Increased sHLA-G Is Associated with Improved COVID-19 Outcome and Reduced Neutrophil Adhesion
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients
2.2. sHLA-G Specific ELISA
2.3. sHLA-E Specific ELISA
2.4. HLA-E Allele Assignment
2.5. HLA-G Allele Assignment
2.6. Endothelial Activation Biomarkers Levels Assay
2.7. Cell Cultures
2.8. Neutrophil Binding Assay
2.9. sE-Selectin and sICAM1 Assay
2.10. FGF2 Expression Assay
2.11. Statistical Analysis
3. Results
3.1. Study Population
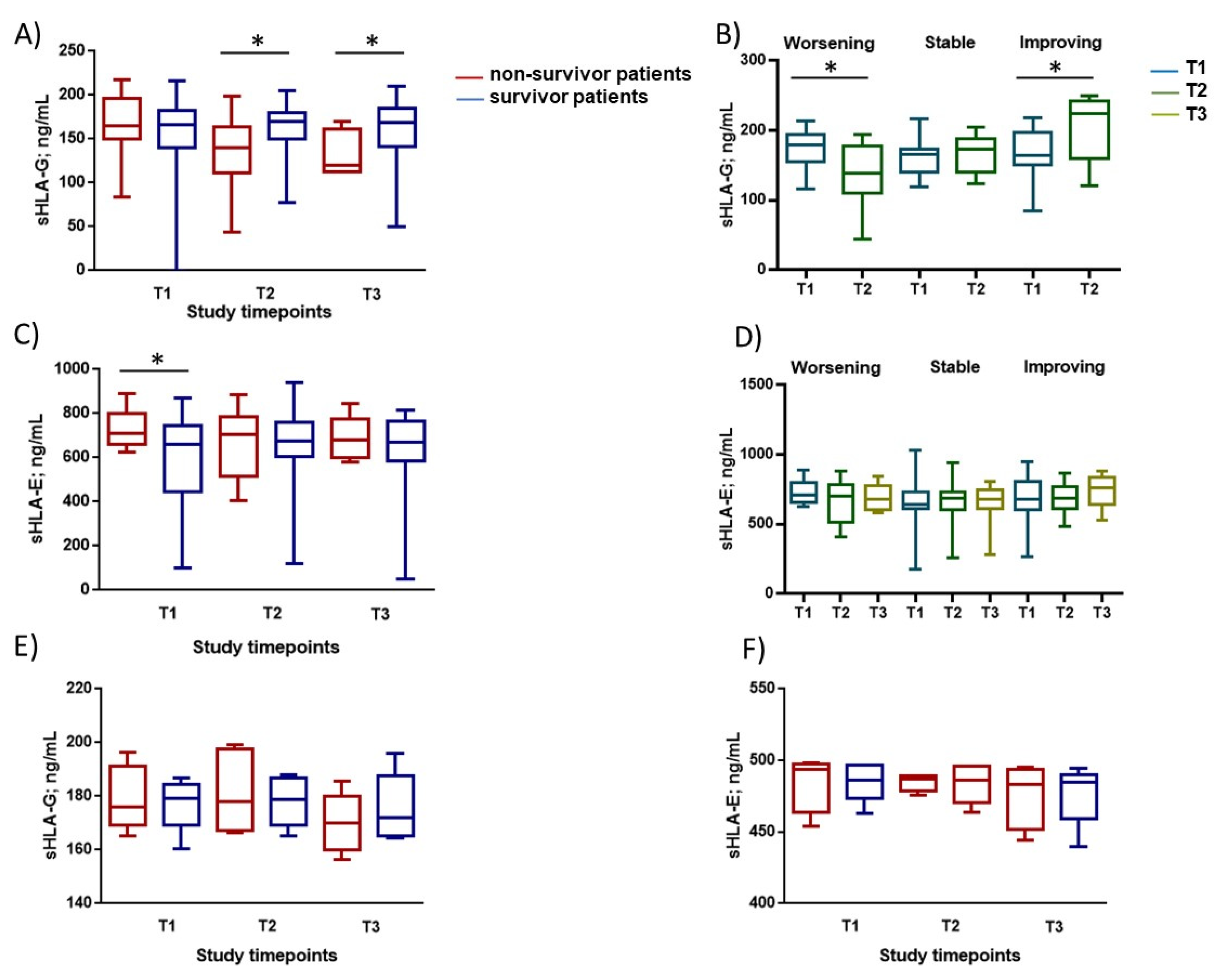
3.2. Immunological Parameters Evaluation
3.3. Allelic Frequencies of HLA-G and HLA-E Genes
3.4. Correlations between Blood sHLA-G Levels and Endothelial Activation Biomarkers in COVID-19 Patients
3.5. Endothelial Cell Response to HLA-G Molecules and Neutrophil Adhesion
3.6. Correlations between Blood sHLA-G Levels and Neutrophil Adhesion to Activated Endothelial Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carosella, E.D.; Paul, P.; Moreau, P.; Rouas-Freiss, N. HLA-G and HLA-E: Fundamental and pathophysiological aspects. Immunol. Today 2000, 21, 532–534. [Google Scholar] [CrossRef]
- Le Bouteiller, P.; Lenfant, F. Antigen-presenting function(s) of the non-classical HLA-E, -F and -G class I molecules: The beginning of a story. Res. Immunol. 1996, 147, 301–313. [Google Scholar] [CrossRef]
- O’Callaghan, C.A.; Bell, J.I. Structure and function of the human MHC class Ib molecules HLA-E, HLA-F and HLA-G. Immunol. Rev. 1998, 163, 129–138. [Google Scholar] [CrossRef]
- Kren, L.; Slaby, O.; Muckova, K.; Lzicarova, E.; Sova, M.; Vybihal, V.; Svoboda, T.; Fadrus, P.; Lakomy, R.; Vanhara, P.; et al. Expression of immune-modulatory molecules HLA-G and HLA-E by tumor cells in glioblastomas: An unexpected prognostic significance? Neuropathology 2011, 31, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Rabot, M.; Tabiasco, J.; Polgar, B.; Aguerre-Girr, M.; Berrebi, A.; Bensussan, A.; Strbo, N.; Rukavina, D.; Le Bouteiller, P. HLA Class I/NK Cell Receptor Interaction in Early Human Decidua basalis: Possible Functional Consequences. Food Allergy Mol. Basis Clin. Pract. 2005, 89, 72–83. [Google Scholar] [CrossRef]
- Amiot, L.; Vu, N.; Samson, M. Biology of the immunomodulatory molecule HLA-G in human liver diseases. J. Hepatol. 2015, 62, 1430–1437. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, R.; Fainardi, E.; Rouas-Freiss, N.; Morandi, F. The Role of HLA-Class Ib Molecules in Immune-Related Diseases, Tumors, and Infections 2016. J. Immunol. Res. 2017, 2017, 1–2. [Google Scholar] [CrossRef]
- Rizzo, R.; Bortolotti, D.; Bolzani, S.; Fainardi, E. HLA-G Molecules in Autoimmune Diseases and Infections. Front. Immunol. 2014, 5, 592. [Google Scholar] [CrossRef] [Green Version]
