] [CrossRef] [PubMed]
- Popova, R.; Zhang, X. The spike but not the hemagglutinin/esterase protein of bovine coronavirus is necessary and sufficient for viral infection. Virology 2002, 294, 222–236. [Google Scholar] [CrossRef][Green Version]
- Storz, J.; Zhang, X.M.; Rott, R. Comparison of hemagglutinating, receptor-destroying, and acetylesterase activities of avirulent and virulent bovine coronavirus strains. Arch. Virol. 1992, 125, 193–204. [Google Scholar] [CrossRef]
- Bakkers, M.J.; Lang, Y.; Feitsma, L.J.; Hulswit, R.J.; de Poot, S.A.; van Vliet, A.L.; Margine, I.; de Groot-Mijnes, J.D.; van Kuppeveld, F.J.; Langereis, M.A.; et al. Betacoronavirus Adaptation to Humans Involved Progressive Loss of Hemagglutinin-Esterase Lectin Activity. Cell Host Microbe 2017, 21, 356–366. [Google Scholar] [CrossRef]
- Langereis, M.A.; Zeng, Q.; Heesters, B.A.; Huizinga, E.G.; de Groot, R.J. The murine coronavirus hemagglutinin-esterase receptor-binding site: A major shift in ligand specificity through modest changes in architecture. PLoS Pathog. 2012, 8, e1002492. [Google Scholar] [CrossRef]
- Bakkers, M.J.; Zeng, Q.; Feitsma, L.J.; Hulswit, R.J.; Li, Z.; Westerbeke, A.; van Kuppeveld, F.J.; Boons, G.J.; Langereis, M.A.; Huizinga, E.G.; et al. Coronavirus receptor switch explained from the stereochemistry of protein-carbohydrate interactions and a single mutation. Proc. Natl. Acad. Sci. USA 2016, 113, E3111–E3119. [Google Scholar] [CrossRef]
- Keha, A.; Xue, L.; Yan, S.; Yue, H.; Tang, C. Prevalence of a novel bovine coronavirus strain with a recombinant hemagglutinin/esterase gene in dairy calves in China. Transbound. Emerg. Dis. 2019, 66, 1971–1981. [Google Scholar] [CrossRef]
- Abi, K.M.; Zhang, Q.; Zhang, B.; Zhou, L.; Yue, H.; Tang, C. An emerging novel bovine coronavirus with a 4-amino-acid insertion in the receptor-binding domain of the hemagglutinin-esterase gene. Arch. Virol. 2020, 165, 3011–3015. [Google Scholar] [CrossRef]
- Decaro, N.; Elia, G.; Campolo, M.; Desario, C.; Mari, V.; Radogna, A.; Colaianni, M.L.; Cirone, F.; Tempesta, M.; Buonavoglia, C. Detection of bovine coronavirus using a TaqMan-based real-time RT-PCR assay. J. Virol. Methods 2008, 151, 167–171. [Google Scholar] [CrossRef]
- Workman, A.M.; Kuehn, L.A.; McDaneld, T.G.; Clawson, M.L.; Loy, J.D. Longitudinal study of humoral immunity to bovine coronavirus, virus shedding, and treatment for bovine respiratory disease in pre-weaned beef calves. BMC Vet. Res. 2019, 15, 161. [Google Scholar] [CrossRef]
- Workman, A.M.; Heaton, M.P.; Harhay, G.P.; Smith, T.P.; Grotelueschen, D.M.; Sjeklocha, D.; Brodersen, B.; Petersen, J.L.; Chitko-McKown, C.G. Resolving Bovine viral diarrhea virus subtypes from persistently infected U.S. beef calves with complete genome sequence. J. Vet. Diagn. Investig. Off. Publ. Am. Assoc. Vet. Lab. Diagn. Inc 2016, 28, 519–528. [Google Scholar] [CrossRef]
- Hause, B.M.; Collin, E.A.; Anderson, J.; Hesse, R.A.; Anderson, G. Bovine rhinitis viruses are common in U.S. cattle with bovine respiratory disease. PLoS ONE 2015, 10, e0121998. [Google Scholar] [CrossRef]
- Huson, D.H. SplitsTree: Analyzing and visualizing evolutionary data. Bioinformatics 1998, 14, 68–73. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Bryant, D.; Moulton, V. Neighbor-net: An agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 2004, 21, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Dress, A.W.; Huson, D.H. Constructing splits graphs. IEEE/ACM Trans. Comput. Biol. Bioinform. 2004, 1, 109–115. [Google Scholar] [CrossRef]
