Serological Cross-Reactions between Expressed VP2 Proteins from Different Bluetongue Virus Serotypes
Abstract
1. Introduction
2. Materials and Methods
2.1. BTV Seg-2 and VP2 Sequence Data and Protein Synthesis
2.1.1. rVP2 Protein Production
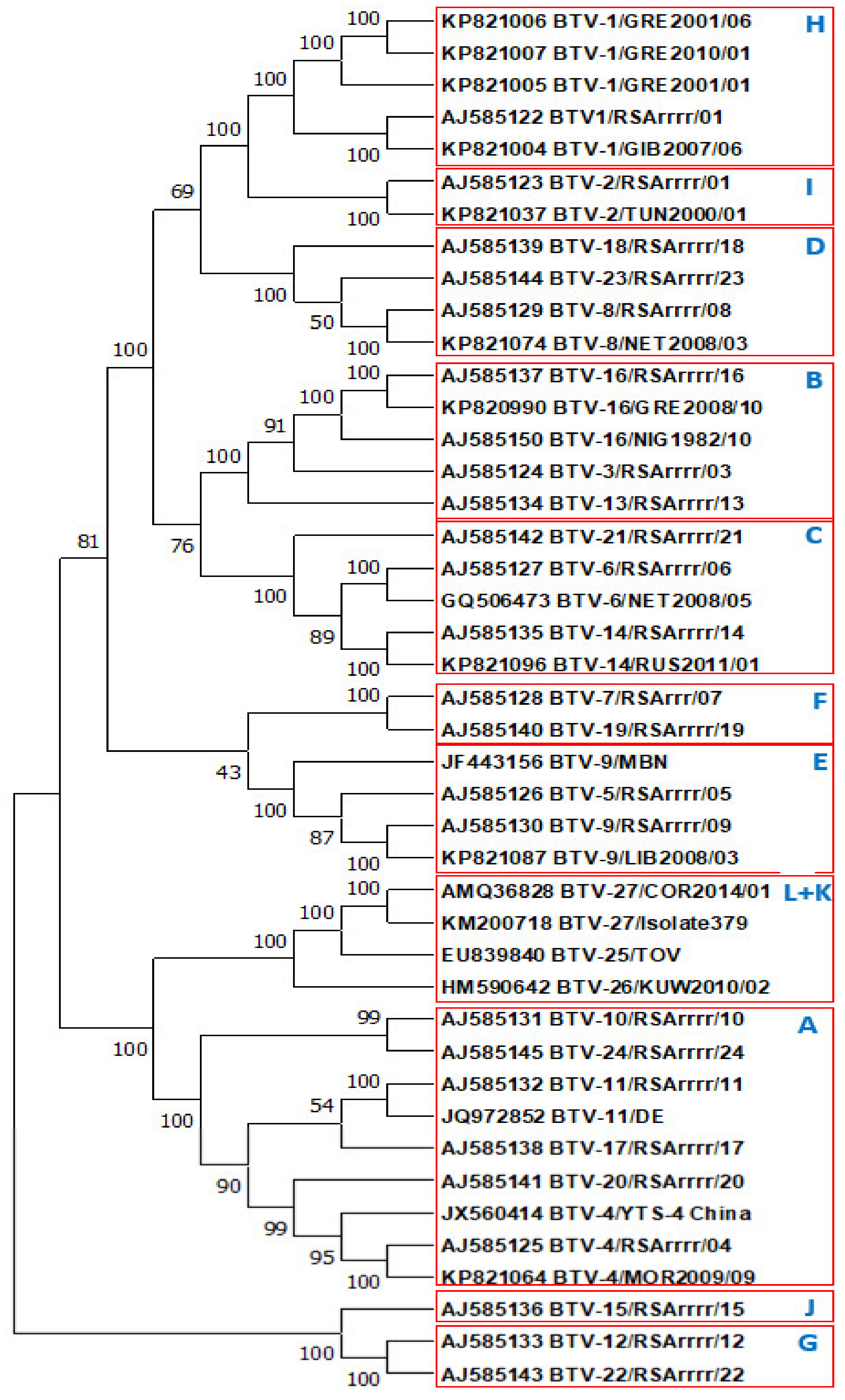
2.1.2. Phylogenetic Comparisons of VP2 Proteins and Subdomains
2.2. Virus Culture and Titration
| BTV Serotype and Country/Region, or Reference-Strain Serotype * [ORC Collection No **] | Serotype, Topotype (Nucleotype ^) | Plant Expression & ELISA Antigen | Used to Immunise Rabbits | Virus Used in SNT & Generate Anti-BTV Sheep Reference-Sera | Seg-2 Acc. No. (Gen Bank) | Reference |
|---|---|---|---|---|---|---|
| BTV-1w Gibraltar [GIB2007/06] | 1w (H) | Yes | Yes | - | KP821004 | [46] |
| BTV-1 * [RSArrrr/01] | 1w (H) | - | - | Yes | AJ585122 | [27] |
| BTV-1e Greece [GRE2001/09] | 1e (H) | - | - | Yes | - | - |
| BTV-1e Greece [GRE2001/06] | 1e (H) | Yes | Yes | - | KP821006 | [46] |
| BTV-2 Tunisia [TUN2000/01] | 2w (I) | Yes | No | - | KP821037 | [45] |
| BTV-2 * [RSArrrr/02] | 2w (I) | - | - | Yes | AJ585123 | [27] |
| BTV-3 * [RSArrrr/03] | 3w (B) | - | - | Yes | AJ585124 | [27] |
| BTV-4e China (1996) YTS-4 | 4e (A) | Yes | No | - | JX560414 | [47] |
| BTV-4w Cyprus [RSArrrr/04] | 4w (A) | - | - | Yes | AJ585125 | [27] |
| BTV-4w Morocco [MOR2009/09] | 4w (A) | Yes | Yes | - | KP821064 | [46] |
| BTV-5 * [RSArrrr/05] | 5w (E) | - | - | Yes | AJ585126 | [27] |
| BTV-6 * [RSArrrr/06] | 6w (C) | - | - | Yes | AJ585127 | [27] |
| BTV-6w Netherlands [NET2008/05] | 6w (C) | Yes | Yes | - | GQ506473 | [40] |
| BTV-7 * [RSArrrr/07] | 7w (F) | - | - | Yes | AJ585128 | [27] |
| BTV-8 * [RSArrrr/08] | 8w (D) | - | - | Yes | AJ585129 | [27] |
| BTV-8w Netherlands [NET2008/03] | 8w (D) | Yes | Yes | - | KP821074 | [45] |
| BTV-9 * [RSArrrr/09] | 9w (E) | - | - | Yes | AJ585130 | [27] |
| BTV-9e India (2002) MBN | 9e (E) | Yes | No | - | JF443156 | [48] |
| BTV-9w Libya [LIB2008/03] | 9w (E) | Yes | No | - | KP821087 | [46] |
| BTV-10 Portugal [RSArrrr/10] | 10w (A) | Yes | No | Yes | AJ585131 | [27] |
| Germany (2010) BTV-11_DE | 11w (A) | Yes | Yes | - | JQ972852 | [49] |
| BTV-11 * [RSArrrr/11] | 11w (A) | - | - | Yes | AJ585132 | [27] |
| BTV-12 * [RSArrrr/12] | 12w (G) | - | - | Yes | AJ585133 | [27] |
| BTV-13 * [RSArrrr/13] | 13w (B) | - | - | Yes | AJ585134 | [27] |
| BTV-14 * [RSArrrr/14] | 14w (C) | - | - | Yes | AJ585135 | [27] |
| BTV-14 Russia [RUS2011/01] | 14w (C) | Yes | Yes | - | KP821096 | [46] |
| BTV-15 * [RSArrrr/15] | 15w (J) | - | - | Yes | AJ585136 | [27] |
| BTV-16 * [RSArrrr/16] | 16e (B) | - | - | Yes | AJ585137 | [27] |
| BTV-16w Nigeria [NIG1982/10] | 16w (B) | Yes | No | - | AJ585150 | [27] |
| BTV-16e Greece [GRE2008/10] | 16e (B) | Yes | No | - | KP820990 | [46] |
| BTV-17w * [RSArrrr/17] | 17w (A) | - | - | Yes | AJ585138 | [27] |
| BTV-18w * [RSArrrr/18] | 18w (D) | - | - | Yes | AJ585139 | [27] |
| BTV-19w * [RSArrrr/19] | 19w (F) | - | - | Yes | AJ585140 | [27] |
| BTV-20e * [RSArrrr/20] | 20e (A) | - | - | Yes | AJ585141 | [27] |
| BTV-21e * [RSArrrr/21] | 21e (C) | - | - | Yes | AJ585142 | [27] |
| BTV-22w * [RSArrrr/22] | 22w (G) | - | - | Yes | AJ585143 | [27] |
| BTV-23e * [RSArrrr/23] | 23e (D) | - | - | Yes | AJ585144 | [27] |
| BTV-24 * [RSArrrr/24] | 24w (A) | - | - | Yes | AJ585145 | [27] |
| BTV-25 Switzerland (TOV **) | 25 (K) | Yes | Yes | - | EU839840 | [10] |
| BTV-26 Kuwait [KUW2010/02] | 26 (K ^^) | Yes | Yes | Yes | HM590642 | [42] |
| BTV-27 Corsica (2015) Strain 379 | 27 (K) | Yes | Yes | - | KM200718 | [50] |
| BTV-27 [COR2014/01] | 27 (K) | - | - | Yes | KU760988 | [51] |
2.3. Animals
Rabbit and Sheep Polyclonal Antisera
2.4. Serological Assays
2.4.1. Antibodies
2.4.2. Indirect-ELISA
2.4.3. Serum Neutralisation Test (SNT)
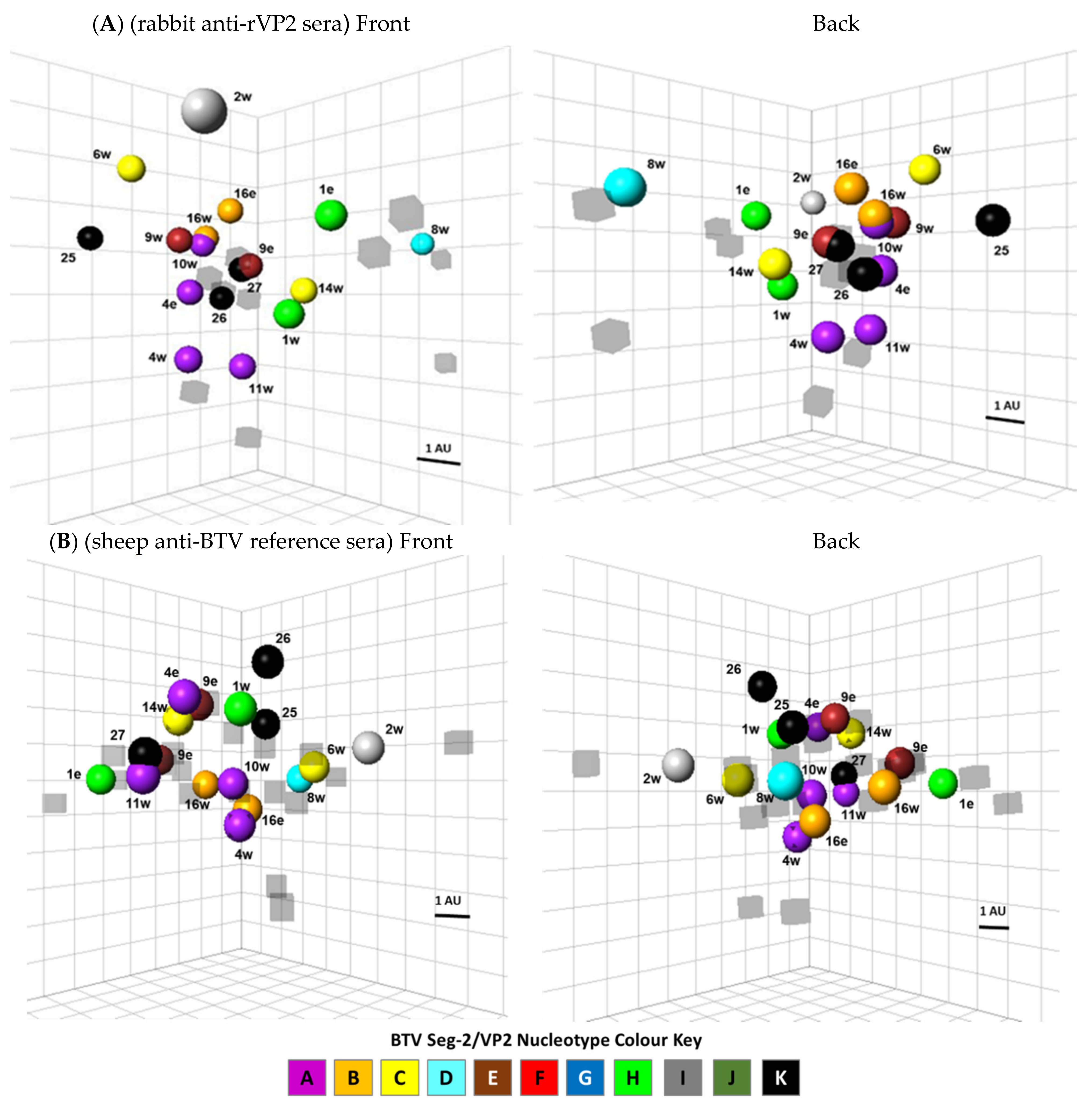
2.5. Antigenic Cartography
3. Results
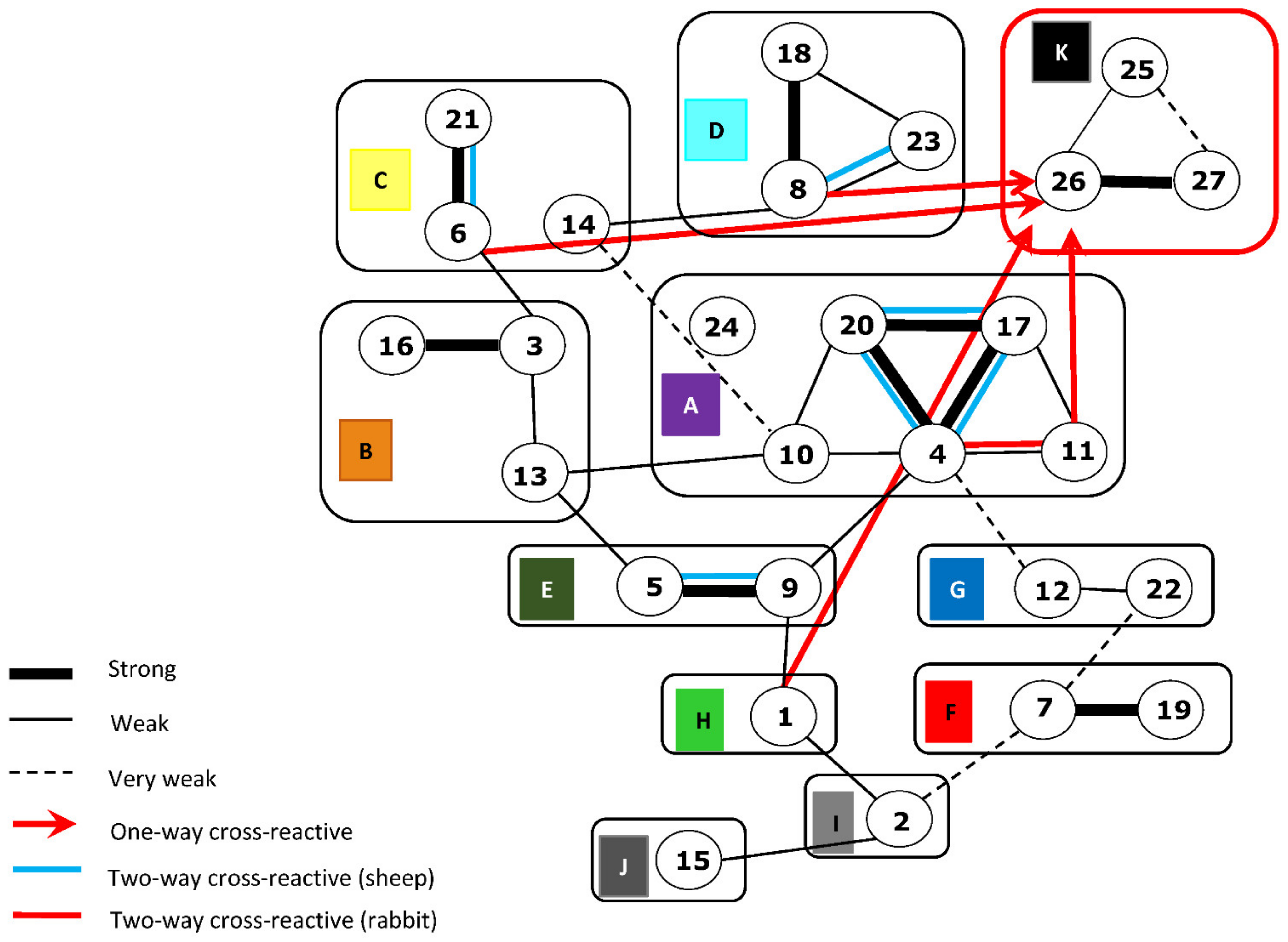
3.1. Antigenic Cross-Reactivity of Rabbit Antisera against rVP2 Proteins by I-ELISA
3.2. Cross-Reactivity of Rabbit Anti-rVP2 in Serum Neutralisation Tests (SNT)
3.3. Cross-Reactivity of BTV-rVP2 Proteins with Sheep Anti-BTV Reference Antisera in I-ELISA
3.4. Cross-Reaction of Sheep BTV Reference-Antisera in SNT with BTV Reference Strains
