Monitoring Natural SARS-CoV-2 Infection in Lions (Panthera leo) at the Barcelona Zoo: Viral Dynamics and Host Responses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Clinical Evaluation and Sample Collection
2.2. Additional Sample Collection
2.3. SARS-CoV-2 Antigen Test
2.4. Cell Culture and SARS-CoV-2
2.5. RNA Extraction and Detection by RT-qPCR
2.6. SARS-CoV-2 Genome Sequencing and Phylogenetic Analyses
2.7. Virus Isolation in Vero E6 Cells
2.8. SARS-CoV-2 RBD and N-ELISA
2.9. SARS-CoV-2 Neutralization Assay
2.10. Data Analysis and Statistics
3. Results
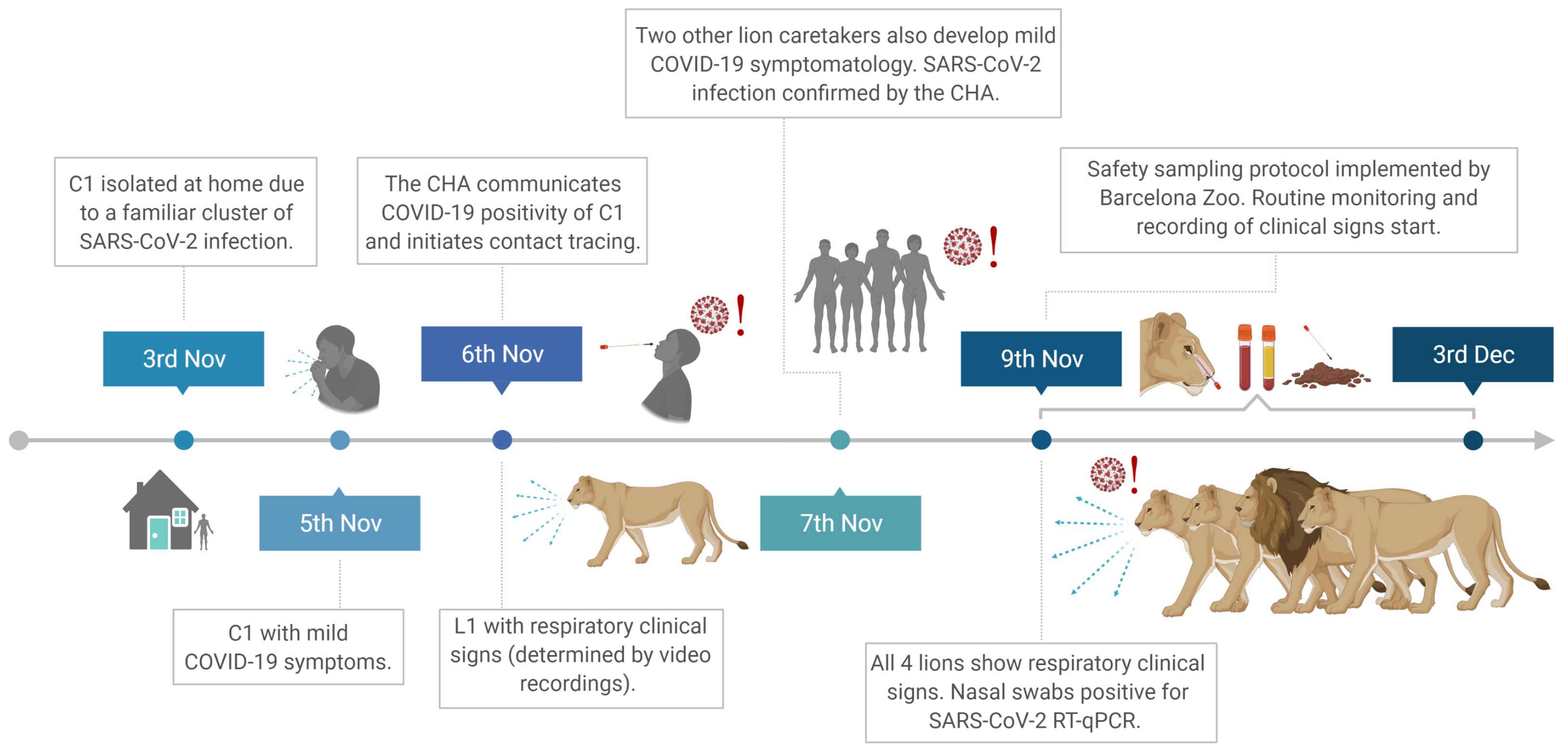
3.1. COVID-19 Confirmation in a Lion Caretaker and Main Chronological Events at the Barcelona Zoo
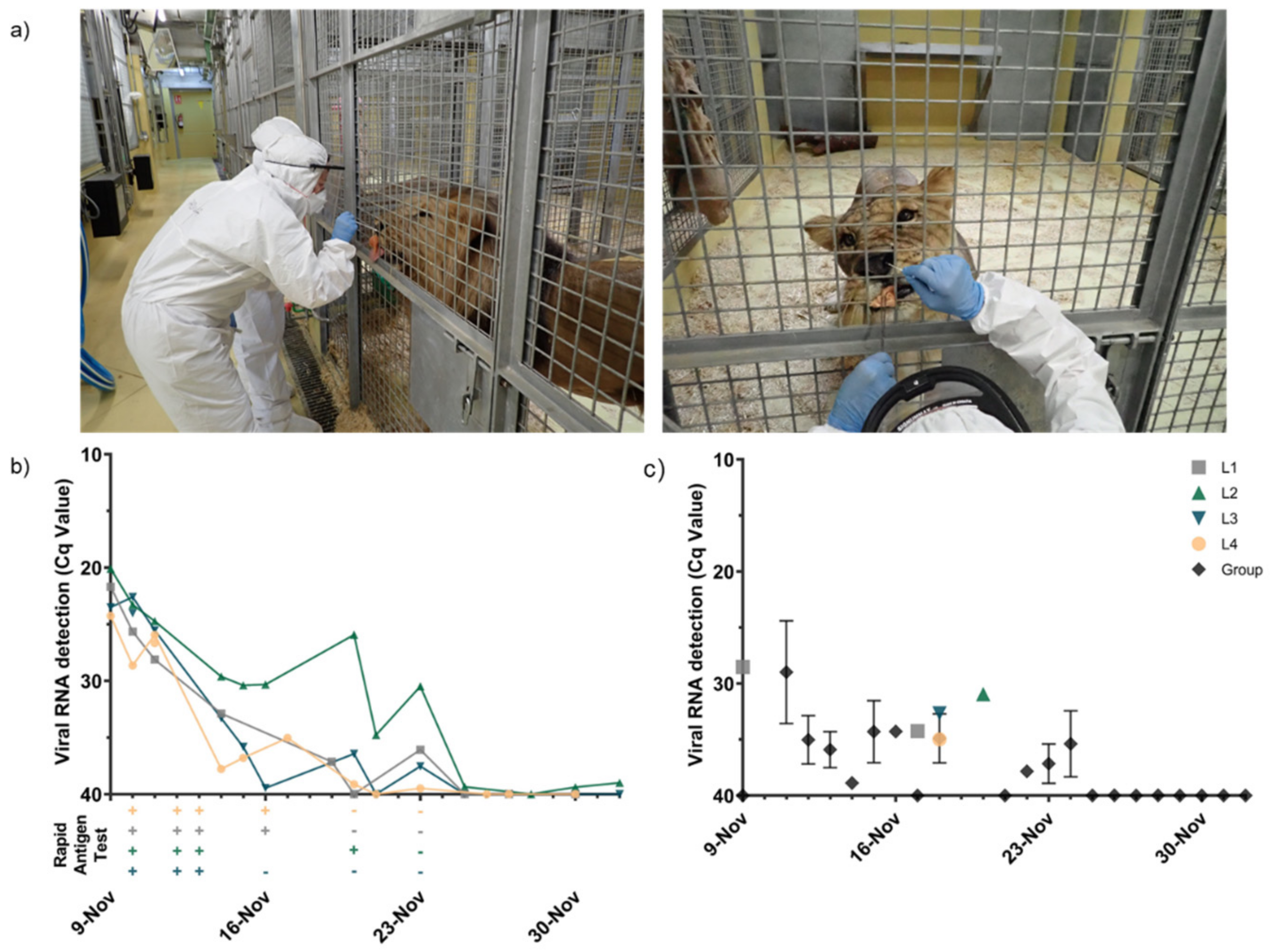
3.2. SARS-CoV-2 Antigen, RNA Detection, and Infectious Virus Isolation
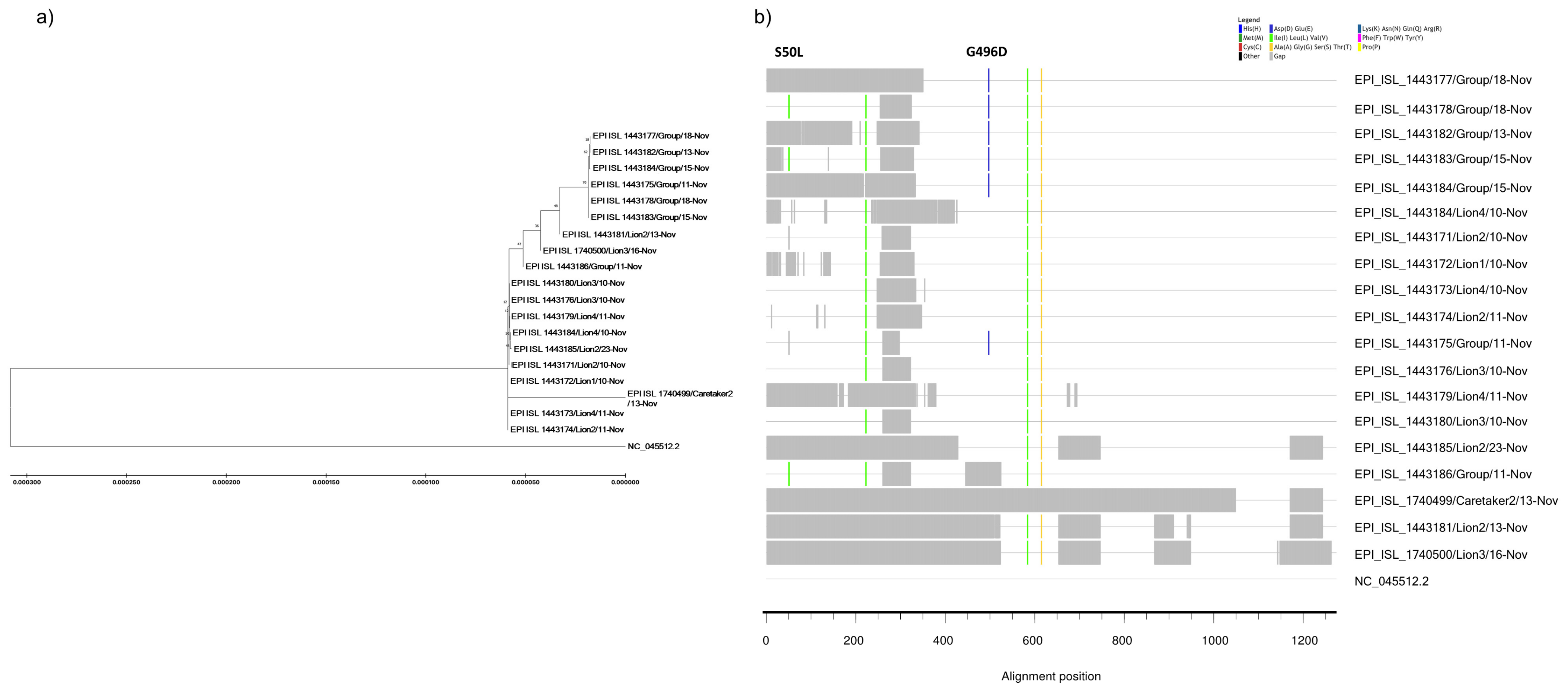
3.3. SARS-CoV-2 Sequencing Confirmed Human-to-Lion Transmission and Genomic Stability in the New Host
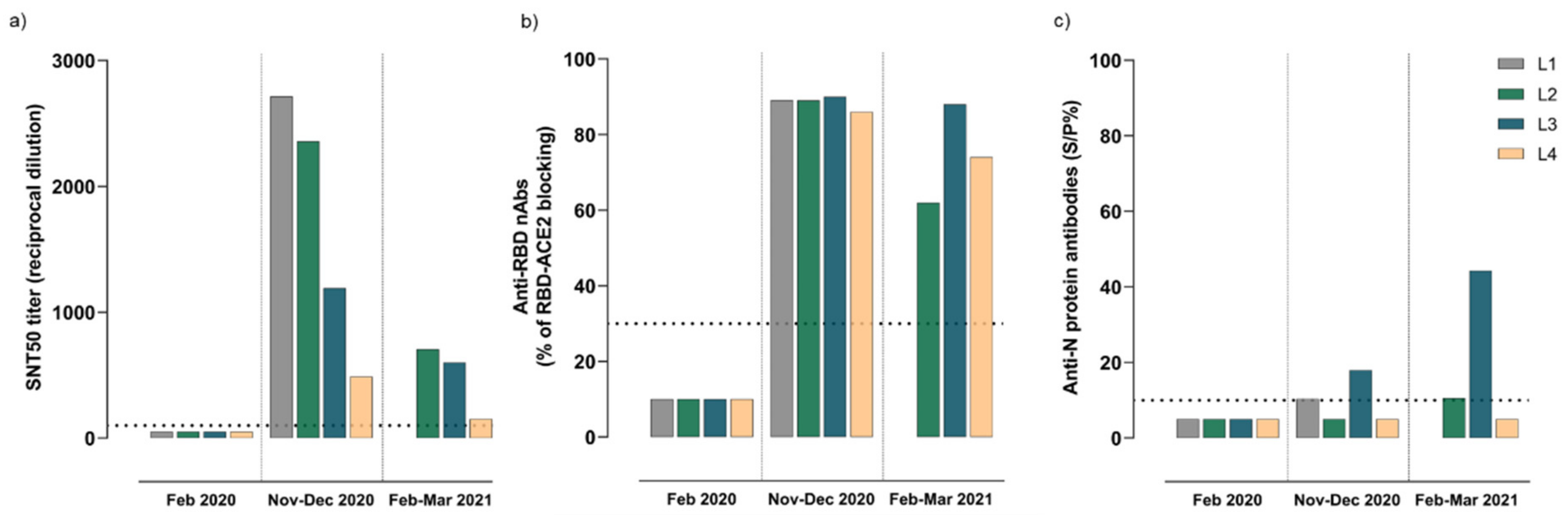
3.4. Kinetics of Humoral Immune Responses Elicited after SARS-CoV-2 Infection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
References
- Santini, J.M.; Edwards, S.J.L. Host range of SARS-CoV-2 and implications for public health. Lancet Microbe 2020, 1, e141–e142. [Google Scholar] [CrossRef]
- Shi, J.; Wen, Z.; Zhong, G.; Yang, H.; Wang, C.; Huang, B.; Liu, R.; He, X.; Shuai, L.; Sun, Z.; et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. Science 2020, 368, 1016–1020. [Google Scholar] [CrossRef] [Green Version]
