The HSV1 Tail-Anchored Membrane Protein pUL34 Contains a Basic Motif That Supports Active Transport to the Inner Nuclear Membrane Prior to Formation of the Nuclear Egress Complex
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells, Viruses and General Methods
2.2. Plasmids
2.3. Immunofluorescence Microscopy
2.4. BAC Mutagenesis
2.5. Formation of Nuclear Transport Complexes
3. Results
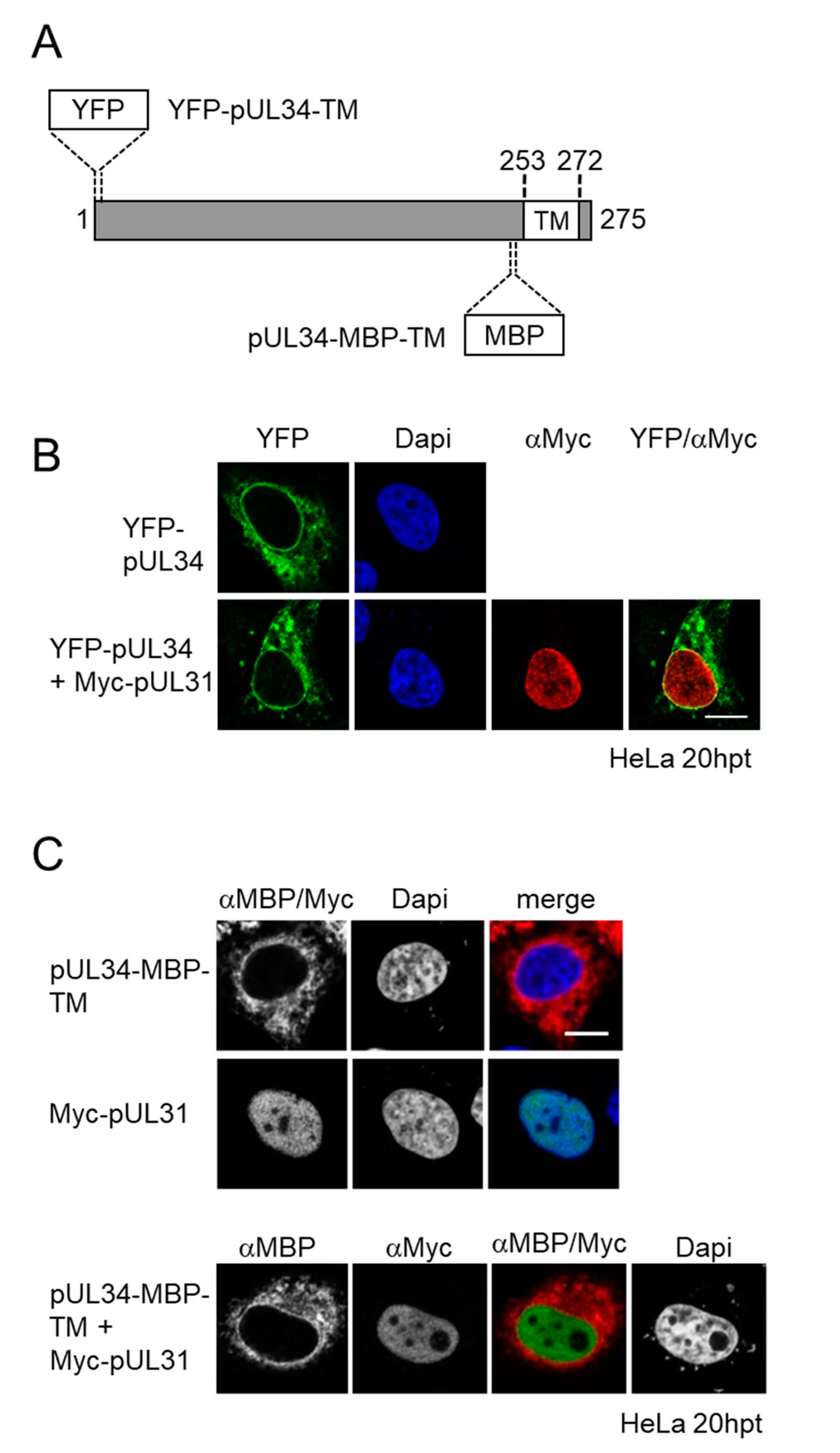
3.1. Membrane Insertion of pUL34 Occurs Prior to Its Nuclear Import
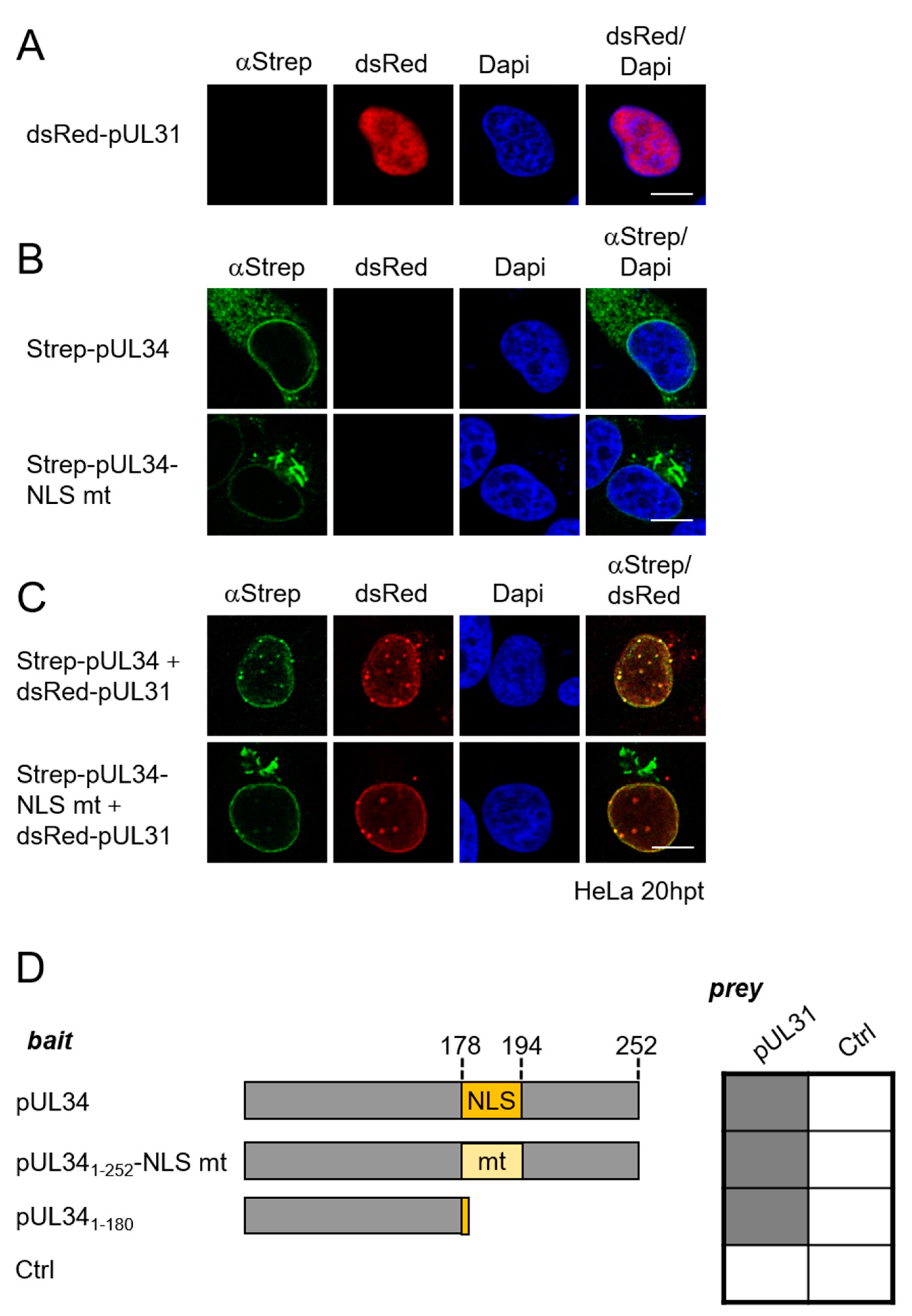
3.2. The NEC Protein pUL34 Contains a Functional Bipartite Nuclear Localization Sequence
3.3. Targeting of pUL34 to the Nuclear Rim Combines Active Transport and Retention by Its NEC Partner pUL31
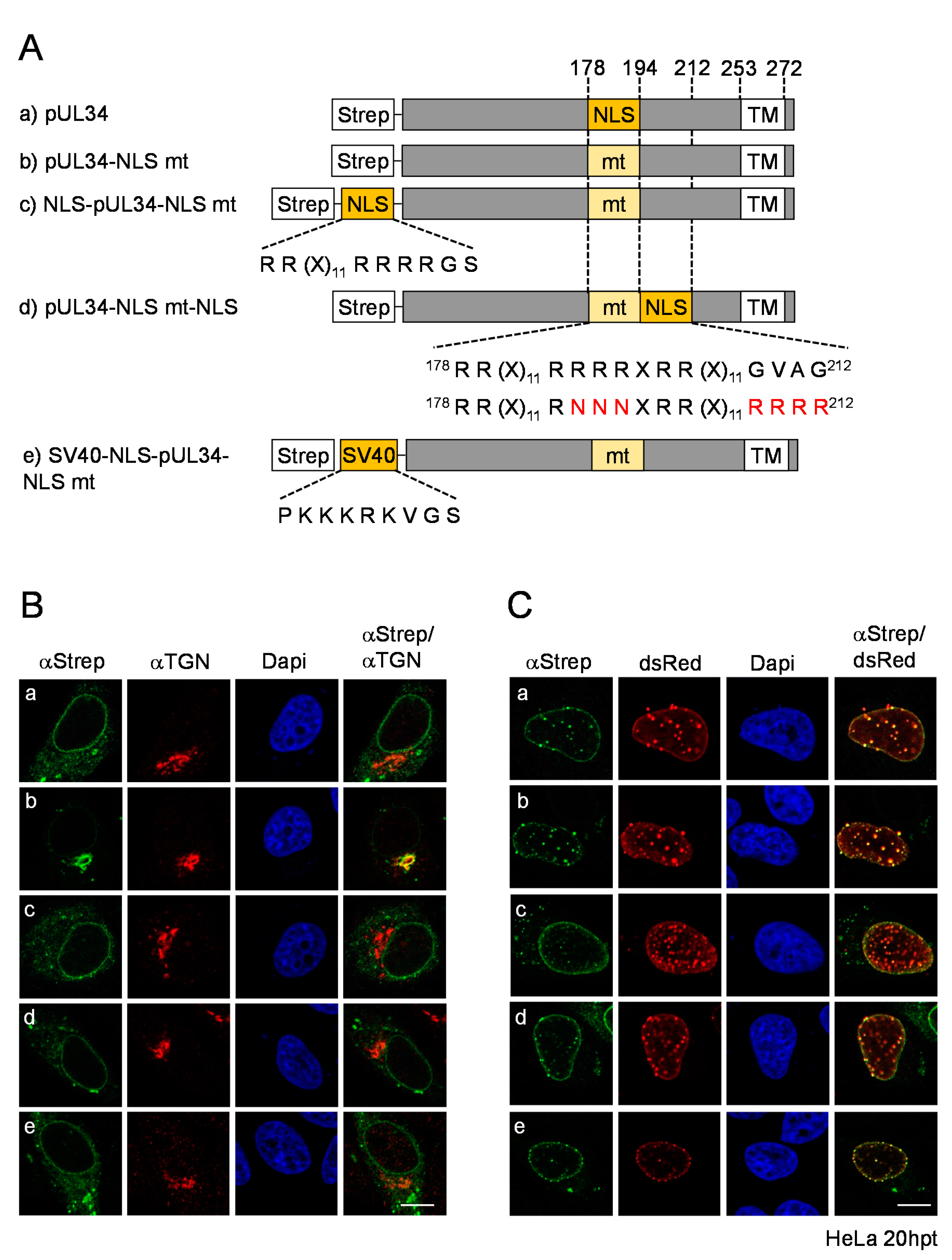
3.4. The pUL34-NLS Functions Independently of Its Position and Can Be Replaced by an SV40-NLS
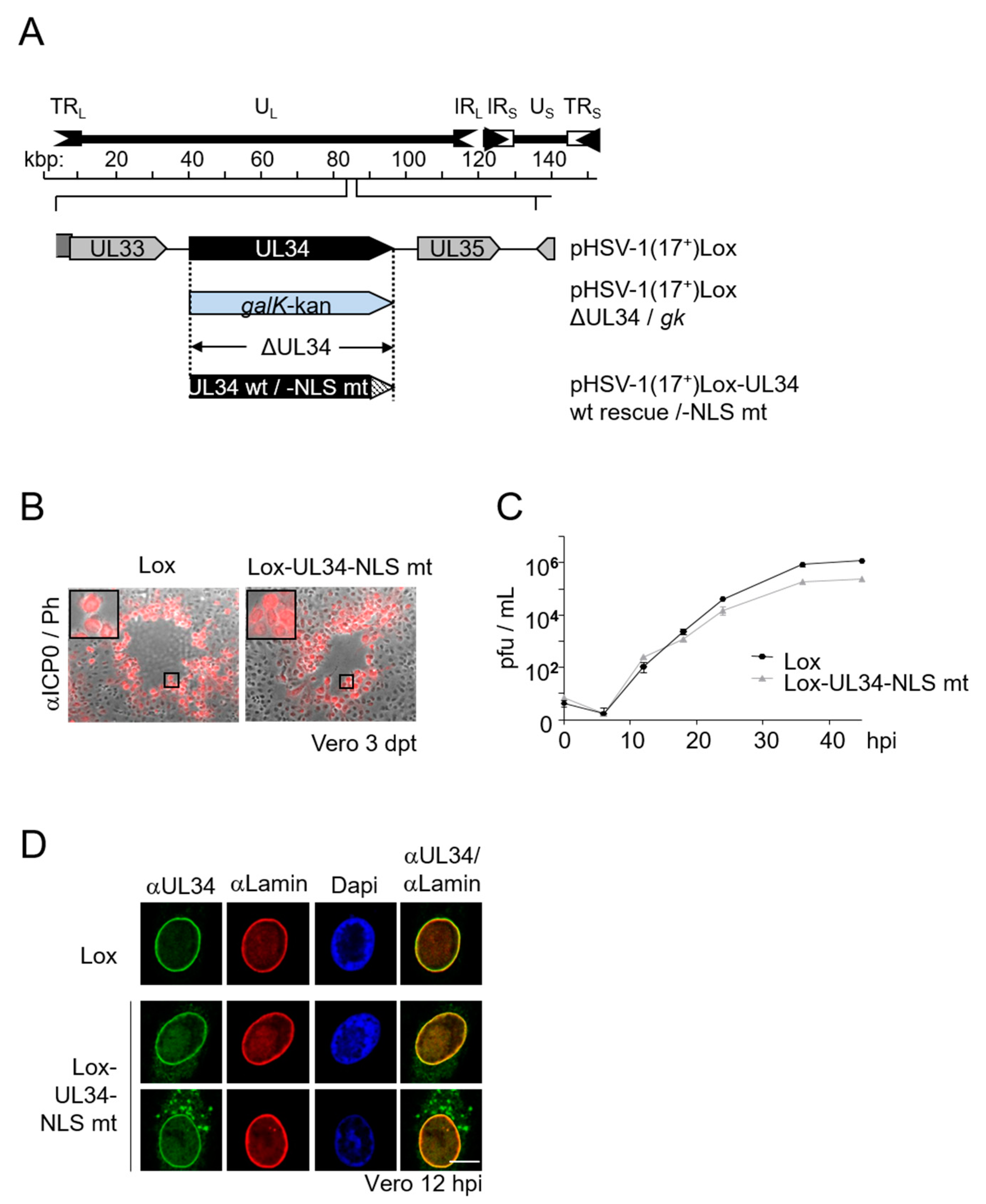
3.5. Viral Replication Is Modestly Compromised in Absence of the pUL34-NLS
4. Discussion
5. Conclusions
- The nuclear import of the HSV1 NEC partners pUL34 and pUL31 ensues separately from each other.
- HSV1 pUL34 contains a functional bipartite NLS that mediates the physical interaction with transport factors of the importin α/β family and the nuclear import of an unrelated cytoplasmic protein.
- In the absence of its NLS, pUL34 is mislocalized to the TGN but retargeted to the authentic localization at the ER upon the insertion of the authentic or a mimic NLS, independent of the insertion site.
- If co-expressed with pUL31, the pUL34-NLS mt is efficiently, although not completely, targeted to the nuclear rim, where NEC formation and membrane budding seemed to occur.
- A viral mutant expressing pUL34-NLS mt is modestly attenuated but viably associated with the partial mislocalization of pUL34-NLS mt in the cytoplasm.
- HSV1 pUL34 reaches the nuclear rim and most likely the INM by an NLS-dependent active process combined with retention by its NEC partner pUL31, thereby promoting viral replication.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BAC | bacterial artificial chromosome |
