Multi-Tissue Transcriptomic-Informed In Silico Investigation of Drugs for the Treatment of Dengue Fever Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Biological Samples, Laboratorial Processing and Raw Data Analysis
2.2. Published Datasets
2.3. Differential Expression Evaluation and Gene Set Enrichment Analysis
2.4. Drug Repurposing
3. Results
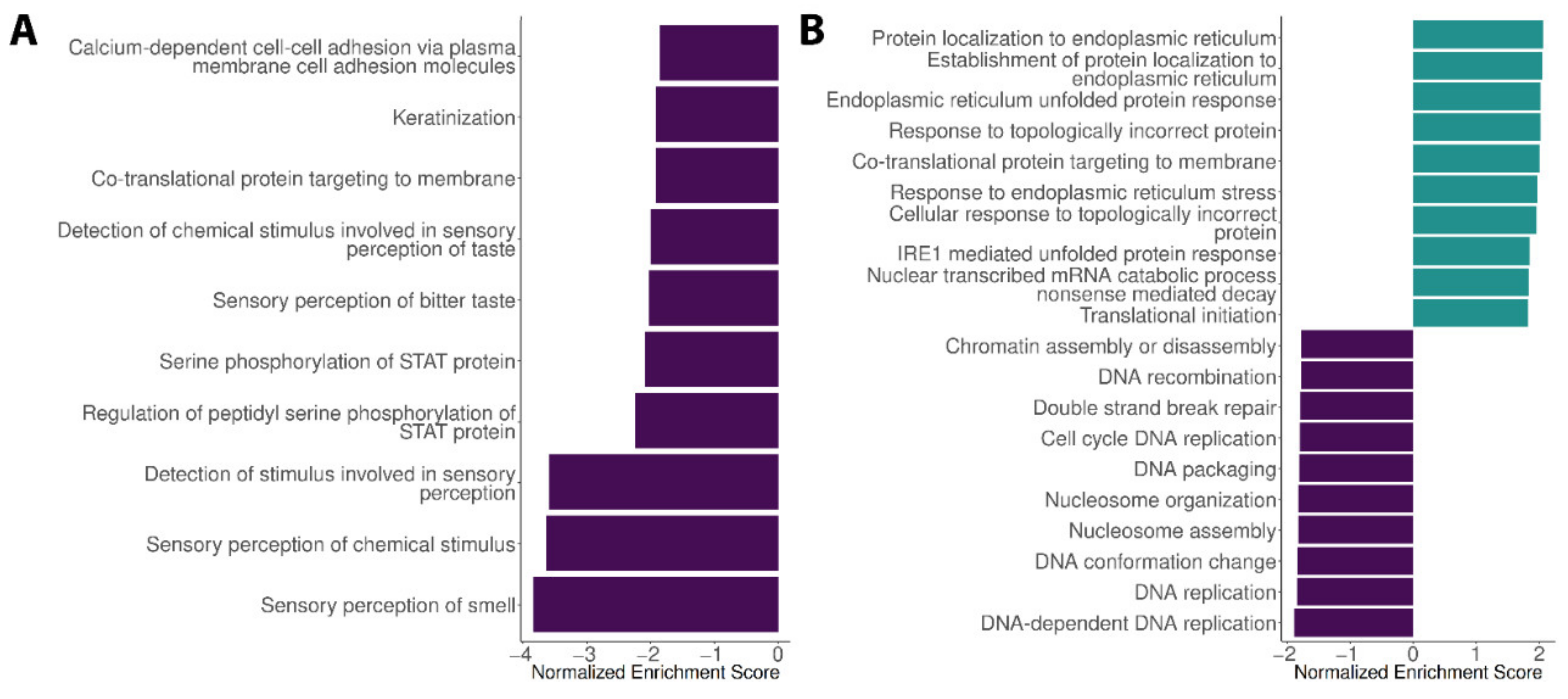
3.1. Expression Profiles in the Liver, Spleen and Encephalon Dengue Cohorts
3.2. Expression Profiles in the Blood Dengue Cohorts
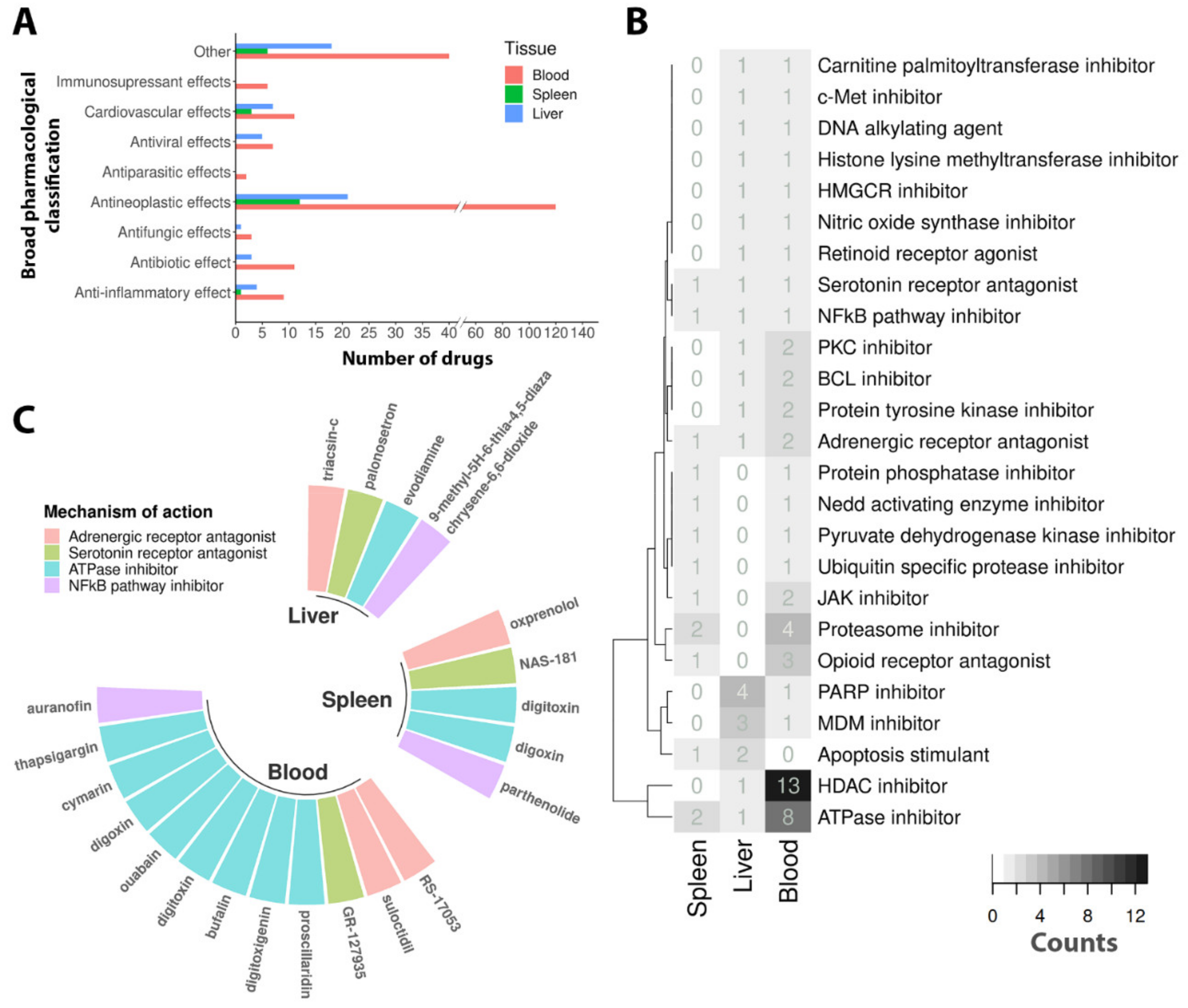
3.3. Drug Repurposing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guzman, M.G.; Gubler, D.J.; Izquierdo, A.; Martinez, E.; Halstead, S.B. Dengue infection. Nat. Rev. Dis. Primers 2016, 2, 16055. [Google Scholar] [CrossRef] [PubMed]
- Mayer, S.V.; Tesh, R.B.; Vasilakis, N. The emergence of arthropod-borne viral diseases: A global prospective on dengue, chikungunya and zika fevers. Acta Trop. 2017, 166, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Shepard, D.S.; Undurraga, E.A.; Halasa, Y.A.; Stanaway, J.D. The global economic burden of dengue: A systematic analysis. Lancet Infect. Dis. 2016, 16, 935–941. [Google Scholar] [CrossRef]
- Kraemer, M.U.; Sinka, M.E.; Duda, K.A.; Mylne, A.Q.; Shearer, F.M.; Barker, C.M.; Moore, C.G.; Carvalho, R.G.; Coelho, G.E.; Van Bortel, W.; et al. The global distribution of the arbovirus vectors Aedes aegypti and Ae. albopictus. Elife 2015, 4, e08347. [Google Scholar] [CrossRef] [PubMed]
- Halstead, S.B.; Mahalingam, S.; Marovich, M.A.; Ubol, S.; Mosser, D.M. Intrinsic antibody-dependent enhancement of microbial infection in macrophages: Disease regulation by immune complexes. Lancet Infect. Dis. 2010, 10, 712–722. [Google Scholar] [CrossRef]
- Oliveira, M.; Ferreira, J.; Fernandes, V.; Sakuntabhai, A.; Pereira, L. Host ancestry and dengue fever: From mapping of candidate genes to prediction of worldwide genetic risk. Future Virol. 2018, 13, 647–655. [Google Scholar] [CrossRef]
- Khor, C.C.; Chau, T.N.; Pang, J.; Davila, S.; Long, H.T.; Ong, R.T.; Dunstan, S.J.; Wills, B.; Farrar, J.; Van Tram, T.; et al. Genome-wide association study identifies susceptibility loci for dengue shock syndrome at MICB and PLCE1. Nat. Genet. 2011, 43, 1139–1141. [Google Scholar] [CrossRef] [PubMed]