- Lakdawala, S.S.; Menachery, V.D. The search for a COVID-19 animal model. Science 2020, 368, 942–943. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Fontela, C.; Dowling, W.E.; Funnell, S.G.P.; Gsell, P.-S.; Riveros-Balta, A.X.; Albrecht, R.A.; Andersen, H.; Baric, R.S.; Carroll, M.W.; Cavaleri, M.; et al. Animal models for COVID-19. Nature 2020, 586, 509–515. [Google Scholar] [CrossRef] [PubMed]
- do Vale, B.; Lopes, A.P.; da Conceição Fontes, M.; Silvestre, M.; Cardoso, L.; Coelho, A.C. Bats, pangolins, minks and other animals-villains or victims of SARS-CoV-2? Vet. Res. Commun. 2021, 45, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Segalés, J.; Puig, M.; Rodon, J.; Avila-Nieto, C.; Carrillo, J.; Cantero, G.; Terrón, M.T.; Cruz, S.; Parera, M.; Noguera-Julián, M.; et al. Detection of SARS-CoV-2 in a cat owned by a COVID-19−affected patient in Spain. Proc. Natl. Acad. Sci. USA 2020, 117, 24790–24793. [Google Scholar] [CrossRef]
- Hamer, S.A.; Pauvolid-Corrêa, A.; Zecca, I.B.; Davila, E.; Auckland, L.D.; Roundy, C.M.; Tang, W.; Torchetti, M.K.; Killian, M.L.; Jenkins-Moore, M.; et al. SARS-CoV-2 Infections and Viral Isolations among Serially Tested Cats and Dogs in Households with Infected Owners in Texas, USA. Viruses 2021, 13, 938. [Google Scholar] [CrossRef] [PubMed]
- Barrs, V.; Peiris, M.; Tam, K.W.S.; Law, P.Y.T.; Brackman, C.; To, E.M.W.; Yu, V.Y.T.; Chu, D.K.W.; Perera, R.A.P.M.; Sit, T.H.C. SARS-CoV-2 in Quarantined Domestic Cats from COVID-19 Households or Close Contacts, Hong Kong, China. Emerg. Infect. Dis. J. 2020, 26, 3071. [Google Scholar] [CrossRef]
- Sit, T.H.C.; Brackman, C.J.; Ip, S.M.; Tam, K.W.S.; Law, P.Y.T.; To, E.M.W.; Yu, V.Y.T.; Sims, L.D.; Tsang, D.N.C.; Chu, D.K.W.; et al. Infection of dogs with SARS-CoV-2. Nature 2020, 586, 776–778. [Google Scholar] [CrossRef]