- Fons, P.; Chabot, S.; Cartwright, J.E.; Lenfant, F.; L’Faqihi, F.; Giustiniani, J.; Herault, J.-P.; Gueguen, G.; Bono, F.; Savi, P.; et al. Soluble HLA-G1 inhibits angiogenesis through an apoptotic pathway and by direct binding to CD160 receptor expressed by endothelial cells. Blood 2006, 108, 2608–2615. [Google Scholar] [CrossRef] [Green Version]
- Rebmann, V.; van der Ven, K.; Pässler, M.; Pfeiffer, K.; Krebs, D.; Grosse-Wilde, H. Association of soluble HLA-G plasma levels with HLA-G alleles. Tissue Antigens 2001, 57, 15–21. [Google Scholar] [CrossRef]
- Braud, V.M.; Allan, D.S.J.; O’Callaghan, C.A.; Söderström, K.; D’Andrea, A.; Ogg, G.S.; Lazetic, S.; Young, N.T.; Bell, J.I.; Phillips, J.H.; et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nat. Cell Biol. 1998, 391, 795–799. [Google Scholar] [CrossRef] [PubMed]
- Braud, V.M.; Allan, D.S.; Wilson, D.; McMichael, A.J. TAP- and tapasin-dependent HLA-E surface expression correlates with the binding of an MHC class I leader peptide. Curr. Biol. 1998, 8, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Llano, M.; Lee, N.; Navarro, F.; García, P.; Albar, J.P.; Geraghty, D.E.; López-Botet, M. HLA-E-bound peptides influence recognition by inhibitory and triggering CD94/NKG2 receptors: Preferential response to an HLA-G-derived nonamer. Eur. J. Immunol. 1998, 28, 2854–2863. [Google Scholar] [CrossRef]
- Allard, M.; Oger, R.; Vignard, V.; Percier, J.-M.; Fregni, G.; Perier, A.; Caignard, A.; Charreau, B.; Bernardeau, K.; Khammari, A.; et al. Serum Soluble HLA-E in Melanoma: A New Potential Immune-Related Marker in Cancer. PLoS ONE 2011, 6, e21118. [Google Scholar] [CrossRef]
- Mociornita, A.G.; Adamson, M.B.; Tumiati, L.C.; Ross, H.J.; Rao, V.; Delgado, D.H. Effects of everolimus and HLA-G on cellular proliferation and neutrophil adhesion in an in vitro model of cardiac allograft vasculopathy. Arab. Archaeol. Epigr. 2018, 18, 3038–3044. [Google Scholar] [CrossRef] [Green Version]
- Coupel, S.; Moreau, A.; Hamidou, M.; Horejsi, V.; Soulillou, J.-P.; Charreau, B. Expression and release of soluble HLA-E is an immunoregulatory feature of endothelial cell activation. Blood 2007, 109, 2806–2814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, H.; Lu, R.; Xie, S.; Wen, X.; Wang, H.; Gao, X.; Guo, L. Human leukocyte antigen-E alleles and expression in patients with serous ovarian cancer. Cancer Sci. 2015, 106, 522–528. [Google Scholar] [CrossRef] [Green Version]