- Sierra, B.; Triska, P.; Soares, P.; Garcia, G.; Perez, A.B.; Aguirre, E.; Oliveira, M. OSBPL10, RXRA and lipid metabolism confer African-ancestry protection against dengue haemorrhagic fever in admixed Cubans. PLoS Pathog. 2017, 13, e1006220. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, M.; Lert-Itthiporn, W.; Cavadas, B.; Fernandes, V.; Chuansumrit, A.; Anunciação, O.; Casademont, I.; Koeth, F.; Penova, M.; Tangnararatchakit, K.; et al. Joint ancestry and association test indicate two distinct pathogenic pathways involved in classical dengue fever and dengue shock syndrome. PLoS Negl. Trop. Dis. 2018, 12, e0006202. [Google Scholar] [CrossRef]
- Oliveira, M.; Saraiva, D.P.; Cavadas, B.; Fernandes, V.; Pedro, N.; Casademont, I.; Koeth, F.; Alshamali, F.; Harich, N.; Cherni, L.; et al. Population genetics-informed meta-analysis in seven genes associated with risk to dengue fever disease. Infect. Genet. Evol. 2018, 62, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Trung, D.T.; Thao, L.T.T.; Hien, T.T.; Hung, N.T.; Vinh, N.N.; Hien, P.T.; Chinh, N.T.; Simmons, C.; Wills, B. Liver involvement associated with dengue infection in adults in Vietnam. Am. J. Trop. Med. Hyg. 2010, 83, 774–780. [Google Scholar] [CrossRef] [PubMed]
- Samanta, J.; Sharma, V. Dengue and its effects on liver. World J. Clin. Cases 2015, 3, 125–131. [Google Scholar] [CrossRef]
- de Silva, W.T.; Gunasekera, M. Spontaneous splenic rupture during the recovery phase of dengue fever. BMC Res. Notes 2015, 8, 286. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nascimento, E.J.; Braga-Neto, U.; Calzavara-Silva, C.E.; Gomes, A.L.; Abath, F.G.; Brito, C.A.; Cordeiro, M.T.; Silva, A.M.; Magalhães, C.; Andrade, R.; et al. Gene expression profiling during early acute febrile stage of dengue infection can predict the disease outcome. PLoS ONE 2009, 4, e7892. [Google Scholar] [CrossRef]
- Loke, P.; Hammond, S.N.; Leung, J.M.; Kim, C.C.; Batra, S.; Rocha, C.; Balmaseda, A.; Harris, E. Gene expression patterns of dengue virus-infected children from nicaragua reveal a distinct signature of increased metabolism. PLoS Negl. Trop. Dis. 2010, 4, e710. [Google Scholar] [CrossRef] [PubMed]
- Popper, S.J.; Gordon, A.; Liu, M.; Balmaseda, A.; Harris, E.; Relman, D.A. Temporal dynamics of the transcriptional response to dengue virus infection in Nicaraguan children. PLoS Negl. Trop. Dis. 2012, 6, e1966. [Google Scholar] [CrossRef] [PubMed]