- Neira, V.; Brito, B.; Agüero, B.; Berrios, F.; Valdés, V.; Gutierrez, A.; Ariyama, N.; Espinoza, P.; Retamal, P.; Holmes, E.C.; et al. A household case evidences shorter shedding of SARS-CoV-2 in naturally infected cats compared to their human owners. Emerg. Microbes Infect. 2021, 10, 376–383. [Google Scholar] [CrossRef]
- Vergara-Alert, J.; Rodon, J.; Carrillo, J.; Te, N.; Izquierdo-Useros, N.; de la Concepción, M.L.R.; Ávila-Nieto, C.; Guallar, V.; Valencia, A.; Cantero, G.; et al. Pigs are not susceptible to SARS-CoV-2 infection but are a model for viral immunogenicity studies. Transbound. Emerg. Dis. 2020. [Google Scholar] [CrossRef]
- Oude Munnink, B.B.; Sikkema, R.S.; Nieuwenhuijse, D.F.; Molenaar, R.J.; Munger, E.; Molenkamp, R.; van der Spek, A.; Tolsma, P.; Rietveld, A.; Brouwer, M.; et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 2021, 371, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Lecu, A.; Bertelsen, M.F.; Walzer, C.; EAZWV Infectious Diseases Working Group. Science-based facts & knowledge about wild animals, zoos and sars-Cov-2 virus. In Transmissible Diseases Handbook; European Association of Zoo and Wildlife Veterinarians: Drenthe, The Netherlands, 2021. [Google Scholar]
- World Organisation for Animal Health (OIE). Infection with SARS-CoV-2 in Animals; World Organisation for Animal Health (OIE): Paris, France, 2020; Volume 2. [Google Scholar]
- Lamberski, N.; Howard, L.; Clancy, M.; Kinney, M.; Sadler, R.; Browning, G.R. Summary of SARS-CoV-2 Episode in Gorillas at the San Diego Zoo Safari Park January–February 2021; San Diego Zoo Safari Park Department of Veterinary Services: San Diego, CA, USA, 2021. [Google Scholar]