3.5. Mapping Antigenic Relationships Using Antigenic Cartography
4. Discussion
4.1. Serological Reactions between BTV Strains
4.2. Multidimensional Mapping of Antigenic Relationships
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mertens, P.P.; Diprose, J.; Maan, S.; Singh, K.P.; Attoui, H.; Samuel, A.R. Bluetongue virus replication, molecular and structural biology. Vet. Ital. 2004, 40, 426–437. [Google Scholar] [PubMed]
- Mertens, P.P.C.; Maan, S.; Samuel, A.; Attoui, H. Orbivirus, Reoviridae. In Virus Taxonomy VIIIth Report of the ICTV; Fauquet, M.A.M., Maniloff, J., Desselberger, U., Ball, L.A., Eds.; Elsevier: Amsterdam, The Netherlands; Academic Press: London, UK, 2004. [Google Scholar]
- Grimes, J.M.; Burroughs, J.N.; Gouet, P.; Diprose, J.M.; Malby, R.; Zientara, S.; Mertens, P.P.; Stuart, D.I. The atomic structure of the bluetongue virus core. Nature 1998, 395, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Roy, P. Bluetongue virus structure and assembly. Curr. Opin. Virol. 2017, 24, 115–123. [Google Scholar] [CrossRef] [PubMed]
- More, S.; Bicout, D.; Bøtner, A.; Butterworth, A.; Depner, K.; Edwards, S.; Garin-Bastuji, B.; Good, M.; Gortazar-Schmidt, C.; Michel, V.; et al. Assessment of listing and categorisation of animal diseases within the framework of the Animal Health Law (Regulation (EU) No 2016/429): Bluetongue. EFSA J. 2017, 15, 4957. [Google Scholar]
- Maclachlan, N.J.; Mayo, C.E.; Daniels, P.W.; Savini, G.; Zientara, S.; Gibbs, E.P. Bluetongue. Rev. Sci. Technol. 2015, 34, 329–340. [Google Scholar] [CrossRef]
- Batten, C.; Darpel, K.; Henstock, M.; Fay, P.; Veronesi, E.; Gubbins, S.; Graves, S.; Frost, L.; Oura, C. Evidence for transmission of bluetongue virus serotype 26 through direct contact. PLoS ONE 2014, 9, e96049. [Google Scholar] [CrossRef]
- Breard, E.; Schulz, C.; Sailleau, C.; Bernelin-Cottet, C.; Viarouge, C.; Vitour, D.; Guillaume, B.; Caignard, G.; Gorlier, A.; Attoui, H.; et al. Bluetongue virus serotype 27: Experimental infection of goats, sheep and cattle with three BTV-27 variants reveal atypical characteristics and likely direct contact transmission BTV-27 between goats. Transbound Emerg. Dis. 2018, 65, e251–e263. [Google Scholar] [CrossRef] [PubMed]
- Bumbarov, V.; Golender, N.; Jenckel, M.; Wernike, K.; Beer, M.; Khinich, E.; Zalesky, O.; Erster, O. Characterization of bluetongue virus serotype 28. Transbound Emerg. Dis. 2020, 67, 171–182. [Google Scholar] [CrossRef]
- Ries, C.; Sharav, T.; Tseren-Ochir, E.-O.; Beer, M.; Hoffmann, B. Putative Novel Serotypes ‘33’ and ‘35’ in Clinically Healthy Small Ruminants in Mongolia Expand the Group of Atypical BTV. Viruses 2021, 13, 42. [Google Scholar] [CrossRef]
- Savini, G.; Puggioni, G.; Meloni, G.; Marcacci, M.; Di Domenico, M.; Rocchigiani, A.M.; Spedicato, M.; Oggiano, A.; Manunta, D.; Teodori, L.; et al. Novel putative Bluetongue virus in healthy goats from Sardinia, Italy. Infect. Genet. Evol. 2017, 51, 108–117. [Google Scholar] [CrossRef]
- Zientara, S.; Sailleau, C.; Viarouge, C.; Hoper, D.; Beer, M.; Jenckel, M.; Hoffmann, B.; Romey, A.; Bakkali-Kassimi, L.; Fablet, A.; et al. Novel bluetongue virus in goats, Corsica, France, 2014. Emerg. Infect. Dis. 2014, 20, 2123–2125. [Google Scholar] [CrossRef] [PubMed]
- Wright, I. Serological and Genetic Characterisation of Putative New Serotypes of Blue-Tongue Virus and Epizootic Haemorrhagic Disease Virus Isolated from Alpaca. Master’s Thesis, North-West University, Potchefstroom, South Africa, 2014. [Google Scholar]
- Pullinger, G.D.; Guimera Busquets, M.; Nomikou, K.; Boyce, M.; Attoui, H.; Mertens, P.P. Identification of the Genome Segments of Bluetongue Virus Serotype 26 (Isolate KUW2010/02) that Restrict Replication in a Culicoides sonorensis Cell Line (KC Cells). PLoS ONE 2016, 11, e0149709. [Google Scholar] [CrossRef]
- Jacquot, M.; Nomikou, K.; Palmarini, M.; Mertens, P.; Biek, R. Bluetongue virus spread in Europe is a consequence of climatic, landscape and vertebrate host factors as revealed by phylogeographic inference. Proc. Biol. Sci. 2017, 284, 1864. [Google Scholar] [CrossRef] [PubMed]