| ER | endoplasmic reticulum |
| HSV1 | Herpes simplex virus type 1 |
| INM | inner nuclear membrane |
| MBP | Maltose binding protein |
| NEC | nuclear egress complex |
| NLS | nuclear localization sequence |
| ONM | outer nuclear membrane |
| POM | pore membrane |
| TA | tail-anchor |
| TGN | trans Golgi network |
| Y2H | yeast two-hybrid |
| YFP | yellow fluorescent protein |
References
- Hellberg, T.; Paßvogel, L.; Schulz, K.S.; Klupp, B.G.; Mettenleiter, T.C. Nuclear Egress of Herpesviruses: The Prototypic Vesicular Nucleocytoplasmic Transport. Adv. Virus Res. 2016, 94, 81–140. [Google Scholar] [PubMed]
- Mettenleiter, T.C.; Klupp, B.G.; Granzow, H. Herpesvirus assembly: A tale of two membranes. Curr. Opin. Microbiol. 2006, 9, 423–429. [Google Scholar] [CrossRef]
- Mettenleiter, T.C.; Müller, F.; Granzow, H.; Klupp, B.G. The way out: What we know and do not know about herpesvirus nuclear egress. Cell Microbiol. 2013, 15, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Roller, R.J.; Baines, J.D. Herpesvirus Nuclear Egress. Adv. Anat. Embryol. Cell Biol. 2017, 223, 143–169. [Google Scholar] [PubMed]
- Crump, C. Virus Assembly and Egress of HSV. Adv. Exp. Med. Biol. 2018, 1045, 23–44. [Google Scholar]
- Bailer, S.M. Venture from the Interior-Herpesvirus pUL31 Escorts Capsids from Nucleoplasmic Replication Compartments to Sites of Primary Envelopment at the Inner Nuclear Membrane. Cells 2017, 6, 46. [Google Scholar] [CrossRef] [Green Version]
- Bigalke, J.M.; Heldwein, E.E. Have NEC Coat, Will Travel: Structural Basis of Membrane Budding during Nuclear Egress in Herpesviruses. Adv. Virus Res. 2017, 97, 107–141. [Google Scholar] [CrossRef] [Green Version]
- Häge, S.; Sonntag, E.; Borst, E.M.; Tannig, P.; Seyler, L.; Bäuerle, T.; Bailer, S.M.; Lee, C.P.; Müller, R.; Wangen, C.; et al. Patterns of Autologous and Nonautologous Interactions Between Core Nuclear Egress Complex (NEC) Proteins of α-, β- and γ-Herpesviruses. Viruses 2020, 12, 303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marschall, M.; Häge, S.; Conrad, M.; Alkhashrom, S.; Kicuntod, J.; Schweininger, J.; Kriegel, M.; Lösing, J.; Tillmanns, J.; Neipel, F.; et al. Nuclear Egress Complexes of HCMV and Other Herpesviruses: Solving the Puzzle of Sequence Coevolution, Conserved Structures and Subfamily-Spanning Binding Properties. Viruses 2020, 12, 683. [Google Scholar] [CrossRef]