- Martin, D.P.; Varsani, A.; Roumagnac, P.; Botha, G.; Maslamoney, S.; Schwab, T.; Kelz, Z.; Kumar, V.; Murrell, B. RDP5: A computer program for analyzing recombination in, and removing signals of recombination from, nucleotide sequence datasets. Virus Evol. 2021, 7, veaa087. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.; Rybicki, E. RDP: Detection of recombination amongst aligned sequences. Bioinformatics 2000, 16, 562–563. [Google Scholar] [CrossRef]
- Padidam, M.; Sawyer, S.; Fauquet, C.M. Possible emergence of new geminiviruses by frequent recombination. Virology 1999, 265, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Posada, D.; Crandall, K.A.; Williamson, C. A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res. Hum. Retrovir. 2005, 21, 98–102. [Google Scholar] [CrossRef]
- Smith, J.M. Analyzing the mosaic structure of genes. J. Mol. Evol. 1992, 34, 126–129. [Google Scholar] [CrossRef]
- Posada, D.; Crandall, K.A. Evaluation of methods for detecting recombination from DNA sequences: Computer simulations. Proc. Natl. Acad. Sci. USA 2001, 98, 13757–13762. [Google Scholar] [CrossRef]
- Gibbs, M.J.; Armstrong, J.S.; Gibbs, A.J. Sister-scanning: A Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 2000, 16, 573–582. [Google Scholar] [CrossRef]
- Lam, H.M.; Ratmann, O.; Boni, M.F. Improved Algorithmic Complexity for the 3SEQ Recombination Detection Algorithm. Mol. Biol. Evol. 2018, 35, 247–251. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- So, R.T.Y.; Chu, D.K.W.; Miguel, E.; Perera, R.; Oladipo, J.O.; Fassi-Fihri, O.; Aylet, G.; Ko, R.L.W.; Zhou, Z.; Cheng, M.S.; et al. Diversity of Dromedary Camel Coronavirus HKU23 in African Camels Revealed Multiple Recombination Events among Closely Related Betacoronaviruses of the Subgenus Embecovirus. J. Virol. 2019, 93, e01236-19. [Google Scholar] [CrossRef] [PubMed]
- Langereis, M.A.; Zeng, Q.; Gerwig, G.J.; Frey, B.; von Itzstein, M.; Kamerling, J.P.; de Groot, R.J.; Huizinga, E.G. Structural basis for ligand and substrate recognition by torovirus hemagglutinin esterases. Proc. Natl. Acad. Sci. USA 2009, 106, 15897–15902. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, P.B.; Zhang, X.; Formanowski, F.; Fitz, W.; Wong, C.H.; Meier-Ewert, H.; Skehel, J.J.; Wiley, D.C. Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus. Nature 1998, 396, 92–96. [Google Scholar] [CrossRef]
- Deregt, D.; Babiuk, L.A. Monoclonal antibodies to bovine coronavirus: Characteristics and topographical mapping of neutralizing epitopes on the E2 and E3 glycoproteins. Virology 1987, 161, 410–420. [Google Scholar] [CrossRef]
- Storz, J.; Herrler, G.; Snodgrass, D.R.; Hussain, K.A.; Zhang, X.M.; Clark, M.A.; Rott, R. Monoclonal antibodies differentiate between the haemagglutinating and the receptor-destroying activities of bovine coronavirus. J. Gen. Virol. 1991, 72 Pt 11, 2817–2820. [Google Scholar] [CrossRef]
- Deregt, D.; Gifford, G.A.; Ijaz, M.K.; Watts, T.C.; Gilchrist, J.E.; Haines, D.M.; Babiuk, L.A. Monoclonal antibodies to bovine coronavirus glycoproteins E2 and E3: Demonstration of in vivo virus-neutralizing activity. J. Gen. Virol. 1989, 70 Pt 4, 993–998. [Google Scholar] [CrossRef]