- Kundlacz, C.; Caignard, G.; Sailleau, C.; Viarouge, C.; Postic, L.; Vitour, D.; Zientara, S.; Breard, E. Bluetongue Virus in France: An Illustration of the European and Mediterranean Context since the 2000s. Viruses 2019, 11, 672. [Google Scholar] [CrossRef] [PubMed]
- Pascall, D.J.; Nomikou, K.; Breard, E.; Zientara, S.; Filipe, A.D.S.; Hoffmann, B.; Jacquot, M.; Singer, J.B.; De Clercq, K.; Botner, A.; et al. “Frozen evolution” of an RNA virus suggests accidental release as a potential cause of arbovirus re-emergence. PLoS Biol. 2020, 18, e3000673. [Google Scholar] [CrossRef]
- Belhouchet, M.; Mohd Jaafar, F.; Firth, A.E.; Grimes, J.M.; Mertens, P.P.; Attoui, H. Detection of a fourth orbivirus non-structural protein. PLoS ONE 2011, 6, e25697. [Google Scholar] [CrossRef]
- Hassan, S.S.; Roy, P. Expression and functional characterization of bluetongue virus VP2 protein: Role in cell entry. J. Virol. 1999, 73, 9832–9842. [Google Scholar] [CrossRef]
- Maan, N.S.; Maan, S.; Belaganahalli, M.N.; Ostlund, E.N.; Johnson, D.J.; Nomikou, K.; Mertens, P.P. Identification and differentiation of the twenty six bluetongue virus serotypes by RT-PCR amplification of the serotype-specific genome segment 2. PLoS ONE 2012, 7, e32601. [Google Scholar] [CrossRef] [PubMed]
- Appleton, J.A.; Letchworth, G.J. Monoclonal antibody analysis of serotype-restricted and unrestricted bluetongue viral antigenic determinants. Virology 1983, 124, 286–299. [Google Scholar] [CrossRef]
- Mertens, P.P.; Pedley, S.; Cowley, J.; Burroughs, J.N.; Corteyn, A.H.; Jeggo, M.H.; Jennings, D.M.; Gorman, B.M. Analysis of the roles of bluetongue virus outer capsid proteins VP2 and VP5 in determination of virus serotype. Virology 1989, 170, 561–565. [Google Scholar] [CrossRef]
- Fay, P.C.; Attoui, H.; Batten, C.; Mohd Jaafar, F.; Lomonossoff, G.P.; Daly, J.M.; Mertens, P.P.C. Bluetongue virus outer-capsid protein VP2 expressed in Nicotiana benthamiana raises neutralising antibodies and a protective immune response in IFNAR (−/−) mice. Vaccine X 2019, 2, 100026. [Google Scholar] [CrossRef]
- Maan, S.; Maan, N.S.; Samuel, A.R.; Rao, S.; Attoui, H.; Mertens, P.P. Analysis and phylogenetic comparisons of full-length VP2 genes of the 24 bluetongue virus serotypes. J. Gen. Virol. 2007, 88, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Howell, P.G.; Kumm, N.A.; Botha, M.J. The application of improved techniques to the identification of strains of bluetongue virus. Onderstepoort J. Vet. Res. 1970, 37, 59–66. [Google Scholar]
- Kahlon, J.; Sugiyama, K.; Roy, P. Molecular basis of bluetongue virus neutralization. J. Virol. 1983, 48, 627–632. [Google Scholar] [CrossRef] [PubMed]
- Maan, S.; Maan, N.S.; Samuel, A.R.; O’Hara, R.; Meyer, A.J.; Rao, S.; Mertens, P.P. Completion of the sequence analysis and comparisons of genome segment 2 (encoding outer capsid protein VP2) from representative isolates of the 24 bluetongue virus serotypes. Vet. Ital. 2004, 40, 484–488. [Google Scholar]
- Capocefalo, A.; Franceschi, V.; Mertens, P.P.; Castillo-Olivares, J.; Cavirani, S.; Di Lonardo, E.; Leni, Z.; Donofrio, G. Expression and secretion of Bluetongue virus serotype 8 (BTV-8)VP2 outer capsid protein by mammalian cells. J. Virol. Methods 2010, 169, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Roy, P.; Boyce, M.; Noad, R. Prospects for improved bluetongue vaccines. Nat. Rev. Microbiol. 2009, 7, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Huismans, H.; Cloete, M. A comparison of different cloned bluetongue virus genome segments as probes for the detection of virus-specified RNA. Virology 1987, 158, 373–380. [Google Scholar] [CrossRef]
- Mohd Jaafar, F.; Belhouchet, M.; Vitour, D.; Adam, M.; Breard, E.; Zientara, S.; Mertens, P.P.; Attoui, H. Immunisation with bacterial expressed VP2 and VP5 of bluetongue virus (BTV) protect alpha/beta interferon-receptor knock-out (IFNAR(−/−)) mice from homologous lethal challenge. Vaccine 2014, 32, 4059–4067. [Google Scholar] [CrossRef]
- Maan, N.S.; Maan, S.; Belaganahalli, M.; Pullinger, G.; Montes, A.J.; Gasparini, M.R.; Guimera, M.; Nomikou, K.; Mertens, P.P. A quantitative real-time reverse transcription PCR (qRT-PCR) assay to detect genome segment 9 of all 26 bluetongue virus serotypes. J. Virol. Methods 2015, 213, 118–126. [Google Scholar] [CrossRef]