- Sessions, O.M.; Tan, Y.; Goh, K.C.; Liu, Y.; Tan, P.; Rozen, S.; Ooi, E.E. Host cell transcriptome profile during wild-type and attenuated dengue virus infection. PLoS Negl. Trop. Dis. 2013, 7, e2107. [Google Scholar] [CrossRef]
- Kwissa, M.; Nakaya, H.I.; Onlamoon, N.; Wrammert, J.; Villinger, F.; Perng, G.C.; Yoksan, S.; Pattanapanyasat, K.; Chokephaibulkit, K.; Ahmed, R.; et al. Dengue virus infection induces expansion of a CD14(+)CD16(+) monocyte population that stimulates plasmablast differentiation. Cell Host Microbe 2014, 16, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Long, H.T.; Hibberd, M.L.; Hien, T.T.; Dung, N.M.; Van Ngoc, T.; Farrar, J.; Wills, B.; Simmons, C.P. Patterns of gene transcript abundance in the blood of children with severe or uncomplicated dengue highlight differences in disease evolution and host response to dengue virus infection. J. Infect. Dis. 2009, 199, 537–546. [Google Scholar] [CrossRef]
- Rajapakse, S.; Rodrigo, C.; Rajapakse, A. Treatment of dengue fever. Infect Drug Resist. 2012, 5, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Gutierrez, M.; Castellanos, J.E.; Gallego-Gómez, J.C. Statins reduce dengue virus production via decreased virion assembly. Intervirology 2011, 54, 202–216. [Google Scholar] [CrossRef]
- Martinez-Gutierrez, M.; Correa-Londoño, L.A.; Castellanos, J.E.; Gallego-Gómez, J.C.; Osorio, J.E. Lovastatin delays infection and increases survival rates in AG129 mice infected with dengue virus serotype 2. PLoS ONE 2014, 9, e87412. [Google Scholar] [CrossRef]
- Whitehorn, J.; Nguyen, C.V.V.; Khanh, L.P.; Kien, D.T.H.; Quyen, N.T.H.; Tran, N.T.T.; Hang, N.T.; Truong, N.T.; Hue Tai, L.T.; Cam Huong, N.T.; et al. Lovastatin for the Treatment of Adult Patients With Dengue: A Randomized, Double-Blind, Placebo-Controlled Trial. Clin. Infect. Dis. 2016, 62, 468–476. [Google Scholar] [CrossRef] [PubMed]
- Chia, P.Y.; Htun, H.L.; Ling, W.P.; Leo, Y.S.; Yeo, T.W.; Lye, D.C.B. Hyperlipidemia, statin use and dengue severity. Sci. Rep. 2018, 8, 17147. [Google Scholar] [CrossRef]
- Wilder-Smith, A.; Flasche, S.; Smith, P.G. Vaccine-attributable severe dengue in the Philippines. Lancet 2019, 394, 2151–2152. [Google Scholar] [CrossRef]
- Loging, W.; Harland, L.; Williams-Jones, B. High-throughput electronic biology: Mining information for drug discovery. Nat. Rev. Drug. Discov. 2007, 6, 220–230. [Google Scholar] [CrossRef]