- Ott, M.; Marques, D.; Funk, C.; Bailer, S.M. Asna1/TRC40 that mediates membrane insertion of tail-anchored proteins is required for efficient release of Herpes simplex virus 1 virions. Virol. J. 2016, 13, 175. [Google Scholar] [CrossRef]
- Ott, M.; Tascher, G.; Hassdenteufel, S.; Zimmermann, R.; Haas, J.; Bailer, S.M. Functional characterization of the essential tail anchor of the herpes simplex virus type 1 nuclear egress protein pUL34. J. Gen. Virol. 2011, 92 Pt 12, 2734–2745. [Google Scholar] [CrossRef]
- Funk, C.; Ott, M.; Raschbichler, V.; Nagel, C.H.; Binz, A.; Sodeik, B.; Bauerfeind, R.; Bailer, S.M. The Herpes Simplex Virus Protein pUL31 Escorts Nucleocapsids to Sites of Nuclear Egress, a Process Coordinated by Its N-Terminal Domain. PLoS Pathog. 2015, 11, e1004957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klupp, B.G.; Granzow, H.; Fuchs, W.; Keil, G.M.; Finke, S.; Mettenleiter, T.C. Vesicle formation from the nuclear membrane is induced by coexpression of two conserved herpesvirus proteins. Proc. Natl. Acad. Sci. USA 2007, 104, 7241–7246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bigalke, J.M.; Heuser, T.; Nicastro, D.; Heldwein, E.E. Membrane deformation and scission by the HSV-1 nuclear egress complex. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Zeev-Ben-Mordehai, T.; Weberruß, M.; Lorenz, M.; Cheleski, J.; Hellberg, T.; Whittle, C.; El Omari, K.; Vasishtan, D.; Dent, K.C.; Harlos, K.; et al. Crystal Structure of the Herpesvirus Nuclear Egress Complex Provides Insights into Inner Nuclear Membrane Remodeling. Cell Rep. 2015, 13, 2645–2652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luitweiler, E.M.; Henson, B.W.; Pryce, E.N.; Patel, V.; Coombs, G.; McCaffery, J.M.; Desai, P.J. Interactions of the Kaposi’s Sarcoma-associated herpesvirus nuclear egress complex: ORF69 is a potent factor for remodeling cellular membranes. J. Virol. 2013, 87, 3915–3929. [Google Scholar] [CrossRef] [Green Version]