- Jacquot, M.; Rao, P.P.; Yadav, S.; Nomikou, K.; Maan, S.; Jyothi, Y.K.; Reddy, N.; Putty, K.; Hemadri, D.; Singh, K.P.; et al. Contrasting selective patterns across the segmented genome of bluetongue virus in a global reassortment hotspot. Virus Evol. 2019, 5, vez027. [Google Scholar] [CrossRef]
- Gould, A.R.; Pritchard, L.I. Relationships amongst bluetongue viruses revealed by comparisons of capsid and outer coat protein nucleotide sequences. Virus Res. 1990, 17, 31–52. [Google Scholar] [CrossRef]
- Erasmus, B.J. Bluetongue virus. In Virus Infections of Ruminants; Dinter, Z., Morein, B., Eds.; Elsevier Science Publishers: New York, NY, USA, 1990; pp. 227–237. [Google Scholar]
- De Maula, C.D.; Bonneau, K.R.; MacLachlan, N.J. Changes in the outer capsid proteins of bluetongue virus serotype ten that abrogate neutralization by monoclonal antibodies. Virus Res. 2000, 67, 59–66. [Google Scholar] [CrossRef]
- Jeggo, M.H.; Wardley, R.C.; Taylor, W.P. Clinical and serological outcome following the simultaneous inoculation of three bluetongue virus types into sheep. Res. Vet. Sci. 1984, 37, 368–370. [Google Scholar] [CrossRef]
- Ma, G.; Eschbaumer, M.; Said, A.; Hoffmann, B.; Beer, M.; Osterrieder, N. An equine herpesvirus type 1 (EHV-1) expressing VP2 and VP5 of serotype 8 bluetongue virus (BTV-8) induces protection in a murine infection model. PLoS ONE 2012, 7, e34425. [Google Scholar] [CrossRef]
- Rybicki, E.P. Plant-based vaccines against viruses. Virol. J. 2014, 11, 205. [Google Scholar] [CrossRef]
- Maan, S.; Maan, N.S.; Ross-Smith, N.; Batten, C.A.; Shaw, A.E.; Anthony, S.J.; Samuel, A.R.; Darpel, K.E.; Veronesi, E.; Oura, C.A.; et al. Sequence analysis of bluetongue virus serotype 8 from the Netherlands 2006 and comparison to other European strains. Virology 2008, 377, 308–318. [Google Scholar] [CrossRef]
- Maan, S.; Maan, N.S.; van Rijn, P.A.; van Gennip, R.G.; Sanders, A.; Wright, I.M.; Batten, C.; Hoffmann, B.; Eschbaumer, M.; Oura, C.A.; et al. Full genome characterisation of bluetongue virus serotype 6 from the Netherlands 2008 and comparison to other field and vaccine strains. PLoS ONE 2010, 5, e10323. [Google Scholar] [CrossRef]
- Maan, S.; Maan, N.S.; Nomikou, K.; Batten, C.; Antony, F.; Belaganahalli, M.N.; Samy, A.M.; Reda, A.A.; Al-Rashid, S.A.; El Batel, M.; et al. Novel bluetongue virus serotype from Kuwait. Emerg. Infect. Dis. 2011, 17, 886–889. [Google Scholar] [CrossRef]
- Peyret, H.; Lomonossoff, G.P. The pEAQ vector series: The easy and quick way to produce recombinant proteins in plants. Plant Mol. Biol. 2013, 83, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Thuenemann, E.C.; Meyers, A.E.; Verwey, J.; Rybicki, E.P.; Lomonossoff, G.P. A method for rapid production of heteromultimeric protein complexes in plants: Assembly of protective bluetongue virus-like particles. Plant Biotechnol. J. 2013, 11, 839–846. [Google Scholar] [CrossRef]
- Kärber, G. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Naunyn-Schmiedebergs. Archiv. Exp. Pathol. Pharmakol. 1931, 162, 480–483. [Google Scholar] [CrossRef]
- Nomikou, K.; Hughes, J.; Wash, R.; Kellam, P.; Breard, E.; Zientara, S.; Palmarini, M.; Biek, R.; Mertens, P. Widespread Reassortment Shapes the Evolution and Epidemiology of Bluetongue Virus following European Invasion. PLoS Pathog. 2015, 11, e1005056. [Google Scholar] [CrossRef]
- Yang, H.; Zhu, J.; Li, H.; Xiao, L.; Wang, J.; Li, N.; Zhang, N.; Kirkland, P.D. Full genome sequence of bluetongue virus serotype 4 from China. J. Virol. 2012, 86, 13122–13123. [Google Scholar] [CrossRef]
- Rao, P.P.; Reddy, Y.N.; Hegde, N.R. Complete genome sequence of bluetongue virus serotype 9: Implications for serotyping. J. Virol. 2012, 86, 8333. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vandenbussche, F.; Sailleau, C.; Rosseel, T.; Desprat, A.; Viarouge, C.; Richardson, J.; Eschbaumer, M.; Hoffmann, B.; De Clercq, K.; Breard, E.; et al. Full-Genome Sequencing of Four Bluetongue Virus Serotype 11 Viruses. Transbound Emerg. Dis. 2015, 62, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Jenckel, M.; Breard, E.; Schulz, C.; Sailleau, C.; Viarouge, C.; Hoffmann, B.; Hoper, D.; Beer, M.; Zientara, S. Complete coding genome sequence of putative novel bluetongue virus serotype 27. Genome Announc. 2015, 3, e00016-15. [Google Scholar] [CrossRef] [PubMed]
