Human APOBEC3 Variations and Viral Infection
Abstract
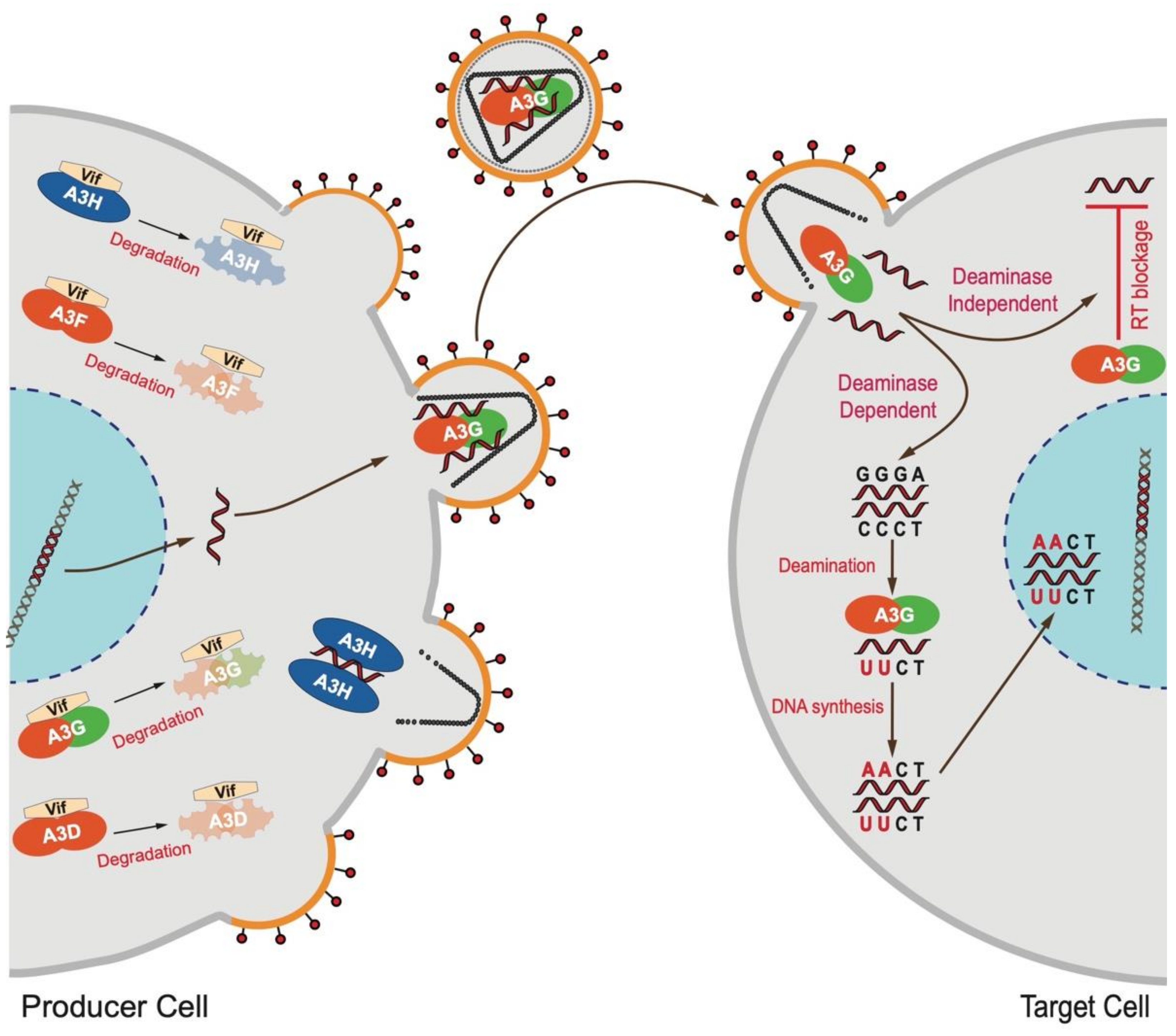
1. Introduction
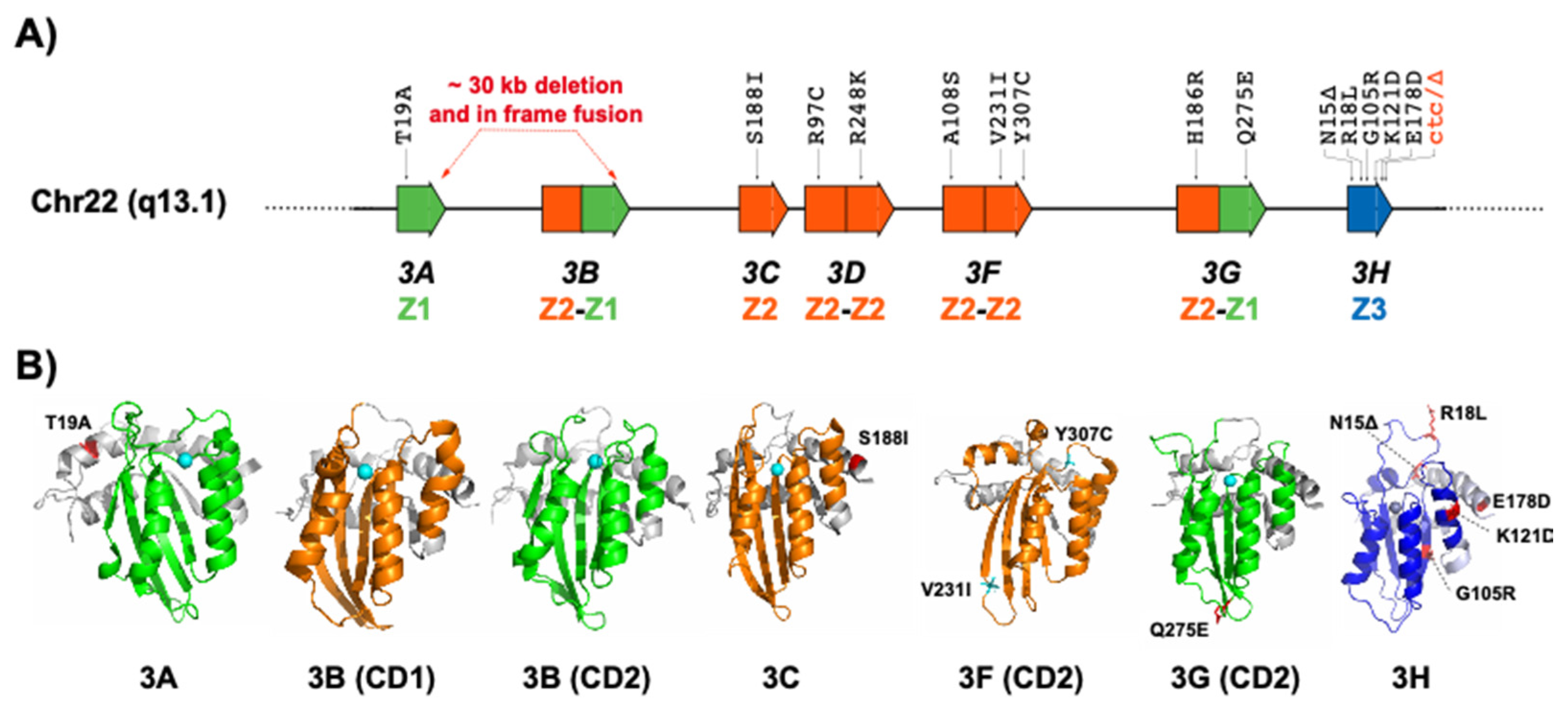
2. APOBEC3 Variations
2.1. APOBEC3A and APOBEC3B
2.2. APOBEC3C
2.3. APOBEC3D
2.4. APOBEC3F
2.5. APOBEC3G
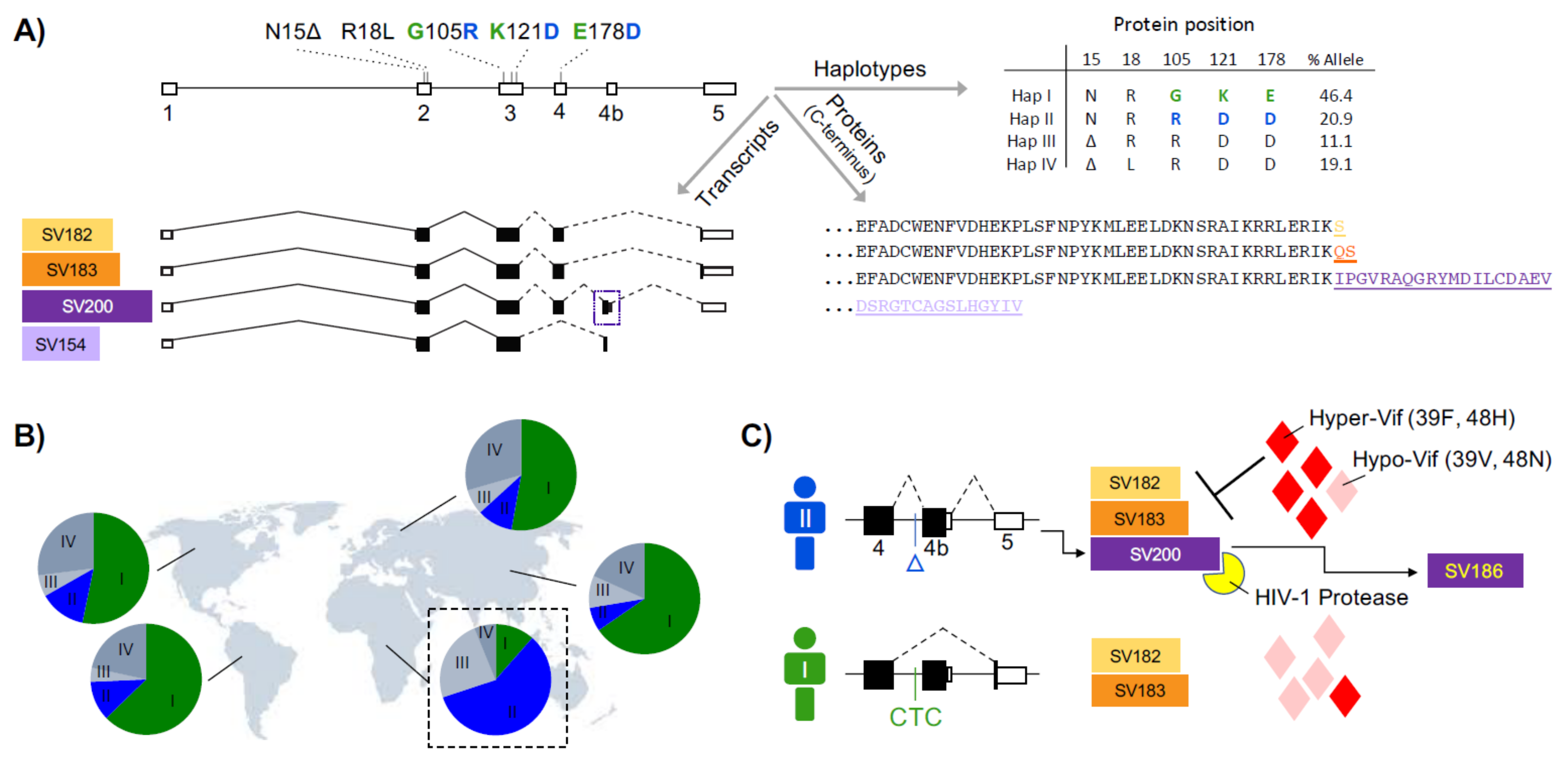
2.6. APOBEC3H
3. Future Directions and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Uriu, K.; Kosugi, Y.; Ito, J.; Sato, K. The Battle between Retroviruses and APOBEC3 Genes: Its Past and Present. Viruses 2021, 13, 124. [Google Scholar] [CrossRef]
- Xu, W.K.; Byun, H.; Dudley, J.P. The Role of APOBECs in Viral Replication. Microorganisms 2020, 8, 1899. [Google Scholar] [CrossRef] [PubMed]
- Stavrou, S.; Ross, S.R. APOBEC3 Proteins in Viral Immunity. J. Immunol. 2015, 195, 4565–4570. [Google Scholar] [CrossRef] [PubMed]
- Goila-Gaur, R.; Strebel, K. HIV-1 Vif, APOBEC, and Intrinsic Immunity. Retrovirology 2008, 5, 51. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.S.; Dudley, J.P. APOBECs and virus restriction. Virology 2015, 479, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.S.; Liddament, M.T. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 2004, 4, 868–877. [Google Scholar] [CrossRef] [PubMed]
- Chiu, Y.-L.; Greene, W.C. The APOBEC3 Cytidine Deaminases: An Innate Defensive Network Opposing Exogenous Retroviruses and Endogenous Retroelements. Annu. Rev. Immunol. 2008, 26, 317–353. [Google Scholar] [CrossRef]
- Okada, A.; Iwatani, Y. APOBEC3G-Mediated G-to-A Hypermutation of the HIV-1 Genome: The Missing Link in Antiviral Molecular Mechanisms. Front. Microbiol. 2016, 7, 2027. [Google Scholar] [CrossRef]
- Hultquist, J.; Lengyel, J.A.; Refsland, E.W.; LaRue, R.S.; Lackey, L.; Brown, W.L.; Harris, R.S. Human and Rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H Demonstrate a Conserved Capacity to Restrict Vif-Deficient HIV-1. J. Virol. 2011, 85, 11220–11234. [Google Scholar] [CrossRef]
- Cuevas, J.; Geller, R.; Garijo, R.; López-Aldeguer, J.; Sanjuán, R. Extremely High Mutation Rate of HIV-1 In Vivo. PLoS Biol. 2015, 13, e1002251. [Google Scholar] [CrossRef]
- Iwatani, Y.; Chan, D.S.; Wang, F.; Stewart-Maynard, K.; Sugiura, W.; Gronenborn, A.M.; Rouzina, I.; Williams, M.C.; Musier-Forsyth, K.; Levin, J.G. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 2007, 35, 7096–7108. [Google Scholar] [CrossRef]
- Holmes, R.K.; Malim, M.H.; Bishop, K.N. APOBEC-mediated viral restriction: Not simply editing? Trends Biochem. Sci. 2007, 32, 118–128. [Google Scholar] [CrossRef]
- Chen, X. Insights into the Structures and Multimeric Status of APOBEC Proteins Involved in Viral Restriction and Other Cellular Functions. Viruses 2021, 13, 497. [Google Scholar] [CrossRef]
- Navarro, F.; Bollman, B.; Chen, H.; König, R.; Yu, Q.; Chiles, K.; Landau, N.R. Complementary function of the two catalytic domains of APOBEC3G. Virology 2005, 333, 374–386. [Google Scholar] [CrossRef]
- Salter, J.D.; Bennett, R.P.; Smith, H.C. The APOBEC Protein Family: United by Structure, Divergent in Function. Trends Biochem. Sci. 2016, 41, 578–594. [Google Scholar] [CrossRef]
- Kitamura, S.; Ode, H.; Iwatani, Y. Structural Features of Antiviral APOBEC3 Proteins Are Linked to Their Functional Activities. Front. Microbiol. 2011, 2, 258. [Google Scholar] [CrossRef]
- Delviks-Frankenberry, K.A.; Desimmie, B.A.; Pathak, V.K. Structural Insights into APOBEC3-Mediated Lentiviral Restriction. Viruses 2020, 12, 587. [Google Scholar] [CrossRef]
- Yang, H.; Ito, F.; Wolfe, A.D.; Li, S.; Mohammadzadeh, N.; Love, R.P.; Yan, M.; Zirkle, B.; Gaba, A.; Chelico, L.; et al. Understanding the structural basis of HIV-1 restriction by the full length double-domain APOBEC3G. Nat. Commun. 2020, 11, 632. [Google Scholar] [CrossRef]
- Ebrahimi, D.; Alinejad-Rokny, H.; Davenport, M.P. Insights into the Motif Preference of APOBEC3 Enzymes. PLoS ONE 2014, 9, e87679. [Google Scholar] [CrossRef]