- Hagen, C.; Dent, K.C.; Zeev-Ben-Mordehai, T.; Grange, M.; Bosse, J.B.; Whittle, C.; Klupp, B.G.; Siebert, C.A.; Vasishtan, D.; Bäuerlein, F.J.; et al. Structural Basis of Vesicle Formation at the Inner Nuclear Membrane. Cell 2015, 163, 1692–1701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desai, P.J.; Pryce, E.N.; Henson, B.W.; Luitweiler, E.M.; Cothran, J. Reconstitution of the Kaposi’s sarcoma-associated herpesvirus nuclear egress complex and formation of nuclear membrane vesicles by coexpression of ORF67 and ORF69 gene products. J. Virol. 2012, 86, 594–598. [Google Scholar] [CrossRef] [Green Version]
- Bigalke, J.M.; Heldwein, E.E. Structural basis of membrane budding by the nuclear egress complex of herpesviruses. Embo. J. 2015, 34, 2921–2936. [Google Scholar] [CrossRef] [PubMed]
- Walzer, S.A.; Egerer-Sieber, C.; Sticht, H.; Sevvana, M.; Hohl, K.; Milbradt, J.; Muller, Y.A.; Marschall, M. Crystal Structure of the Human Cytomegalovirus pUL50-pUL53 Core Nuclear Egress Complex Provides Insight into a Unique Assembly Scaffold for Virus-Host Protein Interactions. J. Biol. Chem. 2015, 290, 27452–27458. [Google Scholar] [CrossRef] [Green Version]
- Lye, M.F.; Sharma, M.; El Omari, K.; Filman, D.J.; Schuermann, J.P.; Hogle, J.M.; Coen, D.M. Unexpected features and mechanism of heterodimer formation of a herpesvirus nuclear egress complex. Embo. J. 2015, 34, 2937–2952. [Google Scholar] [CrossRef] [Green Version]
- Leigh, K.E.; Sharma, M.; Mansueto, M.S.; Boeszoermenyi, A.; Filman, D.J.; Hogle, J.M.; Wagner, G.; Coen, D.M.; Arthanari, H. Structure of a herpesvirus nuclear egress complex subunit reveals an interaction groove that is essential for viral replication. Proc. Natl. Acad. Sci. USA 2015, 112, 9010–9015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marinko, J.T.; Huang, H.; Penn, W.D.; Capra, J.A.; Schlebach, J.P.; Sanders, C.R. Folding and Misfolding of Human Membrane Proteins in Health and Disease: From Single Molecules to Cellular Proteostasis. Chem. Rev. 2019, 119, 5537–5606. [Google Scholar] [CrossRef] [PubMed]