- Bartlett, S.L.; Diel, D.G.; Wang, L.; Zec, S.; Laverack, M.; Martins, M.; Caserta, L.C.; Killian, M.L.; Terio, K.; Olmstead, C.; et al. Sars-Cov-2 Infection and Longitudinal Fecal Screening in Malayan tigers (Panthera tigris jacksoni), Amur tigers (Panthera tigris altaica), and African lions (Panthera leo krugeri) at the Bronx Zoo, New York, USA. J. Zoo Wildl. Med. 2021, 51, 733–744. [Google Scholar] [CrossRef] [PubMed]
- McAloose, D.; Laverack, M.; Wang, L.; Killian, M.L.; Caserta, L.C.; Yuan, F.; Mitchell, P.K.; Queen, K.; Mauldin, M.R.; Cronk, B.D.; et al. From People to Panthera: Natural SARS-CoV-2 Infection in Tigers and Lions at the Bronx Zoo. MBio 2020, 11, e02220-20. [Google Scholar] [CrossRef] [PubMed]
- World Organisation for Animal Health (OIE). SARS-CoV-2 in Animals–Situation Report 1; World Organisation for Animal Health (OIE): Paris, France, 2021. [Google Scholar]
- Mishra, A.; Kumar, N.; Bhatia, S.; Aasdev, A.; Kanniappan, S.; Thayasekhar, A.; Gopinadhan, A.; Silambarasan, R.; Sreekumar, C.; Dubey, C.K.; et al. Natural infection of SARS-CoV-2 delta variant in Asiatic lions (Panthera leo persica) in India. bioRxiv 2021. [Google Scholar] [CrossRef]
- Rodon, J.; Muñoz-Basagoiti, J.; Perez-Zsolt, D.; Noguera-Julian, M.; Paredes, R.; Mateu, L.; Quiñones, C.; Perez, C.; Erkizia, I.; Blanco, I.; et al. Identification of Plitidepsin as Potent Inhibitor of SARS-CoV-2-Induced Cytopathic Effect after a Drug Repurposing Screen. Front. Pharmacol. 2021, 12, 278. [Google Scholar] [CrossRef] [PubMed]