- Armitage, A.; Katzourakis, A.; de Oliveira, T.; Welch, J.J.; Belshaw, R.; Bishop, K.N.; Kramer, B.; McMichael, A.J.; Rambaut, A.; Iversen, A.K.N. Conserved Footprints of APOBEC3G on Hypermutated Human Immunodeficiency Virus Type 1 and Human Endogenous Retrovirus HERV-K(HML2) Sequences. J. Virol. 2008, 82, 8743–8761. [Google Scholar] [CrossRef]
- Salamango, D.J.; Harris, R.S. Dual Functionality of HIV-1 Vif in APOBEC3 Counteraction and Cell Cycle Arrest. Front. Microbiol. 2021, 11, 622012. [Google Scholar] [CrossRef] [PubMed]
- Sheehy, A.M.; Gaddis, N.; Choi, J.D.; Malim, M. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nat. Cell Biol. 2002, 418, 646–650. [Google Scholar] [CrossRef] [PubMed]
- Desimmie, B.A.; Delviks-Frankenberrry, K.A.; Burdick, R.C.; Qi, D.F.; Izumi, T.; Pathak, V.K. Multiple APOBEC3 Restriction Factors for HIV-1 and One Vif to Rule Them All. J. Mol. Biol. 2014, 426, 1220–1245. [Google Scholar] [CrossRef] [PubMed]
- Malim, M.H. Natural resistance to HIV infection: The Vif–APOBEC interaction. Comptes Rendus Biol. 2006, 329, 871–875. [Google Scholar] [CrossRef]
- Liddament, M.T.; Brown, W.L.; Schumacher, A.J.; Harris, R.S. APOBEC3F Properties and Hypermutation Preferences Indicate Activity against HIV-1 In Vivo. Curr. Biol. 2004, 14, 1385–1391. [Google Scholar] [CrossRef]
- Refsland, E.W.; Hultquist, J.; Luengas, E.M.; Ikeda, T.; Shaban, N.M.; Law, E.K.; Brown, W.L.; Reilly, C.; Emerman, M.; Harris, R.S. Natural Polymorphisms in Human APOBEC3H and HIV-1 Vif Combine in Primary T Lymphocytes to Affect Viral G-to-A Mutation Levels and Infectivity. PLoS Genet. 2014, 10, e1004761. [Google Scholar] [CrossRef]
- Ooms, M.; Brayton, B.; Letko, M.; Maio, S.M.; Pilcher, C.D.; Hecht, F.M.; Barbour, J.D.; Simon, V. HIV-1 Vif Adaptation to Human APOBEC3H Haplotypes. Cell Host Microbe 2013, 14, 411–421. [Google Scholar] [CrossRef]
- Nakano, Y.; Misawa, N.; Juarez-Fernandez, G.; Moriwaki, M.; Nakaoka, S.; Funo, T.; Yamada, E.; Soper, A.; Yoshikawa, R.; Ebrahimi, D.; et al. HIV-1 competition experiments in humanized mice show that APOBEC3H imposes selective pressure and promotes virus adaptation. PLoS Pathog. 2017, 13, e1006348. [Google Scholar]
- Wang, X.; Ao, Z.; Chen, L.; Kobinger, G.; Peng, J.; Yao, X. The Cellular Antiviral Protein APOBEC3G Interacts with HIV-1 Reverse Transcriptase and Inhibits Its Function during Viral Replication. J. Virol. 2012, 86, 3777–3786. [Google Scholar] [CrossRef]
- Pollpeter, D.; Parsons, M.; Sobala, A.E.; Coxhead, S.; Lang, R.D.; Bruns, A.M.; Papaioannou, S.; McDonnell, J.M.; Apolonia, L.; Chowdhury, J.A.; et al. Deep sequencing of HIV-1 reverse transcripts reveals the multifaceted antiviral functions of APOBEC3G. Nat. Microbiol. 2018, 3, 220–233. [Google Scholar] [CrossRef]
- Adolph, M.B.; Webb, J.; Chelico, L. Retroviral Restriction Factor APOBEC3G Delays the Initiation of DNA Synthesis by HIV-1 Reverse Transcriptase. PLoS ONE 2013, 8, e64196. [Google Scholar] [CrossRef]
- Ebrahimi, D.; Richards, C.M.; Carpenter, M.A.; Wang, J.; Ikeda, T.; Becker, J.T.; Cheng, A.Z.; McCann, J.L.; Shaban, N.M.; Salamango, D.J.; et al. Genetic and mechanistic basis for APOBEC3H alternative splicing, retrovirus restriction, and counteraction by HIV-1 protease. Nat. Commun. 2018, 9, 4137. [Google Scholar] [CrossRef]
- Ara, A.; Love, R.; Chelico, L. Different Mutagenic Potential of HIV-1 Restriction Factors APOBEC3G and APOBEC3F Is Determined by Distinct Single-Stranded DNA Scanning Mechanisms. PLoS Pathog. 2014, 10, e1004024. [Google Scholar] [CrossRef]
- Delviks-Frankenberry, K.A.; Nikolaitchik, O.A.; Burdick, R.C.; Gorelick, R.J.; Keele, B.F.; Hu, W.-S.; Pathak, V.K. Minimal Contribution of APOBEC3-Induced G-to-A Hypermutation to HIV-1 Recombination and Genetic Variation. PLoS Pathog. 2016, 12, e1005646. [Google Scholar] [CrossRef]