- Schulz, C.; Breard, E.; Sailleau, C.; Jenckel, M.; Viarouge, C.; Vitour, D.; Palmarini, M.; Gallois, M.; Hoper, D.; Hoffmann, B.; et al. Bluetongue virus serotype 27: Detection and characterization of two novel variants in Corsica, France. J. Gen. Virol. 2016, 97, 2073–2083. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.J.; Lapedes, A.S.; de Jong, J.C.; Bestebroer, T.M.; Rimmelzwaan, G.F.; Osterhaus, A.D.; Fouchier, R.A. Mapping the antigenic and genetic evolution of influenza virus. Science 2004, 305, 371–376. [Google Scholar] [CrossRef]
- Maan, S.; Maan, N.S.; Nomikou, K.; Veronesi, E.; Bachanek-Bankowska, K.; Belaganahalli, M.N.; Attoui, H.; Mertens, P.P. Complete genome characterisation of a novel 26th bluetongue virus serotype from Kuwait. PLoS ONE 2011, 6, e26147. [Google Scholar] [CrossRef]
- Cowley, J.A.; Gorman, B.M. Cross-neutralization of genetic reassortants of bluetongue virus serotypes 20 and 21. Vet. Microbiol. 1989, 19, 37–51. [Google Scholar] [CrossRef]
- White, J.R.; Eaton, B.T. Conformation of the VP2 protein of bluetongue virus (BTV) determines the involvement in virus neutralization of highly conserved epitopes within the BTV serogroup. J. Gen. Virol. 1990, 71, 1325–1332. [Google Scholar] [CrossRef]
- Huismans, H.; Erasmus, B.J. Identification of the serotype-specific and group-specific antigens of bluetongue virus. Onderstepoort J. Vet. Res. 1981, 48, 51–58. [Google Scholar]
- Huismans, H.; Bremer, C.W. A comparison of an australian bluetongue virus isolate (CSIRO 19) with other bluetongue virus serotypes by cross-hybridization and cross-immune precipitation. Onderstepoort J. Vet. Res. 1981, 48, 59–67. [Google Scholar]
- Della-Porta, A.J.H.; Sellers, R.F. A serological comparison of the australian isolate of bluetongue virus type 20 (CSIRO 19) with bluetongue group viruses. Vet. Microbiol. 1981, 6, 9–21. [Google Scholar] [CrossRef]
- Rossitto, P.V.; MacLachlan, N.J. Neutralizing epitopes of the serotypes of bluetongue virus present in the United States. J. Gen. Virol. 1992, 73, 1947–1952. [Google Scholar] [CrossRef] [PubMed]
- MacLachlan, N.J.; Rossitto, P.V.; Heidner, H.W.; Iezzi, L.G.; Yilma, T.D.; De Maula, C.D.; Osburn, B.I. Variation amongst the neutralizing epitopes of bluetongue viruses isolated in the United States in 1979–1981. Vet. Microbiol. 1992, 31, 303–316. [Google Scholar] [CrossRef]
- Ristow, S.; Leendersten, L.; Gorham, J.; Yilma, T. Identification of a neutralizing epitope shared by bluetongue virus serotypes 2 and 13. J. Virol. 1988, 62, 2502–2504. [Google Scholar] [CrossRef]
- Ghiasi, H.; Fukusho, A.; Eshita, Y.; Roy, P. Identification and characterization of conserved and variable regions in the neutralization VP2 gene of bluetongue virus. Virology 1987, 160, 100–109. [Google Scholar] [CrossRef]
- Gould, A.R.; Hyatt, A.D. The orbivirus genus. Diversity, structure, replication and phylogenetic relationships. Comp. Immunol. Microbiol. Infect. Dis. 1994, 17, 163–188. [Google Scholar] [CrossRef]
- Letchworth, G.J.; Appleton, J.A. Heterogeneity of neutralization-related epitopes within a bluetongue virus serotype. Virology 1983, 124, 300–307. [Google Scholar] [CrossRef]
- Lobato, Z.I.; Coupar, B.E.; Gray, C.P.; Lunt, R.; Andrew, M.E. Antibody responses and protective immunity to recombinant vaccinia virus-expressed bluetongue virus antigens. Vet. Immunol. Immunopathol. 1997, 59, 293–309. [Google Scholar] [CrossRef]
- Schwartz-Cornil, I.; Mertens, P.P.; Contreras, V.; Hemati, B.; Pascale, F.; Breard, E.; Mellor, P.S.; MacLachlan, N.J.; Zientara, S. Bluetongue virus: Virology, pathogenesis and immunity. Vet. Res. 2008, 39, 46. [Google Scholar] [CrossRef]
- Horton, D.L.; McElhinney, L.M.; Marston, D.A.; Wood, J.L.; Russell, C.A.; Lewis, N.; Kuzmin, I.V.; Fouchier, R.A.; Osterhaus, A.D.; Fooks, A.R.; et al. Quantifying antigenic relationships among the lyssaviruses. J. Virol. 2010, 84, 11841–11848. [Google Scholar] [CrossRef]
- Ludi, A.B.; Horton, D.L.; Li, Y.; Mahapatra, M.; King, D.P.; Knowles, N.J.; Russell, C.A.; Paton, D.J.; Wood, J.L.N.; Smith, D.; et al. Antigenic variation of foot-and-mouth disease virus serotype A. J. Gen. Virol. 2014, 95, 384–392. [Google Scholar] [CrossRef]