- Brustolin, M.; Rodon, J.; Rodríguez de la Concepción, M.L.; Ávila-Nieto, C.; Cantero, G.; Pérez, M.; Te, N.; Noguera-Julián, M.; Guallar, V.; Valencia, A.; et al. Protection against reinfection with D614- or G614-SARS-CoV-2 isolates in golden Syrian hamster. Emerg. Microbes Infect. 2021, 10, 797–809. [Google Scholar] [CrossRef]
- Hodcroft, E.B.; Zuber, M.; Nadeau, S.; Vaughan, T.G.; Crawford, K.H.D.; Althaus, C.L.; Reichmuth, M.L.; Bowen, J.E.; Walls, A.C.; Corti, D.; et al. Emergence and spread of a SARS-CoV-2 variant through Europe in the summer of 2020. medRxiv 2021. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. Subgroup, 1000 Genome Project Data Processing The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Grubaugh, N.D.; Gangavarapu, K.; Quick, J.; Matteson, N.L.; De Jesus, J.G.; Main, B.J.; Tan, A.L.; Paul, L.M.; Brackney, D.E.; Grewal, S.; et al. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019, 20, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasegawa, M.; Kishino, H.; Yano, T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985, 22, 160–174. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Roca, O.; Pacheco, A.; Rodon, J.; Antón, A.; Vergara-Alert, J.; Armadans, L.; Segalés, J.; Pumarola, T.; Campins, M.; Ferrer, R.; et al. Nasal high flow oxygen therapy in COVID-19 patients does not cause environmental surface contamination. J. Hosp. Infect. 2021. [Google Scholar] [CrossRef] [PubMed]
- Trinité, B.; Tarrés-Freixas, F.; Rodon, J.; Pradenas, E.; Urrea, V.; Marfil, S.; Rodríguez de la Concepción, M.L.; Ávila-Nieto, C.; Aguilar-Gurrieri, C.; Barajas, A.; et al. SARS-CoV-2 infection elicits a rapid neutralizing antibody response that correlates with disease severity. Sci. Rep. 2021, 11, 2608. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.K.; Martins, M.; Reilly, T.; Caserta, L.C.; Anderson, R.; Cronk, B.D.; Murphy, J.; Goodrich, E.L.; Diel, D.G. Infection with SARS-CoV-2 Lineage B.1.1.7 in Three Malayan Tigers at the Virginia Zoological Park; Research Square: Durham, NC, USA, 2021; pp. 1–6. [Google Scholar] [CrossRef]
- Plante, J.A.; Liu, Y.; Liu, J.; Xia, H.; Johnson, B.A.; Lokugamage, K.G.; Zhang, X.; Muruato, A.E.; Zou, J.; Fontes-Garfias, C.R.; et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2021, 592, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef]
- Montagutelli, X.; Prot, M.; Levillayer, L.; Salazar, E.B.; Jouvion, G.; Conquet, L.; Donati, F.; Albert, M.; Gambaro, F.; Behillil, S.; et al. The B1.351 and P.1 variants extend SARS-CoV-2 host range to mice. bioRxiv 2021. [Google Scholar] [CrossRef]
- Zhou, H.; Ji, J.; Chen, X.; Bi, Y.; Li, J.; Hu, T.; Song, H.; Chen, Y.; Cui, M.; Zhang, Y.; et al. Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS-CoV-2 and related viruses. bioRxiv 2021. [Google Scholar] [CrossRef]
- Wacharapluesadee, S.; Tan, C.W.; Maneeorn, P.; Duengkae, P.; Zhu, F.; Joyjinda, Y.; Kaewpom, T.; Chia, W.N.; Ampoot, W.; Lim, B.L.; et al. Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia. Nat. Commun. 2021, 12, 972. [Google Scholar] [CrossRef]
- Garigliany, M.; Van Laere, A.-S.; Clercx, C.; Giet, D.; Escriou, N.; Huon, C.; van der Werf, S.; Eloit, M.; Desmecht, D. SARS-CoV-2 Natural Transmission from Human to Cat, Belgium, March 2020. Emerg. Infect. Dis. J. 2020, 26, 3069. [Google Scholar] [CrossRef] [PubMed]