- Armitage, A.E.; Deforche, K.; Welch, J.J.; Van Laethem, K.; Camacho, R.; Rambaut, A.; Iversen, A.K. Possible footprints of APOBEC3F and/or other APOBEC3 deaminases, but not APOBEC3G, on HIV-1 from patients with acute/early and chronic infections. J. Virol. 2014, 88, 12882–12894. [Google Scholar] [CrossRef][Green Version]
- Bélanger, K.; Langlois, M.-A. Comparative analysis of the gene-inactivating potential of retroviral restriction factors APOBEC3F and APOBEC3G. J. Gen. Virol. 2015, 96, 2878–2887. [Google Scholar] [CrossRef]
- Sato, K.; Takeuchi, J.S.; Misawa, N.; Izumi, T.; Kobayashi, T.; Kimura, Y.; Iwami, S.; Takaori-Kondo, A.; Hu, W.-S.; Aihara, K.; et al. APOBEC3D and APOBEC3F Potently Promote HIV-1 Diversification and Evolution in Humanized Mouse Model. PLoS Pathog. 2014, 10, e1004453. [Google Scholar] [CrossRef]
- Adolph, M.B.; Love, R.P.; Chelico, L. Biochemical Basis of APOBEC3 Deoxycytidine Deaminase Activity on Diverse DNA Substrates. ACS Infect. Dis. 2018, 4, 224–238. [Google Scholar] [CrossRef]
- Simon, V.; Zennou, V.; Murray, D.; Huang, Y.; Ho, D.D.; Bieniasz, P.D. Natural Variation in Vif: Differential Impact on APOBEC3G/3F and a Potential Role in HIV-1 Diversification. PLoS Pathog. 2005, 1, e6. [Google Scholar] [CrossRef]
- Anwar, F.; Davenport, M.P.; Ebrahimi, D. Footprint of APOBEC3 on the Genome of Human Retroelements. J. Virol. 2013, 87, 8195–8204. [Google Scholar] [CrossRef]
- Alteri, C.; Surdo, M.; Bellocchi, M.C.; Saccomandi, P.; Continenza, F.; Armenia, D.; Parrotta, L.; Carioti, L.; Costa, G.; Fourati, S.; et al. Incomplete APOBEC3G/F Neutralization by HIV-1 Vif Mutants Facilitates the Genetic Evolution from CCR5 to CXCR4 Usage. Antimicrob. Agents Chemother. 2015, 59, 4870–4881. [Google Scholar] [CrossRef]
- Kim, E.-Y.; Lorenzo-Redondo, R.; Little, S.J.; Chung, Y.-S.; Phalora, P.K.; Berry, I.M.; Archer, J.; Penugonda, S.; Fischer, W.; Richman, D.D.; et al. Human APOBEC3 Induced Mutation of Human Immunodeficiency Virus Type-1 Contributes to Adaptation and Evolution in Natural Infection. PLoS Pathog. 2014, 10, e1004281. [Google Scholar] [CrossRef][Green Version]
- Wood, N.; Bhattacharya, T.; Keele, B.F.; Giorgi, E.; Liu, M.; Gaschen, B.; Daniels, M.; Ferrari, G.; Haynes, B.F.; McMichael, A.; et al. HIV Evolution in Early Infection: Selection Pressures, Patterns of Insertion and Deletion, and the Impact of APOBEC. PLoS Pathog. 2009, 5, e1000414. [Google Scholar] [CrossRef] [PubMed]
- Grant, M.; Larijani, M. Evasion of adaptive immunity by HIV through the action of host APOBEC3G/F enzymes. AIDS Res. Ther. 2017, 14, 44. [Google Scholar] [CrossRef][Green Version]
- Jern, P.; Russell, R.A.; Pathak, V.K.; Coffin, J.M. Likely Role of APOBEC3G-Mediated G-to-A Mutations in HIV-1 Evolution and Drug Resistance. PLoS Pathog. 2009, 5, e1000367. [Google Scholar] [CrossRef] [PubMed]
- Sadler, H.A.; Stenglein, M.D.; Harris, R.S.; Mansky, L.M. APOBEC3G Contributes to HIV-1 Variation through Sublethal Mutagenesis. J. Virol. 2010, 84, 7396–7404. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, M.M.; Fahrny, A.; Jayaprakash, A.; Gers-Huber, G.; Dillon-White, M.; Audigé, A.; Mulder, L.C.F.; Sachidanandam, R.; Speck, R.F.; Simon, V. Impact of Suboptimal APOBEC3G Neutralization on the Emergence of HIV Drug Resistance in Humanized Mice. J. Virol. 2020, 94, 5. [Google Scholar] [CrossRef]
- Mulder, L.C.F.; Harari, A.; Simon, V. Cytidine deamination induced HIV-1 drug resistance. Proc. Natl. Acad. Sci. USA 2008, 105, 5501–5506. [Google Scholar] [CrossRef]
- Fourati, S.; Malet, I.; Binka, M.; Boukobza, S.; Wirden, M.; Sayon, S.; Simon, A.; Katlama, C.; Simon, V.; Calvez, V.; et al. Partially active HIV-1 Vif alleles facilitate viral escape from specific antiretrovirals. AIDS 2010, 24, 2313–2321. [Google Scholar] [CrossRef]
- Tzou, P.L.; Pond, S.L.K.; Avila-Rios, S.; Holmes, S.P.; Kantor, R.; Shafer, R.W. Analysis of unusual and signature APOBEC-mutations in HIV-1 pol next-generation sequences. PLoS ONE 2020, 15, e0225352. [Google Scholar] [CrossRef]