- Katta, S.S.; Smoyer, C.J.; Jaspersen, S.L. Destination: Inner nuclear membrane. Trends Cell Biol. 2014, 24, 221–229. [Google Scholar] [CrossRef]
- Mudumbi, K.C.; Czapiewski, R.; Ruba, A.; Junod, S.L.; Li, Y.; Luo, W.; Ngo, C.; Ospina, V.; Schirmer, E.C.; Yang, W. Nucleoplasmic signals promote directed transmembrane protein import simultaneously via multiple channels of nuclear pores. Nat. Commun. 2020, 11, 2184. [Google Scholar] [CrossRef]
- Sandbaumhuter, M.; Dohner, K.; Schipke, J.; Binz, A.; Pohlmann, A.; Sodeik, B.; Bauerfeind, R. Cytosolic herpes simplex virus capsids not only require binding inner tegument protein pUL36 but also pUL37 for active transport prior to secondary envelopment. Cell Microbiol. 2013, 15, 248–269. [Google Scholar] [CrossRef]
- Nygardas, M.; Paavilainen, H.; Muther, N.; Nagel, C.H.; Roytta, M.; Sodeik, B.; Hukkanen, V. A herpes simplex virus-derived replicative vector expressing LIF limits experimental demyelinating disease and modulates autoimmunity. PLoS ONE 2013, 8, e64200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagel, C.H.; Dohner, K.; Fathollahy, M.; Strive, T.; Borst, E.M.; Messerle, M.; Sodeik, B. Nuclear egress and envelopment of herpes simplex virus capsids analyzed with dual-color fluorescence HSV1(17+). J. Virol. 2008, 82, 3109–3124. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, T.; Striebinger, H.; Haas, J.; Bailer, S.M. The heterogeneous nuclear ribonucleoprotein K is important for Herpes simplex virus-1 propagation. FEBS Lett. 2010, 584, 4361–4365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Striebinger, H.; Koegl, M.; Bailer, S.M. A high-throughput yeast two-hybrid protocol to determine virus-host protein interactions. Methods Mol. Biol. (Clifton N. J.) 2013, 1064, 1–15. [Google Scholar] [CrossRef]