- Gordon, D.E.; Hiatt, J.; Bouhaddou, M.; Rezelj, V.V.; Ulferts, S.; Braberg, H.; Jureka, A.S.; Obernier, K.; Guo, J.Z.; Batra, J.; et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science 2020, 370, 6521. [Google Scholar] [CrossRef]
- Lamb, J.; Crawford, E.D.; Peck, D.; Modell, J.W.; Blat, I.C.; Wrobel, M.J.; Lerner, J.; Brunet, J.P.; Subramanian, A.; Ross, K.N.; et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science 2006, 313, 1929–1935. [Google Scholar] [CrossRef]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171, 1437–1452.e1417. [Google Scholar] [CrossRef]
- Agren, R.; Mardinoglu, A.; Asplund, A.; Kampf, C.; Uhlen, M.; Nielsen, J. Identification of anticancer drugs for hepatocellular carcinoma through personalized genome-scale metabolic modeling. Mol. Syst. Biol. 2014, 10, 721. [Google Scholar] [CrossRef]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, 6352. [Google Scholar] [CrossRef]
- Amemiya, T.; Gromiha, M.M.; Horimoto, K.; Fukui, K. Drug repositioning for dengue haemorrhagic fever by integrating multiple omics analyses. Sci. Rep. 2019, 9, 523. [Google Scholar] [CrossRef]
- Lanciotti, R.S.; Calisher, C.H.; Gubler, D.J.; Chang, G.J.; Vorndam, A.V. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J. Clin. Microbiol. 1992, 30, 545–551. [Google Scholar] [CrossRef]
- Santiago, G.A.; Vergne, E.; Quiles, Y.; Cosme, J.; Vazquez, J.; Medina, J.F.; Medina, F.; Colón, C.; Margolis, H.; Muñoz-Jordán, J.L. Analytical and clinical performance of the CDC real time RT-PCR assay for detection and typing of dengue virus. PLoS Negl. Trop. Dis. 2013, 7, e2311. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets--update. Nucleic Acids Res. 2013, 41, D991-995. [Google Scholar] [CrossRef] [PubMed]
- Zanini, F.; Pu, S.Y.; Bekerman, E.; Einav, S.; Quake, S.R. Single-cell transcriptional dynamics of flavivirus infection. Elife 2018, 7, e32942. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 2019, 47, D1102–D1109. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Walker, D.G.; Whetzel, A.M.; Serrano, G.; Sue, L.I.; Lue, L.F.; Beach, T.G. Characterization of RNA isolated from eighteen different human tissues: Results from a rapid human autopsy program. Cell Tissue Bank 2016, 17, 361–375. [Google Scholar] [CrossRef] [PubMed]
- Russell, J.A.; Walley, K.R. Vasopressin and its immune effects in septic shock. J. Innate Immun. 2010, 2, 446–460. [Google Scholar] [CrossRef] [PubMed]