- Poulain, F.; Lejeune, N.; Willemart, K.; Gillet, N.A. Footprint of the host restriction factors APOBEC3 on the genome of human viruses. PLoS Pathog. 2020, 16, e1008718. [Google Scholar] [CrossRef]
- Turelli, P.; Liagre-Quazzola, A.; Mangeat, B.; Verp, S.; Jost, S.; Trono, D. APOBEC3-Independent Interferon-Induced Viral Clearance in Hepatitis B Virus Transgenic Mice. J. Virol. 2008, 82, 6585–6590. [Google Scholar] [CrossRef]
- Turelli, P.; Mangeat, B.; Jost, S.; Vianin, S.; Trono, D. Inhibition of hepatitis B virus replication by APOBEC3G. Science 2004, 303, 1829. [Google Scholar] [CrossRef]
- Chen, Y.; Hu, J.; Cai, X.; Huang, Y.; Zhou, X.; Tu, Z.; Hu, J.; Tavis, J.E.; Tang, N.; Huang, A.; et al. APOBEC3B edits HBV DNA and inhibits HBV replication during reverse transcription. Antivir. Res. 2018, 149, 16–25. [Google Scholar] [CrossRef]
- Kanagaraj, A.; Sakamoto, N.; Que, L.; Li, Y.; Mohiuddin, M.; Koura, M.; Wakae, K.; Kurachi, M.; Muramatsu, M.; Kitamura, K. Different antiviral activities of natural APOBEC3C, APOBEC3G, and APOBEC3H variants against hepatitis B virus. Biochem. Biophys. Res. Commun. 2019, 518, 26–31. [Google Scholar] [CrossRef]
- Peretti, A.; Geoghegan, E.M.; Pastrana, D.V.; Smola, S.; Feld, P.; Sauter, M.; Lohse, S.; Ramesh, M.; Lim, E.S.; Wang, D.; et al. Characterization of BK Polyomaviruses from Kidney Transplant Recipients Suggests a Role for APOBEC3 in Driving In-Host Virus Evolution. Cell Host Microbe 2018, 23, 628–635.E7. [Google Scholar] [CrossRef]
- Verhalen, B.; Starrett, G.J.; Harris, R.S.; Jiang, M. Functional Upregulation of the DNA Cytosine Deaminase APOBEC3B by Polyomaviruses. J. Virol. 2016, 90, 6379–6386. [Google Scholar] [CrossRef]
- Fan, J.; Ma, G.; Nosaka, K.; Tanabe, J.; Satou, Y.; Koito, A.; Wain-Hobson, S.; Vartanian, J.P.; Matsuoka, M. APOBEC3G Generates Nonsense Mutations in HTLV-1 Proviral Genomes In Vivo. J. Virol. 2010, 84, 7278–7287. [Google Scholar] [CrossRef]
- Sasada, A.; Takaori-Kondo, A.; Shirakawa, K.; Kobayashi, M.; Abudu, A.; Hishizawa, M.; Imada, K.; Tanaka, Y.; Uchiyama, T. APOBEC3G targets human T-cell leukemia virus type 1. Retrovirology 2005, 2, 32. [Google Scholar] [CrossRef][Green Version]
- Argyris, P.P.; Wilkinson, P.E.; Jarvis, M.C.; Magliocca, K.R.; Patel, M.R.; Vogel, R.I.; Gopalakrishnan, R.; Koutlas, I.G.; Harris, R.S. Endogenous APOBEC3B overexpression characterizes HPV-positive and HPV-negative oral epithelial dysplasias and head and neck cancers. Mod. Pathol. 2021, 34, 280–290. [Google Scholar] [CrossRef]
- Zhu, B.; Xiao, Y.; Yeager, M.; Clifford, G.; Wentzensen, N.; Cullen, M.; Boland, J.F.; Bass, S.; Steinberg, M.K.; Raine-Bennett, T.; et al. Mutations in the HPV16 genome induced by APOBEC3 are associated with viral clearance. Nat. Commun. 2020, 11, 886. [Google Scholar] [CrossRef] [PubMed]
- Warren, C.; Xu, T.; Guo, K.; Griffin, L.M.; Westrich, J.A.; Lee, D.; Lambert, P.F.; Santiago, M.L.; Pyeon, D. APOBEC3A Functions as a Restriction Factor of Human Papillomavirus. J. Virol. 2014, 89, 688–702. [Google Scholar] [CrossRef]
- Cheng, A.; Moraes, S.; Shaban, N.; Fanunza, E.; Bierle, C.; Southern, P.; Bresnahan, W.; Rice, S.; Harris, R. APOBECs and Herpesviruses. Viruses 2021, 13, 390. [Google Scholar] [CrossRef]
- Cheng, A.Z.; Moraes, S.N.; Attarian, C.; Yockteng-Melgar, J.; Jarvis, M.C.; Biolatti, M.; Galitska, G.; Dell’Oste, V.; Frappier, L.; Bierle, C.J.; et al. A Conserved Mechanism of APOBEC3 Relocalization by Herpesviral Ribonucleotide Reductase Large Subunits. J. Virol. 2019, 93, 64. [Google Scholar] [CrossRef]
- Suspène, R.; Aynaud, M.-M.; Koch, S.; Pasdeloup, D.; Labetoulle, M.; Gaertner, B.; Vartanian, J.-P.; Meyerhans, A.; Wain-Hobson, S. Genetic Editing of Herpes Simplex Virus 1 and Epstein-Barr Herpesvirus Genomes by Human APOBEC3 Cytidine Deaminases in Culture and In Vivo. J. Virol. 2011, 85, 7594–7602. [Google Scholar] [CrossRef]