- Fossum, E.; Friedel, C.C.; Rajagopala, S.V.; Titz, B.; Baiker, A.; Schmidt, T.; Kraus, T.; Stellberger, T.; Rutenberg, C.; Suthram, S.; et al. Evolutionarily conserved herpesviral protein interaction networks. PLoS Pathog. 2009, 5, e1000570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Striebinger, H.; Zhang, J.; Ott, M.; Funk, C.; Radtke, K.; Duron, J.; Ruzsics, Z.; Haas, J.; Lippé, R.; Bailer, S.M. Subcellular trafficking and functional importance of herpes simplex virus type 1 glycoprotein M domains. J. Gen. Virol. 2015, 96, 3313–3325. [Google Scholar] [CrossRef]
- Damelin, M.; Silver, P.A.; Corbett, A.H. Nuclear protein transport. Methods Enzymol. 2002, 351, 587–607. [Google Scholar]
- Timney, B.L.; Raveh, B.; Mironska, R.; Trivedi, J.M.; Kim, S.J.; Russel, D.; Wente, S.R.; Sali, A.; Rout, M.P. Simple rules for passive diffusion through the nuclear pore complex. J. Cell Biol. 2016, 215, 57–76. [Google Scholar] [CrossRef]
- Reynolds, A.E.; Ryckman, B.J.; Baines, J.D.; Zhou, Y.; Liang, L.; Roller, R.J. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 2001, 75, 8803–8817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamauchi, Y.; Shiba, C.; Goshima, F.; Nawa, A.; Murata, T.; Nishiyama, Y. Herpes simplex virus type 2 UL34 protein requires UL31 protein for its relocation to the internal nuclear membrane in transfected cells. J. Gen. Virol. 2001, 82, 1423–1428. [Google Scholar] [CrossRef]
- Ryckman, B.J.; Roller, R.J. Herpes simplex virus type 1 primary envelopment: UL34 protein modification and the US3-UL34 catalytic relationship. J. Virol. 2004, 78, 399–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bjerke, S.L.; Roller, R.J. Roles for herpes simplex virus type 1 UL34 and US3 proteins in disrupting the nuclear lamina during herpes simplex virus type 1 egress. Virology 2006, 347, 261–276. [Google Scholar] [CrossRef] [Green Version]
- Bjerke, S.L.; Cowan, J.M.; Kerr, J.K.; Reynolds, A.E.; Baines, J.D.; Roller, R.J. Effects of charged cluster mutations on the function of herpes simplex virus type 1 UL34 protein. J. Virol. 2003, 77, 7601–7610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmeiser, C.; Borst, E.; Sticht, H.; Marschall, M.; Milbradt, J. The cytomegalovirus egress proteins pUL50 and pUL53 are translocated to the nuclear envelope through two distinct modes of nuclear import. J. Gen. Virol. 2013, 94 Pt 9, 2056–2069. [Google Scholar] [CrossRef]