- Zidi, I. Puzzling out the COVID-19: Therapy targeting HLA-G and HLA-E. Hum. Immunol. 2020, 81, 697–701. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, R.; Neri, L.M.; Simioni, C.; Bortolotti, D.; Occhionorelli, S.; Zauli, G.; Secchiero, P.; Semprini, C.M.; Laface, I.; Sanz, J.M.; et al. SARS-COV2 nucleocapsid protein and ultrastructural modifications in small bowel of a 4-week-negative COVID19 patient. Clin. Microbiol. Infect. 2021, 27, 936–937. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://covid19.who.int (accessed on 5 August 2021).
- Leeuwenberg, J.F.; Smeets, E.F.; Neefjes, J.; Shaffer, M.A.; Cinek, T.; Jeunhomme, T.M.; Ahern, T.J.; Buurman, W.A. E-selectin and intercellular adhesion molecule-1 are released by activated human endothelial cells in vitro. Immunology 1992, 77, 543–549. [Google Scholar] [PubMed]
- Barnes, B.J.; Adrover, J.M.; Baxter-Stoltzfus, A.; Borczuk, A.; Cools-Lartigue, J.; Crawford, J.M.; Daßler-Plenker, J.; Guerci, P.; Huynh, C.; Knight, J.S.; et al. Targeting potential drivers of COVID-19: Neutrophil extracellular traps. J. Exp. Med. 2020, 6, 217. [Google Scholar] [CrossRef] [PubMed]
- Parackova, Z.; Zentsova, I.; Bloomfield, M.; Vrabcova, P.; Smetanova, J.; Klocperk, A.; Mesežnikov, G.; Mendez, L.F.C.; Vymazal, T.; Sediva, A. Disharmonic Inflammatory Signatures in COVID-19: Augmented Neutrophils’ but Impaired Monocytes’ and Dendritic Cells’ Responsiveness. Cells 2020, 9, 2206. [Google Scholar] [CrossRef] [PubMed]
- Contoli, M.; Papi, A.; Tomassetti, L.; Rizzo, P.; Sega, F.V.D.; Fortini, F.; Torsani, F.; Morandi, L.; Ronzoni, L.; Zucchetti, O.; et al. Blood Interferon-α Levels and Severity, Outcomes, and Inflammatory Profiles in Hospitalized COVID-19 Patients. Front. Immunol. 2021, 12, 648004. [Google Scholar] [CrossRef]
- Spadaro, S.; Fogagnolo, A.; Campo, G.; Zucchetti, O.; Verri, M.; Ottaviani, I.; Tunstall, T.; Grasso, S.; Scaramuzzo, V.; Murgolo, F.; et al. Markers of endothelial and epithelial pulmonary injury in mechanically ventilated COVID19 ICU patients. Crit. Care. 2021, 25, 74. [Google Scholar] [CrossRef] [PubMed]
- Campo, G.; Contoli, M.; Fogagnolo, A.; Sega, F.V.D.; Zucchetti, O.; Ronzoni, L.; Verri, M.; Fortini, F.; Pavasini, R.; Morandi, L.; et al. Over time relationship between platelet reactivity, myocardial injury and mortality in patients with SARS-COV2-associated respiratory failure. Platelets 2021, 32, 560–567. [Google Scholar] [CrossRef]
- Fainardi, E.; Rizzo, R.; Melchiorri, L.; Stignani, M.; Castellazzi, M.; Caniatti, M.L.; Baldi, E.; Tola, M.R.; Granieri, E.; Baricordi, O.R. Soluble HLA-G molecules are released as HLA-G5 and not as soluble HLA-G1 isoforms in CSF of patients with relapsing–remitting Multiple Sclerosis. J. Neuroimmunol. 2007, 192, 219–225. [Google Scholar] [CrossRef]