- Cheng, A.Z.; Yockteng-Melgar, J.; Jarvis, M.; Malik-Soni, N.; Borozan, I.; Carpenter, M.A.; McCann, J.L.; Ebrahimi, D.; Shaban, N.M.; Marcon, E.; et al. Epstein–Barr virus BORF2 inhibits cellular APOBEC3B to preserve viral genome integrity. Nat. Microbiol. 2019, 4, 78–88. [Google Scholar] [CrossRef]
- Martinez, T.; Shapiro, M.; Bhaduri-McIntosh, S.; MacCarthy, T. Evolutionary effects of the AID/APOBEC family of mutagenic enzymes on human gamma-herpesviruses. Virus Evol. 2019, 5, vey040. [Google Scholar] [CrossRef] [PubMed]
- Nakaya, Y.; Stavrou, S.; Blouch, K.; Tattersall, P.; Ross, S.R. In Vivo Examination of Mouse APOBEC3- and Human APOBEC3A- and APOBEC3G-Mediated Restriction of Parvovirus and Herpesvirus Infection in Mouse Models. J. Virol. 2016, 90, 8005–8012. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.A.; Holland, T.C.; Bhagwat, A.S. Human Herpes Simplex Virus-1 depletes APOBEC3A from nuclei. Virology 2019, 537, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Chaurasiya, K.R.; McCauley, M.J.; Wang, W.; Qualley, M.F.; Wu, T.; Kitamura, S.; Geertsema, H.; Chan, D.S.B.; Hertz, A.; Iwatani, Y.; et al. Oligomerization transforms human APOBEC3G from an efficient enzyme to a slowly dissociating nucleic acid-binding protein. Nat. Chem. 2013, 6, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Morse, M.; Huo, R.; Feng, Y.; Rouzina, I.; Chelico, L.; Williams, M.C. Dimerization regulates both deaminase-dependent and deaminase-independent HIV-1 restriction by APOBEC3G. Nat. Commun. 2017, 8, 597. [Google Scholar] [CrossRef]
- Guo, F.; Cen, S.; Niu, M.; Saadatmand, J.; Kleiman, L. Inhibition of tRNA3Lys-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J. Virol. 2006, 80, 11710–11722. [Google Scholar] [CrossRef]
- Guo, F.; Cen, S.; Niu, M.; Yang, Y.; Gorelick, R.J.; Kleiman, L. The Interaction of APOBEC3G with Human Immunodeficiency Virus Type 1 Nucleocapsid Inhibits tRNA 3 Lys Annealing to Viral RNA. J. Virol. 2007, 81, 11322–11331. [Google Scholar] [CrossRef]
- Sawyer, S.; Emerman, M.; Malik, H.S. Ancient Adaptive Evolution of the Primate Antiviral DNA-Editing Enzyme APOBEC3G. PLoS Biol. 2004, 2, e275. [Google Scholar] [CrossRef]
- Ito, J.; Gifford, R.J.; Sato, K. Retroviruses drive the rapid evolution of mammalianAPOBEC3genes. Proc. Natl. Acad. Sci. USA 2020, 117, 610–618. [Google Scholar] [CrossRef]
- Uriu, K.; Kosugi, Y.; Suzuki, N.; Ito, J.; Sato, K. Elucidation of the Complicated Scenario of Primate APOBEC3 Gene Evolution. J. Virol. 2021, 95, 12. [Google Scholar] [CrossRef]
- Oh Ainle, M.; Kerns, J.A.; Malik, H.S.; Emerman, M. Adaptive Evolution and Antiviral Activity of the Conserved Mammalian Cytidine Deaminase APOBEC3H. J. Virol. 2006, 80, 3853–3862. [Google Scholar] [CrossRef]
- Ortiz, M.; Bleiber, G.; Martinez, R.; Kaessmann, H.; Telenti, A. Patterns of evolution of host proteins involved in retroviral pathogenesis. Retrovirology 2006, 3, 11. [Google Scholar] [CrossRef]
- An, P.; Bleiber, G.; Duggal, P.; Nelson, G.; May, M.; Mangeat, B.; Alobwede, I.; Trono, D.; Vlahov, D.; Donfield, S.; et al. APOBEC3G Genetic Variants and Their Influence on the Progression to AIDS. J. Virol. 2004, 78, 11070–11076. [Google Scholar] [CrossRef]
- Matume, N.D.; Tebit, D.; Gray, L.R.; Turner, S.D.; Rekosh, D.; Bessong, P.O.; Hammarskjöld, M.-L. Characterization of APOBEC3 variation in a population of HIV-1 infected individuals in northern South Africa. BMC Med. Genet. 2019, 20, 21. [Google Scholar] [CrossRef]
- Li, M.M.H.; Emerman, M. Polymorphism in Human APOBEC3H Affects a Phenotype Dominant for Subcellular Localization and Antiviral Activity. J. Virol. 2011, 85, 8197–8207. [Google Scholar] [CrossRef] [PubMed]
- Zhen, A.; Wang, T.; Zhao, K.; Xiong, Y.; Yu, X.-F. A Single Amino Acid Difference in Human APOBEC3H Variants Determines HIV-1 Vif Sensitivity. J. Virol. 2009, 84, 1902–1911. [Google Scholar] [CrossRef]