- Sundin, D.R.; Dean, V.C.; DuBard, K.M.; Mecham, J.O. In vitro neutralization of antigenic variants of bluetongue virus is related to in vivo protection. Viral Immunol. 1989, 2, 195–203. [Google Scholar] [CrossRef]
- Zhang, X.; Patel, A.; Celma, C.C.; Yu, X.; Roy, P.; Zhou, Z.H. Atomic model of a nonenveloped virus reveals pH sensors for a coordinated process of cell entry. Nat. Struct. Mol. Biol. 2016, 23, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Belaganahalli, M.N.; Maan, S.; Maan, N.S.; Brownlie, J.; Tesh, R.; Attoui, H.; Mertens, P.P. Genetic characterization of the tick-borne orbiviruses. Viruses 2015, 7, 2185–2209. [Google Scholar] [CrossRef] [PubMed]
- Jeggo, M.H.; Gumm, I.D.; Taylor, W.P. Clinical and serological response of sheep to serial challenge with different bluetongue virus types. Res. Vet. Sci. 1983, 34, 205–211. [Google Scholar] [CrossRef]
- Dungu, B.; Gerdes, T.; Smit, T. The use of vaccination in the control of bluetongue in southern Africa. Vet. Ital. 2004, 40, 616–622. [Google Scholar] [PubMed]
- Savini, G.; Monaco, F.; Conte, A.; Migliaccio, P.; Casaccia, C.; Salucci, S.; Di Ventura, M. Virological and serological response of sheep following field vaccination with bivalent modified-live vaccine against bluetongue virus serotypes 2 and 9. Vet. Ital. 2004, 40, 631–634. [Google Scholar]
- Wahala, W.M.; Silva, A.M. The human antibody response to dengue virus infection. Viruses 2011, 3, 2374–2395. [Google Scholar] [CrossRef]
- Chan, K.R.; Zhang, S.L.; Tan, H.C.; Chan, Y.K.; Chow, A.; Lim, A.P.; Vasudevan, S.G.; Hanson, B.J.; Ooi, E.E. Ligation of Fc gamma receptor IIB inhibits antibody-dependent enhancement of dengue virus infection. Proc. Natl. Acad. Sci. USA 2011, 108, 12479–12484. [Google Scholar] [CrossRef]
- Du Plessis, D. Serological differentiation of five bluetongue virus serotypes in indirect ELISA. Onderstepoort J. Vet. Res. 1992, 59, 119–122. [Google Scholar] [PubMed]
- Letchworth, G.J., III; Appleton, J.A. Passive protection of mice and sheep against bluetongue virus by a neutralizing monoclonal antibody. Infect. Immun. 1983, 39, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Afshar, A.T.F.; Wright, P.F.; Shapiro, J.L.; Shettigara, P.T.; Anderson, J. Comparison of competitive and indirect enzyme-linked immunosorbent assays for detection of bluetongue virus antibodies in serum and whole blood. J. Clin. Microbiol. 1987, 25, 1705–1710. [Google Scholar] [CrossRef] [PubMed]
| BTV Strain Providing Sequence Data for rVP2 Expression | Rabbit Anti-BTV-rVP2 Sera | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1w | 1e | 4w | 6w | 8w | 11w | 14w | 25 | 26 | 27 | |
| BTV-1w [GIB2007/06] | 40,960 | 20,480 | 40,960 | 2560 | 640 | 10,240 | 1280 | 1280 | 1280 | 2560 |
| BTV-1e [GRE2001/06] | 40,960 | 40,960 | 320 | 5120 | 1280 | 1280 | - | 640 | 640 | 2560 |
| BTV-2w [TUN2000/01] | 10,240 | - | 80 | 160 | 40 | 1280 | 40 | 40 | 40 | 640 |
| BTV-4w [MOR2009/09] | 1280 | - | 40,960 | 2560 | 1280 | 40,960 | 2560 | 1280 | 2560 | 5120 |
| BTV-4e China YTS-4 | 1280 | - | 5120 | 2560 | 320 | 10,240 | 320 | 5120 | 5120 | 10,240 |
| BTV-6w [ NET2008/05] | 640 | - | 640 | 2560 | 160 | 1280 | 160 | 320 | 320 | 2560 |
| BTV-8w [NET2008/03] | - | - | - | 640 | 40,960 | 160 | - | - | - | 160 |
| BTV-9w [LIB2008/03] | 1280 | - | 1280 | 2560 | 320 | 2560 | 320 | 2560 | 2560 | 10,240 |
| BTV-9e India MBN | 2560 | - | 5120 | 5120 | 2560 | 5120 | 640 | 2560 | 5120 | 5120 |
| BTV-10w [RSArrrr/10] | 1280 | - | 2560 | 2560 | 1280 | 2560 | 320 | 2560 | 2560 | 10,240 |
| BTV-11w Germany (BTV-11_DE) | 1280 | - | 20,480 | 1280 | 160 | 40,960 | 640 | 1280 | 2560 | 10,240 |
| BTV-14w [RUS2011/01] | 10,240 | - | - | 5120 | 1280 | 2560 | 40,960 | 2560 | 2560 | 10,240 |
| BTV-16w [NIG1982/10] | 1280 | - | 1280 | 5120 | 640 | 2560 | 320 | 2560 | 5120 | 5120 |
| BTV-16e [GRE2008/10] | 1280 | - | 320 | 5120 | 640 | 5120 | 1280 | 1280 | 2560 | 2560 |
| BTV-25 Switzerland (TOV) | 80 | - | 640 | 2560 | 40 | 1280 | - | 640 | 640 | 5120 |
| BTV-26 [KUW2010/02] | 2560 | 40 | 5120 | 5120 | 2560 | 10,240 | 1280 | 2560 | 5120 | 10,240 |
| BTV-27 Corsica (379) | 2560 | - | 2560 | 5120 | 1280 | 5120 | 1280 | 2560 | 10,240 | 10,240 |