- Morandi, F.; Venturi, C.; Rizzo, R.; Castellazzi, M.; Baldi, E.; Caniatti, M.L.; Tola, M.R.; Granieri, E.; Fainardi, E.; Uccelli, A.; et al. Intrathecal Soluble HLA-E Correlates with Disease Activity in Patients with Multiple Sclerosis and may Cooperate with Soluble HLA-G in the Resolution of Neuroinflammation. J. Neuroimmune Pharmacol. 2013, 8, 944–955. [Google Scholar] [CrossRef]
- Nishizawa, A.; Kumada, K.; Tateno, K.; Wagata, M.; Saito, S.; Katsuoka, F.; Mizuno, S.; Ogishima, S.; Yamamoto, M.; Yasuda, J.; et al. Analysis of HLA-G long-read genomic sequences in mother–offspring pairs with preeclampsia. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef]
- Rizzo, R.; D’Accolti, M.; Bortolotti, D.; Caccuri, F.; Caruso, A.; Di Luca, D.; Caselli, E. Human Herpesvirus 6A and 6B inhibit in vitro angiogenesis by induction of Human Leukocyte Antigen G. Sci. Rep. 2018, 8, 17683. [Google Scholar] [CrossRef]
- Kjaergaard, A.G.; Dige, A.; Krog, J.; Tønnesen, E.; Wogensen, L. Soluble Adhesion Molecules Correlate with Surface Expression in anIn VitroModel of Endothelial Activation. Basic Clin. Pharmacol. Toxicol. 2013, 113, 273–279. [Google Scholar] [CrossRef]
- Rizzo, R.; Hviid, T.V.F.; Stignani, M.; Balboni, A.; Grappa, M.T.; Melchiorri, L.; Baricordi, O.R. The HLA-G genotype is associated with IL-10 levels in activated PBMCs. Immunogenetics 2005, 57, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Wilhelmsen, K.; Farrar, K.; Hellman, J. Quantitative In vitro Assay to Measure Neutrophil Adhesion to Activated Primary Human Microvascular Endothelial Cells under Static Conditions. J. Vis. Exp. 2013, 78, e50677. [Google Scholar] [CrossRef] [Green Version]
- Zittermann, S.I.; Issekutz, A.C. Basic Fibroblast Growth Factor (bFGF, FGF-2) Potentiates Leukocyte Recruitment to Inflammation by Enhancing Endothelial Adhesion Molecule Expression. Am. J. Pathol. 2006, 168, 835–846. [Google Scholar] [CrossRef] [Green Version]
- Al-Bayatee, N.T.; Ad’hiah, A.H. Soluble HLA-G is upregulated in serum of patients with severe COVID19. Human Immunol 2021. [Google Scholar] [CrossRef]
- Morandi, F.; Rizzo, R.; Fainardi, E.; Rouas-Freiss, N.; Pistoia, V. Recent Advances in Our Understanding of HLA-G Biology: Lessons from a Wide Spectrum of Human Diseases. J. Immunol. Res. 2016, 2016, 4326495. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Gan, J.; Chen, B.; Zheng, D.; Zhang, J.; Lin, R.; Zhou, Y.; Yang, W.; Lin, A.; Yan, W. Dynamics of peripheral immune cells and their HLA-G and receptor expressions in a patient suffering from critical COVID-19 pneumonia to convalescence. Clin. Transl. Immunol. 2020, 9, e1128. [Google Scholar] [CrossRef]