- Milbradt, J.; Sonntag, E.; Wagner, S.; Strojan, H.; Wangen, C.; Lenac Rovis, T.; Lisnic, B.; Jonjic, S.; Sticht, H.; Britt, W.J.; et al. Human Cytomegalovirus Nuclear Capsids Associate with the Core Nuclear Egress Complex and the Viral Protein Kinase pUL97. Viruses 2018, 10, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| No. | Name | Sequence | Use |
|---|---|---|---|
| 1 | UL34_vec_fw | GCAGACTGGAAGCTGGCACCAGGCCCTGCGG | Gibson |
| 2 | UL34_vec_rv | TGCTTCCCGGGCGGGAGGGCCCTTGGGTTAAC | Gibson |
| 3 | MBP_fw | TCCCGCCCGGGAAGCAAAATCGAAGAAGGTAAA | Gibson |
| 4 | MBP_rev | CCTGGTGCCAGCTTCCAGTCTGCGCGTCTTTCAGG | Gibson |
| 5 | UL34-NLS-mt_fw | CGCCGAGCAGGCTATTACCCGTAACAACAACACCCGGCGGTCCCGGGG | mutagenesis |
| 6 | UL34-NLS-mt_rev | CCCCGGGACCGCCGGGTGTTGTTGTTACGGGTAATAGCCTGCTCGGCG | mutagenesis |
| 7 | NLS-UL34-NLS-mt_vector_fw | GCGGGACTGGGCAAGCCCTACAC | Gibson |
| 8 | NLS-UL34-NLS-mt_vector_rev | CCTCGAGGGATCCCCGGGTACCGAG | Gibson |
| 9 | NLS-UL34-NLS-mt_fw | CCGGGGATCCCTCGAGGAGACGGATCCTGTGC | Gibson |
| 10 | NLS-UL34-NLS-mt_rev1 | AGTCCCGCGGATCCTCGGCGGCGACGGGTAATAG | Gibson |
| 11 | NLS-UL34-NLS-mt_rev2 | TGTAGGGCTTGCCCAGTCCCGCGGATCCTC | Gibson |
| 12 | UL34-NLS-mt-NLS_fw | CCGAGGCCGGGCTGCGGCGGCGCCGAACGGGTTTC | mutagenesis |
| 13 | UL34-NLS-mt-NLS_rev | CGGAAACCCGTTCGGCGCCGCCGCAGCCCGGCCTC | mutagenesis |
| 14 | SV40NLS-UL34-NLS-mt_fw1 | GCGGTCCCGAATTCGAGCTCGGTAC | Gibson |
| 15 | SV40NLS-UL34-NLS-mt_rev1 | CCTACCTTTCTCTTCTTTTTTGGCCTCGAGGGATCCC | Gibson |
| 16 | SV40NLS-UL34-NLS-mt_fw2 | GCCGAGACCGCGGTCCCGAATTCGAG | Gibson |
| 17 | SV40NLS-UL34-NLS-mt_rev2 | CTTGCCCAGTCCCGCGGATCCTACCTTTCTCTTCTTTTTTG | Gibson |
| 18 | EYFP-UL34-NLS_fw | TCAGATCCGCTAGCGCTACCGGTCGCCACCATGGTGAGCAAGGGCGAG | restriction cloning |
| 19 | EYFP-UL34-NLS_rev1 | CTGCTCGGCGGCGCGGCACAGAATCCGTCTCGTAGCTCGAGATCTGAG | restriction cloning |
| 20 | EYFP-UL34-NLS_rev2 | TTTGGATCCTAAGGTTCGGCGGCGACGGGTAATAGCCTGCTCGGCGGCGC | restriction cloning |
| 21 | UL34/gk_fw | GAACCCTTTGGTGGGTTTACGCGGGCACGCACGCTCCCATCGCGGGCGCCCCTGTTGACAATTAATCATCGGCA | BAC |
| 22 | UL34/gk_rev | GCGAAGGCGTCCGGAACGCACTGGCGATTAGGGCGGCGGTGCGTCCTTTTGCCAGTGTTACAACCAATTAACC | BAC |