| BTV Reference Strain [ORC Number] | Rabbit Anti-BTV-rVP2 Sera | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1w | 1e | 4w | 6w | 8w | 11w | 14w | 25w | 26e | 27w | |
| BTV-1w [RSArrrr/01] | 140 | 60 | - | - | - | - | - | - | - | - |
| BTV-1e [GRE2001/09] | 120 | 60 | - | - | - | - | - | - | - | - |
| BTV-2w [RSArrrr/02] | - | - | - | - | - | - | - | - | - | - |
| BTV-3w [RSArrrr/03] | - | - | - | - | - | - | - | - | - | - |
| BTV-4w [RSArrrr/04] | - | - | 240 | - | - | 15 | 10 | - | - | - |
| BTV-5w [RSArrrr/05] | - | - | - | - | - | - | - | - | - | - |
| BTV-6w [RSArrrr/06] | - | - | - | 690 | - | - | - | - | - | - |
| BTV-7w [RSArrrr/07] | - | - | - | - | - | - | - | - | - | - |
| BTV-8w [RSArrrr/08] | - | - | - | - | 90 | - | - | - | - | - |
| BTV-9w [RSArrrr/09] | - | - | - | - | - | - | - | - | - | - |
| BTV-10w [RSArrrr/10] | - | - | - | - | - | - | - | - | - | - |
| BTV-11w [RSArrrr/11] | - | - | 20 | - | - | 120 | - | - | - | - |
| BTV-12w [RSArrrr/12] | - | - | - | - | - | - | - | - | - | - |
| BTV-13w [RSArrrr/13] | - | - | - | - | - | - | - | - | - | - |
| BTV-14w [RSArrrr/14] | 40 | - | - | - | - | - | 600 | - | - | - |
| BTV-15w [RSArrrr/15] | - | - | - | - | - | - | - | - | - | - |
| BTV-16e [RSArrrr/16] | - | - | - | - | - | - | - | - | - | - |
| BTV-17w [RSArrrr/17] | - | - | 70 | - | - | 30 | - | - | - | 20 |
| BTV-18w [RSArrrr/18] | - | - | - | - | - | - | - | - | - | - |
| BTV-19w [RSArrrr/19] | - | - | - | - | - | - | - | - | - | - |
| BTV-20e [RSArrrr/20] | - | - | 160 | - | - | 80 | 10 | - | - | - |
| BTV-21e [RSArrrr/21] | - | - | - | - | - | - | 10 | - | - | - |
| BTV-22w [RSArrrr/22] | - | - | - | - | - | - | - | - | - | - |
| BTV-23e [RSArrrr/23] | - | - | - | - | 30 | - | 10 | - | - | - |
| BTV-24w [RSArrrr/24] | - | - | 15 | - | - | 10 | - | - | - | - |
| BTV-26e [KUW2010/01] | - | 30 | - | 20 | 10 | 20 | - | 30 | 40 | 40 |
| BTV-27w [COR2014/01] | - | - | - | - | - | - | - | 10 | 15 | 15 |
| Sheep Reference Antisera * | BTV rVP2 Antigens | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1w | 1e | 2w | 4w | 4e | 6w | 8w | 9w | 9e | 10w | 11w | 14w | 16w | 16e | 25 | 26 | 27 | |
| BTV-1w | 5120 | 1280 | - | - | - | 40 | - | - | - | - | - | - | - | - | - | - | - |
| BTV-2w | 10 | 160 | 10,240 | 80 | 40 | 10,240 | 80 | 20 | 40 | 160 | 20 | 160 | 80 | 40 | 160 | 160 | 20 |
| BTV-3w | - | 20 | - | 10 | - | 80 | - | - | - | - | - | - | - | - | 20 | - | - |
| BTV-4w | - | - | - | 10,240 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| BTV-5w | 10 | 60 | 20 | 80 | 40 | 320 | 80 | 40 | 80 | 160 | 20 | 80 | 20 | 160 | 320 | 80 | 20 |
| BTV-6w | 5120 | 10 | - | - | - | 20 | - | - | - | - | - | - | - | - | - | - | - |
| BTV-8w | - | 40 | - | 80 | 20 | 640 | 2560 | 20 | 40 | 80 | - | - | 160 | 320 | 640 | 80 | - |
| BTV-9w | - | - | - | 640 | - | - | - | 5120 | 10,240 | - | - | - | - | - | - | - | - |
| BTV-10w | 320 | 1280 | 1280 | 5120 | 320 | 2560 | 1280 | 640 | 1280 | 1280 | 320 | 160 | 320 | 5120 | 640 | 1280 | 320 |
| BTV-11w | - | 40 | - | 80 | 20 | 320 | 40 | 10 | 20 | 160 | 40 | 10 | - | - | 80 | 10 | 20 |
| BTV-12w | - | 20 | - | - | - | 40 | - | 40 | 80 | - | - | - | - | - | 80 | - | - |
| BTV-13w | - | 10 | - | - | 10 | - | - | - | 10 | - | - | - | - | - | - | - | - |
| BTV-14w | - | - | - | - | - | - | - | - | - | - | - | 1280 | - | - | - | - | - |
| BTV-15w | - | - | - | - | - | - | - | - | - | - | - | 160 | - | - | - | - | - |
| BTV-16e | 40 | 80 | 40 | 320 | 40 | 320 | 160 | 40 | 40 | 320 | 40 | - | 160 | 2560 | 80 | 40 | 40 |
| BTV-17w | 20 | 40 | 160 | 320 | 80 | 80 | 20 | 40 | 40 | 320 | 40 | 80 | 160 | 320 | 80 | 20 | 80 |
| BTV-18w | - | - | - | 10 | - | - | - | 80 | - | - | - | - | - | - | - | - | 10 |
| BTV-19w | - | 20 | 10 | 10 | 10 | 40 | 20 | 320 | - | 80 | 20 | - | - | - | 20 | 10 | 20 |
| BTV-20e | 10 | 40 | 40 | 1280 | 20 | 80 | 40 | 20 | 20 | 80 | - | - | 20 | 40 | 20 | 20 | 40 |
| BTV-21e | - | 10 | - | 10 | - | - | - | 40 | 10 | - | - | 40 | - | - | - | - | 20 |
| BTV-22w | - | - | - | 20 | - | - | - | - | - | - | - | - | - | - | - | - | |
| BTV-23e | - | - | - | - | - | - | - | 160 | - | - | - | - | - | - | - | - | 10 |
| BTV-24w | - | 10 | - | 80 | - | - | - | 160 | - | - | - | - | - | - | - | - | 10 |