- Bortolotti, D.; Gentili, V.; Rizzo, S.; Rotola, A.; Rizzo, R. SARS-COV2 Spike 1 Protein Controls Natural Killer Cell Activation via the HLA-E/NKG2A Pathway. Cells 2020, 9, 1975. [Google Scholar] [CrossRef] [PubMed]
- Lovren, F.; Pan, Y.; Quan, A.; Teoh, H.; Wang, G.; Shukla, P.C.; Levitt, K.S.; Oudit, G.Y.; Al-Omran, M.; Stewart, D.J.; et al. Angiotensin converting enzyme-2 confers endothelial protection and attenuates atherosclerosis. Am. J. Physiol. Circ. Physiol. 2008, 295, H1377–H1384. [Google Scholar] [CrossRef] [Green Version]
- Sluimer, J.; Gasc, J.M.; Hamming, I.; van Goor, H.; Michaud, A.; Akker, L.H.V.D.; Jutten, B.; Cleutjens, J.; Bijnens, A.P.J.J.; Corvol, P.; et al. Angiotensin-converting enzyme 2 (ACE2) expression and activity in human carotid atherosclerotic lesions. J. Pathol. 2008, 215, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Godo, S.; Shimokawa, H. Endothelial Functions. Arter. Thromb. Vasc. Biol. 2017, 37, e108–e114. [Google Scholar] [CrossRef] [Green Version]
- Aird, W.C. The role of the endothelium in severe sepsis and multiple organ dysfunction syndrome. Blood 2003, 101, 3765–3777. [Google Scholar] [CrossRef]
- Vejlsgaard, G.L.; Ralfkiaer, E.; Avnstorp, C.; Czajkowski, M.; Marlin, S.D.; Rothlein, R. Kinetics and characterization of intercellular adhesion molecule-1 (ICAM-1) expression on keratinocytes in various inflammatory skin lesions and malignant cutaneous lymphomas. J. Am. Acad. Dermatol. 1989, 20, 782–790. [Google Scholar] [CrossRef]
- Johnson, J.P.; Stade, B.G.; Holzmann, B.; Schwable, W.; Riethmuller, G. De novo expression of intercellular-adhesion molecule 1 in melanoma correlates with increased risk of metastasis. Proc. Natl. Acad. Sci. USA 1989, 86, 641–644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Admas, D.H.; Hubscher, S.G.; Shaw, J.; Rothlein, R.; Neuberger, J.M. Intercellular adhesion molecule 1 on liver allografts dur-ing rejection. Lancet 1989, 334, 1122–1125. [Google Scholar] [CrossRef]
- Leeuwenberg, J.F.M.; Jeunhomme, G.M.A.A.; Buurman, W.A. Adhesion of polymorphonuclear cells to human endothelial cells. Adhesion-molecule-dependent, and Fc receptor-mediated adhesion-molecule-independent mechanisms. Clin. Exp. Immunol. 2008, 81, 496–500. [Google Scholar] [CrossRef]
- Ley, K.; Laudanna, C.; Cybulsky, M.I.; Nourshargh, S. Getting to the site of inflammation: The leukocyte adhesion cascade updated. Nat. Rev. Immunol. 2007, 7, 678–689. [Google Scholar] [CrossRef]
- Zarbock, A.; Lowell, C.A.; Ley, K. Spleen tyrosine ki-nase Syk is necessary for E-selectin-inducedL2integrin-mediated rolling on intercellular ad-hesion molecule-1. Immunity 2007, 26, 773–783. [Google Scholar] [CrossRef] [Green Version]