- Gaudreault, N.N.; Trujillo, J.D.; Carossino, M.; Meekins, D.A.; Morozov, I.; Madden, D.W.; Indran, S.V.; Bold, D.; Balaraman, V.; Kwon, T.; et al. SARS-CoV-2 infection, disease and transmission in domestic cats. Emerg. Microbes Infect. 2020, 9, 2322–2332. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández-Bellon, H.; Rodon, J.; Fernández-Bastit, L.; Almagro, V.; Padilla-Solé, P.; Lorca-Oró, C.; Valle, R.; Roca, N.; Grazioli, S.; Trogu, T.; et al. Monitoring Natural SARS-CoV-2 Infection in Lions (Panthera leo) at the Barcelona Zoo: Viral Dynamics and Host Responses. Viruses 2021, 13, 1683. https://doi.org/10.3390/v13091683
Fernández-Bellon H, Rodon J, Fernández-Bastit L, Almagro V, Padilla-Solé P, Lorca-Oró C, Valle R, Roca N, Grazioli S, Trogu T, et al. Monitoring Natural SARS-CoV-2 Infection in Lions (Panthera leo) at the Barcelona Zoo: Viral Dynamics and Host Responses. Viruses. 2021; 13(9):1683. https://doi.org/10.3390/v13091683
Chicago/Turabian StyleFernández-Bellon, Hugo, Jordi Rodon, Leira Fernández-Bastit, Vanessa Almagro, Pilar Padilla-Solé, Cristina Lorca-Oró, Rosa Valle, Núria Roca, Santina Grazioli, Tiziana Trogu, and et al. 2021. "Monitoring Natural SARS-CoV-2 Infection in Lions (Panthera leo) at the Barcelona Zoo: Viral Dynamics and Host Responses" Viruses 13, no. 9: 1683. https://doi.org/10.3390/v13091683
APA StyleFernández-Bellon, H., Rodon, J., Fernández-Bastit, L., Almagro, V., Padilla-Solé, P., Lorca-Oró, C., Valle, R., Roca, N., Grazioli, S., Trogu, T., Bensaid, A., Carrillo, J., Izquierdo-Useros, N., Blanco, J., Parera, M., Noguera-Julián, M., Clotet, B., Moreno, A., Segalés, J., & Vergara-Alert, J. (2021). Monitoring Natural SARS-CoV-2 Infection in Lions (Panthera leo) at the Barcelona Zoo: Viral Dynamics and Host Responses. Viruses, 13(9), 1683. https://doi.org/10.3390/v13091683