| BTV-26 | - | 10 | - | - | - | - | - | 1280 | - | 40 | - | - | - | - | - | 10 | - |
| BTV Strain * | Sheep Anti-BTV Reference Sera ** | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1w | 2w | 3w | 4w | 5w | 6w | 8w | 9w | 10w | 11w | 12w | 13w | 14w | 15w | 16e | 17w | 18w | 19w | 20e | 21e | 22w | 23e | 24w | 26 | |
| BTV-1w | 10,240 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| BTV-1e | 40 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| BTV-2w | - | 160 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 10 | - | - |
| BTV-3w | - | - | 560 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| BTV-4w | - | - | - | 320 | - | - | - | - | - | - | - | - | - | - | - | 5 | - | - | 32 | - | - | - | - | - |
| BTV-5w | - | - | - | - | 3160 | - | - | 10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| BTV-6w | 3.2 | - | 10 | - | - | 1780 | - | - | - | - | - | 32 | - | - | - | - | - | - | - | 32 | - | - | - | - |
| BTV-7w | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| BTV-8w | - | - | - | - | - | - | 1000 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 10 | - | - |
| BTV-9w | - | - | - | - | 10 | - | - | 1780 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| BTV-10w | - | - | - | - | 320 | - | - | - | 10k | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| BTV-11w | - | - | - | - | - | - | - | - | - | 10k | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| BTV-12w | - | - | 10 | - | - | - | - | - | - | - | 1000 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| BTV-13w | - | - | - | - | - | - | - | - | - | - | - | 1780 | - | - | - | - | - | - | - | - | - | - | - | - |
| BTV-14w | - | - | - | - | - | 60 | - | - | - | - | - | - | 560 | - | - | - | - | - | - | - | - | - | - | - |
| BTV-15w | - | - | - | - | - | - | 20 | - | - | - | - | - | - | 560 | - | - | - | - | - | - | - | - | - | - |
| BTV-16e | - | - | 100 | - | - | - | - | - | - | - | - | - | - | - | 320 | - | - | - | - | 32 | - | - | - | - |
| BTV-17w | - | - | - | 32 | - | - | - | - | - | - | - | - | - | - | - | 100 | - | - | 5 | - | - | - | - | - |
| BTV-18w | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1000 | - | - | - | - | - | - | - |
| BTV-19w | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 10k | - | - | - | - | - | - |
| BTV-20e | - | - | - | 3.1 | - | - | - | - | - | - | - | - | - | - | - | 32 | - | - | 100 | - | - | - | - | - |
| BTV-21e | - | - | - | - | - | 32 | - | - | - | - | - | - | - | - | - | - | - | - | - | 180 | - | - | - | - |
| BTV-22w | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1780 | - | - | - |
| BTV-23e | - | - | - | - | - | - | 32 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1000 | - | - |
| BTV-24w | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1000 | - |
| BTV-26 * | - | 15 | 10 | 10 | 10 | - | - | - | - | - | 10 | 10 | - | 10 | - | - | - | - | - | - | - | - | - | 60 |
| BTV-27 * | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fay, P.C.; Mohd Jaafar, F.; Batten, C.; Attoui, H.; Saunders, K.; Lomonossoff, G.P.; Reid, E.; Horton, D.; Maan, S.; Haig, D.; et al. Serological Cross-Reactions between Expressed VP2 Proteins from Different Bluetongue Virus Serotypes. Viruses 2021, 13, 1455. https://doi.org/10.3390/v13081455
Fay PC, Mohd Jaafar F, Batten C, Attoui H, Saunders K, Lomonossoff GP, Reid E, Horton D, Maan S, Haig D, et al. Serological Cross-Reactions between Expressed VP2 Proteins from Different Bluetongue Virus Serotypes. Viruses. 2021; 13(8):1455. https://doi.org/10.3390/v13081455
Chicago/Turabian StyleFay, Petra C., Fauziah Mohd Jaafar, Carrie Batten, Houssam Attoui, Keith Saunders, George P. Lomonossoff, Elizabeth Reid, Daniel Horton, Sushila Maan, David Haig, and et al. 2021. "Serological Cross-Reactions between Expressed VP2 Proteins from Different Bluetongue Virus Serotypes" Viruses 13, no. 8: 1455. https://doi.org/10.3390/v13081455
APA StyleFay, P. C., Mohd Jaafar, F., Batten, C., Attoui, H., Saunders, K., Lomonossoff, G. P., Reid, E., Horton, D., Maan, S., Haig, D., Daly, J. M., & Mertens, P. P. C. (2021). Serological Cross-Reactions between Expressed VP2 Proteins from Different Bluetongue Virus Serotypes. Viruses, 13(8), 1455. https://doi.org/10.3390/v13081455