- Bonaventura, A.; Vecchié, A.; Dagna, L.; Martinod, K.; Dixon, D.L.; Van Tassell, B.W.; Dentali, F.; Montecucco, F.; Massberg, S.; Levi, M.; et al. Endothelial dysfunction and immunothrombosis as key pathogenic mechanisms in COVID-19. Nat. Rev. Immunol. 2021, 21, 319–329. [Google Scholar] [CrossRef]
- Matsunami, K.; Miyagawa, S.; Nakai, R.; Murase, A.; Shirakura, R. The possible use of HLA-G1 and G3 in the inhibition of NK cell-mediated swine endothelial cell lysis. Clin. Exp. Immunol. 2001, 126, 165–172. [Google Scholar] [CrossRef]
- Tong, M.; Jiang, Y.; Xia, D.; Xiong, Y.; Zheng, Q.; Chen, F.; Zou, L.; Xiao, W.; Zhu, Y. Elevated Expression of Serum Endothelial Cell Adhesion Molecules in COVID-19 Patients. J. Infect. Dis. 2020, 222, 894–898. [Google Scholar] [CrossRef]



| Study Population (n = 165) | COVID-19 Patients (n = 54) | ||||||
|---|---|---|---|---|---|---|---|
| COVID-19 Patients, n = 54 | Control Patients’ Respiratory Failure n = 11 | Control Patients n = 100 | p-Value * | Non-Survivor n = 16 | Survivors n = 38 | p-Value | |
| Gender N (%) | >0.9 | >0.9 | |||||
| Male | 40 (74%) | 8 (73%) | 74 (74%) | 12 (75%) | 28 (74%) | ||
| Female | 14 (26%) | 3 (27%) | 26 (26%) | 4 (25%) | 10 (26%) | ||
| Age | 65 (57, 73) | 70 (66, 76) | 67 (56, 74) | 0.2 | 72 (65, 78) | 62 (55, 71) | 0.004 |
| Smoking habit N (%) | |||||||
| Active smoker | 0 (0) | 3 (27%) | 1 (1%) | 0.003 | 0 (0) | 0 (0) | NA |
| Former smoker | 16 (30%) | 4 (36%) | 29 (29%) | 0.725 | 7 (44%) | 9 (24%) | 0.2 |
| BMI (kg/m2) | 26.4 (24.2, 30.0) | 24.8 (22.0, 27.1) | 25.3 (23.1, 28.6) | 0.13 | 28.5 (26.4, 30.9) | 26.0 (24.1, 29.4) | 0.2 |
| Number of Comorbidities/patients | 1.00 (0.00, 3.00) | 2.00 (1.50, 3.00) | 0 (0.00, 0,00) | 0.12 | 3.00 (1.75, 4.00) | 1.00 (0.00, 2.00) | 0.004 |
| Respiratory support at recruitment N (%) | |||||||
| O2 only | 11 (20%) | 2 (18%) | NA | 2 (12%) | 9 (24%) | ||
| HFNC or NIV | 16 (30%) | 6 (54%) | NA | 4 (25%) | 12 (31%) | ||
| IV | 27 (50%) | 3 (27%) | NA | 10 (62%) | 17 (45%) | ||
| Days from symptoms onset to recruitment | 9 (5–14) | 5 (2–8) | NA | 10 (5–14) | 8 (5–15) | 0.60 | |
| Treatments N (%) | |||||||
| Low molecular weight heparin | 54 (100%) | 11 (100%) | NA | >0.9 | 16 (100%) | 38 (100%) | >0.9 |
| Antibiotics | 47 (87%) | 10 (90%) | NA | >0.9 | 14 (88%) | 33 (87%) | >0.9 |
| Systemic corticosterods | 37 (69%) | 9 (81%) | NA | >0.9 | 12 (75%) | 25 (66%) | 0.7 |
| Antivirals | 29 (54%) | NA | NA | NA | 7 (44%) | 22 (58%) | 0.5 |
| Hydroxychloroquine | 40 (74%) | NA | NA | NA | 11 (69%) | 29 (76%) | 0.7 |