- Lemke, G.; Rothlin, C.V. Immunobiology of the TAM receptors. Nat. Rev. Immunol. 2008, 8, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Dalrymple, N.A.; Cimica, V.; Mackow, E.R. Dengue Virus NS Proteins Inhibit RIG-I/MAVS Signaling by Blocking TBK1/IRF3 Phosphorylation: Dengue Virus Serotype 1 NS4A Is a Unique Interferon-Regulating Virulence Determinant. MBio 2015, 6, e00553-15. [Google Scholar] [CrossRef]
- Mishra, R.; Lahon, A.; Banerjea, A.C. Dengue Virus Degrades USP33-ATF3 Axis via Extracellular Vesicles to Activate Human Microglial Cells. J. Immunol. 2020, 205, 1787–1798. [Google Scholar] [CrossRef]
- Hai, T.; Wolford, C.C.; Chang, Y.S. ATF3, a hub of the cellular adaptive-response network, in the pathogenesis of diseases: Is modulation of inflammation a unifying component? Gene. Expr. 2010, 15, 1–11. [Google Scholar] [CrossRef]
- Phoolcharoen, W.; Smith, D.R. Internalization of the dengue virus is cell cycle modulated in HepG2, but not Vero cells. J. Med. Virol. 2004, 74, 434–441. [Google Scholar] [CrossRef]
- Tokunaga, M.; Miyamoto, Y.; Suzuki, T.; Otani, M.; Inuki, S.; Esaki, T.; Nagao, C.; Mizuguchi, K.; Ohno, H.; Yoneda, Y.; et al. Novel anti-flavivirus drugs targeting the nucleolar distribution of core protein. Virology 2020, 541, 41–51. [Google Scholar] [CrossRef]
- Diamond, M.S.; Zachariah, M.; Harris, E. Mycophenolic acid inhibits dengue virus infection by preventing replication of viral RNA. Virology 2002, 304, 211–221. [Google Scholar] [CrossRef] [PubMed]
- He, J.C.; Lu, T.C.; Fleet, M.; Sunamoto, M.; Husain, M.; Fang, W.; Neves, S.; Chen, Y.; Shankland, S.; Iyengar, R.; et al. Retinoic acid inhibits HIV-1-induced podocyte proliferation through the cAMP pathway. J. Am. Soc. Nephrol. 2007, 18, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Machado, C.R.; Machado, E.S.; Rohloff, R.D.; Azevedo, M.; Campos, D.P.; de Oliveira, R.B.; Brasil, P. Is pregnancy associated with severe dengue? A review of data from the Rio de Janeiro surveillance information system. PLoS Negl. Trop. Dis. 2013, 7, e2217. [Google Scholar] [CrossRef] [PubMed]
- Srisutthisamphan, K.; Jirakanwisal, K.; Ramphan, S.; Tongluan, N.; Kuadkitkan, A.; Smith, D.R. Hsp90 interacts with multiple dengue virus 2 proteins. Sci. Rep. 2018, 8, 4308. [Google Scholar] [CrossRef]