- Harari, A.; Ooms, M.; Mulder, L.C.F.; Simon, V. Polymorphisms and Splice Variants Influence the Antiretroviral Activity of Human APOBEC3H. J. Virol. 2009, 83, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Lassen, K.G.; Wissing, S.; Lobritz, M.A.; Santiago, M.L.; Greene, W.C. Identification of Two APOBEC3F Splice Variants Displaying HIV-1 Antiviral Activity and Contrasting Sensitivity to Vif*. J. Biol. Chem. 2010, 285, 29326–29335. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Chen, Q.; Xiao, X.; Ito, F.; Wolfe, A.; Chen, X.S. Biochemical Characterization of APOBEC3H Variants: Implications for Their HIV-1 Restriction Activity and mC Modification. J. Mol. Biol. 2016, 428, 4626–4638. [Google Scholar] [CrossRef] [PubMed]
- Doehle, B.P.; Schafer, A.; Cullen, B.R. Human APOBEC3B is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 Vif. Virology 2005, 339, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Bogerd, H.P.; Wiegand, H.L.; Doehle, B.P.; Cullen, B.R. The intrinsic antiretroviral factor APOBEC3B contains two enzymatically active cytidine deaminase domains. Virology 2007, 364, 486–493. [Google Scholar] [CrossRef] [PubMed]
- Berger, G.; Durand, S.; Fargier, G.; Nguyen, X.-N.; Cordeil, S.; Bouaziz, S.; Muriaux, D.; Darlix, J.-L.; Cimarelli, A. APOBEC3A Is a Specific Inhibitor of the Early Phases of HIV-1 Infection in Myeloid Cells. PLoS Pathog. 2011, 7, e1002221. [Google Scholar] [CrossRef]
- Suspène, R.; Guétard, D.; Henry, M.; Sommer, P.; Wain-Hobson, S.; Vartanian, J.-P. Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc. Natl. Acad. Sci. USA 2005, 102, 8321–8326. [Google Scholar] [CrossRef]
- Warren, C.; Westrich, J.A.; Van Doorslaer, K.; Pyeon, D. Roles of APOBEC3A and APOBEC3B in Human Papillomavirus Infection and Disease Progression. Viruses 2017, 9, 233. [Google Scholar] [CrossRef]
- Narvaiza, I.; Linfesty, D.C.; Greener, B.N.; Hakata, Y.; Pintel, D.J.; Logue, E.; Landau, N.R.; Weitzman, M.D. Deaminase-Independent Inhibition of Parvoviruses by the APOBEC3A Cytidine Deaminase. PLoS Pathog. 2009, 5, e1000439. [Google Scholar] [CrossRef]
- Bogerd, H.P.; Wiegand, H.L.; Hulme, A.E.; Garcia-Perez, J.L.; O’Shea, K.S.; Moran, J.V.; Cullen, B.R. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl. Acad. Sci. USA 2006, 103, 8780–8785. [Google Scholar] [CrossRef]
- Chen, H.; Lilley, C.E.; Yu, Q.; Lee, D.V.; Chou, J.; Narvaiza, I.; Landau, N.R.; Weitzman, M.D. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr. Biol. 2006, 16, 480–485. [Google Scholar] [CrossRef]
- Esnault, C.; Priet, S.; Ribet, D.; Heidmann, O.; Heidmann, T. Restriction by APOBEC3 proteins of endogenous retroviruses with an extracellular life cycle: Ex vivo effects and in vivo “traces” on the murine IAPE and human HERV-K elements. Retrovirology 2008, 5, 75. [Google Scholar] [CrossRef]
- Refsland, E.W.; Stenglein, M.D.; Shindo, K.; Albin, J.S.; Brown, W.L.; Harris, R.S. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: Implications for HIV-1 restriction. Nucleic Acids Res. 2010, 38, 4274–4284. [Google Scholar] [CrossRef]
- A Roberts, S.; Lawrence, M.S.; Klimczak, L.J.; A Grimm, S.; Fargo, D.; Stojanov, P.; Kiezun, A.; Kryukov, G.; Carter, S.L.; Saksena, G.; et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 2013, 45, 970–976. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Borresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Kim, J.; Haradhvala, N.J.; Huang, M.N.; Ng, A.W.T.; Wu, Y.; Boot, A.; Covington, K.R.; Gordenin, D.A.; Bergstrom, E.N.; et al. The repertoire of mutational signatures in human cancer. Nature 2020, 578, 94–101. [Google Scholar] [CrossRef]
- Klonowska, K.; Kluzniak, W.; Rusak, B.; Jakubowska, A.; Ratajska, M.; Krawczynska, N.; Vasilevska, D.; Czubak, K.; Wojciechowska, M.; Cybulski, C.; et al. The 30 kb deletion in the APOBEC3 cluster decreases APOBEC3A and APOBEC3B expression and creates a transcriptionally active hybrid gene but does not associate with breast cancer in the European population. Oncotarget 2017, 8, 76357–76374. [Google Scholar] [CrossRef]