| COVID-19 Patients, n = 54 | Control Patients’ Respiratory Failure, n = 11 | Control Patients, n = 100 | p-Value * | p-Value ** | p-Value *** | |
|---|---|---|---|---|---|---|
| sHLA-G (ng/mL) | 165.87 (44.3, 218.03) | 49.54 (18.3, 54.9) | 20.51 (0.0, 43.53) | 0.01 | <0.001 | 0.01 |
| sHLA-E (ng/mL) | 672.22 (173.9, 890.9) | 224.63 (98.6, 310.4) | 10.23 (0.0, 21.51) | 0.001 | <0.001 | <0.001 |
| Total blood leucocytes (cells × 103/μL) | 9.1 (6.8, 12.6) | 12.0 (9.1, 14.7) | 5.0 (4.1, 11.0) | 0.2 | 0.023 | 0.021 |
| Blood lymphocites (cells × 103/μL) | 0.83 (0.59, 1.04) | 1.12 (0.52, 1.73) | 0.96 (0.54, 1.29) | 0.3 | 0.23 | 0.12 |
| Blood Neutrophils (cells × 103/μL) | 7.9 (5.6, 10.2) | 10.1 (5.8, 12.1) | 3.2 (2.0–7.4) | 0.2 | 0.01 | 0.01 |
| Blood eosinophils (cells × 103/μL) | 0.04 (0.00, 0.14) | 0.00 (0.00, 0.06) | 0.00 (0.00, 0.02) | 0.074 | 0.069 | 0.12 |
| COVID-19 Patients n = 54 | Control Patients’ Respiratory Failure n = 11 | Control Patients n = 100 | p-Value ** | p-Value *** | |
|---|---|---|---|---|---|
| HLA-E* alleles | |||||
| 0101 N (%) | 25 (47) | 5 (46) | 48 (48) | 0.59 | 0.53 |
| 0103 | 28 (53) | 6 (54) | 52 (52) | ||
| HLA-G* alleles | |||||
| 0101 N (%) | 47 (87) | 9 (86) | 85 (85) | 0.63 | 0.76 |
| 0103 | 1 (1) | 0 (0) | 1 (1) | ||
| 0104 | 4 (8) | 1 (7) | 8 (8) | ||
| 0105N | 2 (4) | 1 (6) | 6 (6) | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bortolotti, D.; Gentili, V.; Rizzo, S.; Schiuma, G.; Beltrami, S.; Spadaro, S.; Strazzabosco, G.; Campo, G.; Carosella, E.D.; Papi, A.; et al. Increased sHLA-G Is Associated with Improved COVID-19 Outcome and Reduced Neutrophil Adhesion. Viruses 2021, 13, 1855. https://doi.org/10.3390/v13091855
Bortolotti D, Gentili V, Rizzo S, Schiuma G, Beltrami S, Spadaro S, Strazzabosco G, Campo G, Carosella ED, Papi A, et al. Increased sHLA-G Is Associated with Improved COVID-19 Outcome and Reduced Neutrophil Adhesion. Viruses. 2021; 13(9):1855. https://doi.org/10.3390/v13091855
Chicago/Turabian StyleBortolotti, Daria, Valentina Gentili, Sabrina Rizzo, Giovanna Schiuma, Silvia Beltrami, Savino Spadaro, Giovanni Strazzabosco, Gianluca Campo, Edgardo D. Carosella, Alberto Papi, and et al. 2021. "Increased sHLA-G Is Associated with Improved COVID-19 Outcome and Reduced Neutrophil Adhesion" Viruses 13, no. 9: 1855. https://doi.org/10.3390/v13091855
APA StyleBortolotti, D., Gentili, V., Rizzo, S., Schiuma, G., Beltrami, S., Spadaro, S., Strazzabosco, G., Campo, G., Carosella, E. D., Papi, A., Rizzo, R., & Contoli, M. (2021). Increased sHLA-G Is Associated with Improved COVID-19 Outcome and Reduced Neutrophil Adhesion. Viruses, 13(9), 1855. https://doi.org/10.3390/v13091855