- Hughes, T.R.; Marton, M.J.; Jones, A.R.; Roberts, C.J.; Stoughton, R.; Armour, C.D.; Bennett, H.A.; Coffey, E.; Dai, H.; He, Y.D.; et al. Functional discovery via a compendium of expression profiles. Cell 2000, 102, 109–126. [Google Scholar] [CrossRef]
- Ferreira, P.G.; Muñoz-Aguirre, M.; Reverter, F.; CP, S.G.; Sousa, A.; Amadoz, A.; Sodaei, R.; Hidalgo, M.R.; Pervouchine, D.; Carbonell-Caballero, J.; et al. The effects of death and post-mortem cold ischemia on human tissue transcriptomes. Nat. Commun. 2018, 9, 490. [Google Scholar] [CrossRef]
- Cheng, Y.L.; Lin, Y.S.; Chen, C.L.; Wan, S.W.; Ou, Y.D.; Yu, C.Y.; Tsai, T.T.; Tseng, P.C.; Lin, C.F. Dengue Virus Infection Causes the Activation of Distinct NF-κB Pathways for Inducible Nitric Oxide Synthase and TNF-α Expression in RAW264.7 Cells. Mediat. Inflamm. 2015, 2015, 274025. [Google Scholar] [CrossRef]
- Barrows, N.J.; Campos, R.K.; Powell, S.T.; Prasanth, K.R.; Schott-Lerner, G.; Soto-Acosta, R.; Galarza-Muñoz, G.; McGrath, E.L.; Urrabaz-Garza, R.; Gao, J.; et al. A Screen of FDA-Approved Drugs for Inhibitors of Zika Virus Infection. Cell Host Microbe 2016, 20, 259–270. [Google Scholar] [CrossRef]
- Mazzon, M.; Ortega-Prieto, A.M.; Imrie, D.; Luft, C.; Hess, L.; Czieso, S.; Grove, J.; Skelton, J.K.; Farleigh, L.; Bugert, J.J.; et al. Identification of Broad-Spectrum Antiviral Compounds by Targeting Viral Entry. Viruses 2019, 11, 176. [Google Scholar] [CrossRef] [PubMed]
- Ko, H.C.; Wang, Y.H.; Liou, K.T.; Chen, C.M.; Chen, C.H.; Wang, W.Y.; Chang, S.; Hou, Y.C.; Chen, K.T.; Chen, C.F.; et al. Anti-inflammatory effects and mechanisms of the ethanol extract of Evodia rutaecarpa and its bioactive components on neutrophils and microglial cells. Eur. J. Pharmacol. 2007, 555, 211–217. [Google Scholar] [CrossRef]
- Dai, J.P.; Li, W.Z.; Zhao, X.F.; Wang, G.F.; Yang, J.C.; Zhang, L.; Chen, X.X.; Xu, Y.X.; Li, K.S. A drug screening method based on the autophagy pathway and studies of the mechanism of evodiamine against influenza A virus. PLoS ONE 2012, 7, e42706. [Google Scholar] [CrossRef] [PubMed]
- Pollard, B.S.; JC, B.L.; Pollard, J.R. Classical Drug Digitoxin Inhibits Influenza Cytokine Storm, With Implications for Covid-19 Therapy. In Vivo 2020, 34, 3723–3730. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.L.; Stein, D.A.; Shum, D.; Fischer, M.A.; Radu, C.; Bhinder, B.; Djaballah, H.; Nelson, J.A.; Früh, K.; Hirsch, A.J. Inhibition of dengue virus replication by a class of small-molecule compounds that antagonize dopamine receptor d4 and downstream mitogen-activated protein kinase signaling. J. Virol. 2014, 88, 5533–5542. [Google Scholar] [CrossRef]