- Kidd, J.; Newman, T.L.; Tuzun, E.; Kaul, R.; E Eichler, E. Population Stratification of a Common APOBEC Gene Deletion Polymorphism. PLoS Genet. 2007, 3, e63. [Google Scholar] [CrossRef]
- Prasetyo, A.A.; Sariyatun, R.; Reviono; Sari, Y.; Hudiyono; Haryati, S.; Adnan, Z.A.; Hartono; Kageyama, S. The APOBEC3B deletion polymorphism is associated with prevalence of hepatitis B virus, hepatitis C virus, Torque Teno virus, and Toxoplasma gondii co-infection among HIV-infected individuals. J. Clin. Virol. 2015, 70, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Ochi, H.; Maekawa, T.; Hatakeyama, T.; Tsuge, M.; Kitamura, S.; Kimura, T.; Miki, D.; Mitsui, F.; Hiraga, N.; et al. Effects of structural variations of APOBEC3A and APOBEC3B genes in chronic hepatitis B virus infection. Hepatol. Res. 2009, 39, 1159–1168. [Google Scholar] [CrossRef] [PubMed]
- Ezzikouri, S.; Kitab, B.; Rebbani, K.; Marchio, A.; Wain-Hobson, S.; Dejean, A.; Vartanian, J.-P.; Pineau, P.; Benjelloun, S. Polymorphic APOBEC3 modulates chronic hepatitis B in Moroccan population. J. Viral Hepat. 2013, 20, 678–686. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.; Marathe, S.D.; Nain, S.; Nema, V.; Ghate, M.V.; Gangakhedkar, R.R. APOBEC3B deletion impacts on susceptibility to acquire HIV-1 and its advancement among individuals in western India. APMIS 2016, 124, 881–887. [Google Scholar] [CrossRef] [PubMed]
- An, P.; Johnson, R.; Phair, J.; Kirk, G.D.; Yu, X.F.; Donfield, S.; Buchbinder, S.; Goedert, J.J.; Winkler, C.A. APOBEC3B deletion and risk of HIV-1 acquisition. J. Infect. Dis. 2009, 200, 1054–1058. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, T.; E Nakayama, E.; Kaur, G.; Terunuma, H.; Mimaya, J.-I.; Ohtani, H.; Mehra, N.; Shioda, T.; Kimura, A. Impact of novel TRIM5α variants, Gly110Arg and G176del, on the anti-HIV-1 activity and the susceptibility to HIV-1 infection. AIDS 2009, 23, 2091–2100. [Google Scholar] [CrossRef] [PubMed]
- Itaya, S.; Nakajima, T.; Kaur, G.; Terunuma, H.; Ohtani, H.; Mehra, N.; Kimura, A. No evidence of an association between the APOBEC3B deletion polymorphism and susceptibility to HIV infection and AIDS in Japanese and Indian populations. J. Infect. Dis. 2010, 202, 815–816. [Google Scholar] [CrossRef][Green Version]
- Imahashi, M.; Izumi, T.; Watanabe, D.; Imamura, J.; Matsuoka, K.; Ode, H.; Masaoka, T.; Sato, K.; Kaneko, N.; Ichikawa, S.; et al. Lack of association between intact/deletion polymorphisms of the APOBEC3B gene and HIV-1 risk. PLoS ONE 2014, 9, e92861. [Google Scholar] [CrossRef]
- Koning, F.A.; Newman, E.N.; Kim, E.Y.; Kunstman, K.J.; Wolinsky, S.M.; Malim, M.H. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J. Virol. 2009, 83, 9474–9485. [Google Scholar] [CrossRef]
- Muckenfuss, H.; Hamdorf, M.; Held, U.; Perković, M.; Löwer, J.; Cichutek, K.; Flory, E.; Schumann, G.G.; Münk, C. APOBEC3 Proteins Inhibit Human LINE-1 Retrotransposition. J. Biol. Chem. 2006, 281, 22161–22172. [Google Scholar] [CrossRef]
- Horn, A.V.; Klawitter, S.; Held, U.; Berger, A.; Vasudevan, A.A.J.; Bock, A.; Hofmann, H.; Hanschmann, K.-M.O.; Trösemeier, J.-H.; Flory, E.; et al. Human LINE-1 restriction by APOBEC3C is deaminase independent and mediated by an ORF1p interaction that affects LINE reverse transcriptase activity. Nucleic Acids Res. 2014, 42, 396–416. [Google Scholar] [CrossRef]
- Wittkopp, C.J.; Adolph, M.B.; Wu, L.I.; Chelico, L.; Emerman, M. A Single Nucleotide Polymorphism in Human APOBEC3C Enhances Restriction of Lentiviruses. PLoS Pathog. 2016, 12, e1005865. [Google Scholar] [CrossRef] [PubMed]
- Anderson, B.D.; Ikeda, T.; Moghadasi, S.A.; Martin, A.S.; Brown, W.L.; Harris, R.S. Natural APOBEC3C variants can elicit differential HIV-1 restriction activity. Retrovirology 2018, 15, 78. [Google Scholar] [CrossRef] [PubMed]
- Refsland, E.W.; Hultquist, J.F.; Harris, R.S. Endogenous Origins of HIV-1 G-to-A Hypermutation and Restriction in the Nonpermissive T Cell Line CEM2n. PLoS Pathog. 2012, 8, e1002800. [Google Scholar] [CrossRef] [PubMed]