| 23 | UL34_fw | GAACCCTTTGGTGGGTTTACGCGGGCACGCACGCTCCCATCGCGGGCGCCATGGCGGGACTGGGCAAGCCCTAC | BAC |
| 24 | UL34_rev | GCGAAGGCGTCCGGAACGCACTGGCGATTAGGGCGGCGGTGCGTCCTTTTTTATAGGCGCGCGCCAGCACCAAC | BAC |
| No. | BAC | Inserted Fragment | Recipient BAC |
|---|---|---|---|
| 1 | pHSV1(17+)Lox | ||
| 2 | pHSV1(17+)Lox ΔUL34/gK | galK-kan in UL34 locus (bp 1-825); PCR with oligos no. 17 and 18 and pGPS-galK-kan as template | No. 1 |
| 3 | pHSV1(17+)Lox-UL34 wt rescue | UL34 wt; PCR with oligos no. 19 and 20 and pDONR207-UL341-275 as template | No. 2 |
| 4 | pHSV1(17+)Lox-UL34-NLS mt | UL34-NLS mt; PCR with no. 19 and 20 and pDONR207-UL341-275-NLS mt as template | No. 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Funk, C.; Marques da Silveira e Santos, D.; Ott, M.; Raschbichler, V.; Bailer, S.M. The HSV1 Tail-Anchored Membrane Protein pUL34 Contains a Basic Motif That Supports Active Transport to the Inner Nuclear Membrane Prior to Formation of the Nuclear Egress Complex. Viruses 2021, 13, 1544. https://doi.org/10.3390/v13081544
Funk C, Marques da Silveira e Santos D, Ott M, Raschbichler V, Bailer SM. The HSV1 Tail-Anchored Membrane Protein pUL34 Contains a Basic Motif That Supports Active Transport to the Inner Nuclear Membrane Prior to Formation of the Nuclear Egress Complex. Viruses. 2021; 13(8):1544. https://doi.org/10.3390/v13081544
Chicago/Turabian StyleFunk, Christina, Débora Marques da Silveira e Santos, Melanie Ott, Verena Raschbichler, and Susanne M. Bailer. 2021. "The HSV1 Tail-Anchored Membrane Protein pUL34 Contains a Basic Motif That Supports Active Transport to the Inner Nuclear Membrane Prior to Formation of the Nuclear Egress Complex" Viruses 13, no. 8: 1544. https://doi.org/10.3390/v13081544
APA StyleFunk, C., Marques da Silveira e Santos, D., Ott, M., Raschbichler, V., & Bailer, S. M. (2021). The HSV1 Tail-Anchored Membrane Protein pUL34 Contains a Basic Motif That Supports Active Transport to the Inner Nuclear Membrane Prior to Formation of the Nuclear Egress Complex. Viruses, 13(8), 1544. https://doi.org/10.3390/v13081544