- Wang, P.; Wang, Z.; Liu, J. Role of HDACs in normal and malignant hematopoiesis. Mol. Cancer 2020, 19, 5. [Google Scholar] [CrossRef]
- Delgado, F.G.; Cárdenas, P.; Castellanos, J.E. Valproic Acid Downregulates Cytokine Expression in Human Macrophages Infected with Dengue Virus. Diseases 2018, 6, 59. [Google Scholar] [CrossRef] [PubMed]
- Choy, M.M.; Zhang, S.L.; Costa, V.V.; Tan, H.C.; Horrevorts, S.; Ooi, E.E. Proteasome Inhibition Suppresses Dengue Virus Egress in Antibody Dependent Infection. PLoS Negl. Trop. Dis. 2015, 9, e0004058. [Google Scholar] [CrossRef]
- Mukhopadhyay, P.; Rajesh, M.; Cao, Z.; Horváth, B.; Park, O.; Wang, H.; Erdelyi, K.; Holovac, E.; Wang, Y.; Liaudet, L.; et al. Poly (ADP-ribose) polymerase-1 is a key mediator of liver inflammation and fibrosis. Hepatology 2014, 59, 1998–2009. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sierra, B.; Magalhães, A.C.; Soares, D.; Cavadas, B.; Perez, A.B.; Alvarez, M.; Aguirre, E.; Bracho, C.; Pereira, L.; Guzman, M.G. Multi-Tissue Transcriptomic-Informed In Silico Investigation of Drugs for the Treatment of Dengue Fever Disease. Viruses 2021, 13, 1540. https://doi.org/10.3390/v13081540
Sierra B, Magalhães AC, Soares D, Cavadas B, Perez AB, Alvarez M, Aguirre E, Bracho C, Pereira L, Guzman MG. Multi-Tissue Transcriptomic-Informed In Silico Investigation of Drugs for the Treatment of Dengue Fever Disease. Viruses. 2021; 13(8):1540. https://doi.org/10.3390/v13081540
Chicago/Turabian StyleSierra, Beatriz, Ana Cristina Magalhães, Daniel Soares, Bruno Cavadas, Ana B. Perez, Mayling Alvarez, Eglis Aguirre, Claudia Bracho, Luisa Pereira, and Maria G. Guzman. 2021. "Multi-Tissue Transcriptomic-Informed In Silico Investigation of Drugs for the Treatment of Dengue Fever Disease" Viruses 13, no. 8: 1540. https://doi.org/10.3390/v13081540
APA StyleSierra, B., Magalhães, A. C., Soares, D., Cavadas, B., Perez, A. B., Alvarez, M., Aguirre, E., Bracho, C., Pereira, L., & Guzman, M. G. (2021). Multi-Tissue Transcriptomic-Informed In Silico Investigation of Drugs for the Treatment of Dengue Fever Disease. Viruses, 13(8), 1540. https://doi.org/10.3390/v13081540