- Suzuki, T.; Otake, Y.; Uchimoto, S.; Hasebe, A.; Goto, Y. Genomic Characterization and Phylogenetic Classification of Bovine Coronaviruses Through Whole Genome Sequence Analysis. Viruses 2020, 12, 183. [Google Scholar] [CrossRef]
- Park, S.J.; Kim, G.Y.; Choy, H.E.; Hong, Y.J.; Saif, L.J.; Jeong, J.H.; Park, S.I.; Kim, H.H.; Kim, S.K.; Shin, S.S.; et al. Dual enteric and respiratory tropisms of winter dysentery bovine coronavirus in calves. Arch. Virol. 2007, 152, 1885–1900. [Google Scholar] [CrossRef]
- Bidokhti, M.R.; Traven, M.; Ohlson, A.; Baule, C.; Hakhverdyan, M.; Belak, S.; Liu, L.; Alenius, S. Tracing the transmission of bovine coronavirus infections in cattle herds based on S gene diversity. Vet. J. 2012, 193, 386–390. [Google Scholar] [CrossRef]
- Beuttemmuller, E.A.; Alfieri, A.F.; Headley, S.A.; Alfieri, A.A. Brazilian strain of bovine respiratory coronavirus is derived from dual enteric and respiratory tropism. Genet. Mol. Res. 2017, 16, 28387879. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Hasoksuz, M.; Spiro, D.; Halpin, R.; Wang, S.; Vlasova, A.; Janies, D.; Jones, L.R.; Ghedin, E.; Saif, L.J. Quasispecies of bovine enteric and respiratory coronaviruses based on complete genome sequences and genetic changes after tissue culture adaptation. Virology 2007, 363, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J. What is the evidence that bovine coronavirus is a biologically significant respiratory pathogen in cattle? Can. Vet. J. La Rev. Vet. Can. 2019, 60, 147–152. [Google Scholar]
- Vlasova, A.N.; Saif, L.J. Bovine Coronavirus and the Associated Diseases. Front. Vet. Sci. 2021, 8, 643220. [Google Scholar] [CrossRef]
- Vijgen, L.; Keyaerts, E.; Moes, E.; Thoelen, I.; Wollants, E.; Lemey, P.; Vandamme, A.M.; Van Ranst, M. Complete genomic sequence of human coronavirus OC43: Molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J. Virol. 2005, 79, 1595–1604. [Google Scholar] [CrossRef] [PubMed]
- Vijgen, L.; Lemey, P.; Keyaerts, E.; Van Ranst, M. Genetic variability of human respiratory coronavirus OC43. J. Virol. 2005, 79, 3223–3224, author reply 3224–3225. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Workman, A.M.; McDaneld, T.G.; Harhay, G.P.; Das, S.; Loy, J.D.; Hause, B.M. Recent Emergence of Bovine Coronavirus Variants with Mutations in the Hemagglutinin-Esterase Receptor Binding Domain in U.S. Cattle. Viruses 2022, 14, 2125. https://doi.org/10.3390/v14102125
Workman AM, McDaneld TG, Harhay GP, Das S, Loy JD, Hause BM. Recent Emergence of Bovine Coronavirus Variants with Mutations in the Hemagglutinin-Esterase Receptor Binding Domain in U.S. Cattle. Viruses. 2022; 14(10):2125. https://doi.org/10.3390/v14102125
Chicago/Turabian StyleWorkman, Aspen M., Tara G. McDaneld, Gregory P. Harhay, Subha Das, John Dustin Loy, and Benjamin M. Hause. 2022. "Recent Emergence of Bovine Coronavirus Variants with Mutations in the Hemagglutinin-Esterase Receptor Binding Domain in U.S. Cattle" Viruses 14, no. 10: 2125. https://doi.org/10.3390/v14102125
APA StyleWorkman, A. M., McDaneld, T. G., Harhay, G. P., Das, S., Loy, J. D., & Hause, B. M. (2022). Recent Emergence of Bovine Coronavirus Variants with Mutations in the Hemagglutinin-Esterase Receptor Binding Domain in U.S. Cattle. Viruses, 14(10), 2125. https://doi.org/10.3390/v14102125