- Chaipan, C.; Smith, J.L.; Hu, W.-S.; Pathak, V.K. APOBEC3G Restricts HIV-1 to a Greater Extent than APOBEC3F and APOBEC3DE in Human Primary CD4+ T Cells and Macrophages. J. Virol. 2012, 87, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Gillick, K.; Pollpeter, D.; Phalora, P.; Kim, E.-Y.; Wolinsky, S.; Malim, M.H. Suppression of HIV-1 Infection by APOBEC3 Proteins in Primary Human CD4+ T Cells Is Associated with Inhibition of Processive Reverse Transcription as Well as Excessive Cytidine Deamination. J. Virol. 2013, 87, 1508–1517. [Google Scholar] [CrossRef] [PubMed]
- Duggal, N.K.; Fu, W.; Akey, J.M.; Emerman, M. Identification and antiviral activity of common polymorphisms in the APOBEC3 locus in human populations. Virology 2013, 443, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Holmes, R.K.; Koning, F.A.; Bishop, K.N.; Malim, M.H. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation—Comparisons with APOBEC3G. J. Biol. Chem. 2007, 282, 2587–2595. [Google Scholar] [CrossRef]
- Krisko, J.F.; Begum, N.; Baker, C.E.; Foster, J.L.; Garcia, J.V. APOBEC3G and APOBEC3F Act in Concert to Extinguish HIV-1 Replication. J. Virol. 2016, 90, 4681–4695. [Google Scholar] [CrossRef]
- Jonsson, S.R.; Hache, G.; Stenglein, M.D.; Fahrenkrug, S.C.; Andresdottir, V.; Harris, R.S. Evolutionarily conserved and non-conserved retrovirus restriction activities of artiodactyl APOBEC3F proteins. Nucleic Acids Res. 2006, 34, 5683–5694. [Google Scholar] [CrossRef]
- Ara, A.; Love, R.P.; Follack, T.B.; Ahmed, K.A.; Adolph, M.B.; Chelico, L. Mechanism of Enhanced HIV Restriction by Virion Coencapsidated Cytidine Deaminases APOBEC3F and APOBEC3G. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Mbisa, J.L.; Bu, W.; Pathak, V.K. APOBEC3F and APOBEC3G Inhibit HIV-1 DNA Integration by Different Mechanisms. J. Virol. 2010, 84, 5250–5259. [Google Scholar] [CrossRef]
- Adolph, M.B.; Ara, A.; Chelico, L. APOBEC3 Host Restriction Factors of HIV-1 Can Change the Template Switching Frequency of Reverse Transcriptase. J. Mol. Biol. 2019, 431, 1339–1352. [Google Scholar] [CrossRef] [PubMed]
- Mohammadzadeh, N.; Love, R.P.; Gibson, R.; Arts, E.J.; Poon, A.F.; Chelico, L. Role of co-expressed APOBEC3F and APOBEC3G in inducing HIV-1 drug resistance. Heliyon 2019, 5, e01498. [Google Scholar] [CrossRef]
- An, P.; Penugonda, S.; Thorball, C.W.; Bartha, I.; Goedert, J.J.; Donfield, S.; Buchbinder, S.; Binns-Roemer, E.; Kirk, G.D.; Zhang, W.; et al. Role of APOBEC3F Gene Variation in HIV-1 Disease Progression and Pneumocystis Pneumonia. PLoS Genet. 2016, 12, e1005921. [Google Scholar] [CrossRef]
- Mohammadzadeh, N.; Follack, T.B.; Love, R.P.; Stewart, K.; Sanche, S.; Chelico, L. Polymorphisms of the cytidine deaminase APOBEC3F have different HIV-1 restriction efficiencies. Virology 2019, 527, 21–31. [Google Scholar] [CrossRef]
- Malim, M.H. APOBEC proteins and intrinsic resistance to HIV-1 infection. Philos. Trans. R. Soc. B Biol. Sci. 2008, 364, 675–687. [Google Scholar] [CrossRef]
- Noguchi, C.; Hiraga, N.; Mori, N.; Tsuge, M.; Imamura, M.; Takahashi, S.; Fujimoto, Y.; Ochi, H.; Abe, H.; Maekawa, T.; et al. Dual effect of APOBEC3G on Hepatitis B virus. J. Gen. Virol. 2007, 88, 432–440. [Google Scholar] [CrossRef]
- Lei, Y.C.; Hao, Y.H.; Zhang, Z.M.; Tian, Y.J.; Wang, B.J.; Yang, Y.; Zhao, X.P.; Lu, M.J.; Gong, F.L.; Yang, D.L. Inhibition of hepatitis B virus replication by APOBEC3G in vitro and in vivo. World J. Gastroenterol. 2006, 12, 4492–4497. [Google Scholar] [CrossRef]
- Hulme, A.E.; Bogerd, H.P.; Cullen, B.R.; Moran, J.V. Selective inhibition of Alu retrotransposition by APOBEC3G. Gene 2007, 390, 199–205. [Google Scholar] [CrossRef]
- Reddy, K.; Ooms, M.; Letko, M.; Garrett, N.; Simon, V.; Ndung’U, T. Functional characterization of Vif proteins from HIV-1 infected patients with different APOBEC3G haplotypes. AIDS 2016, 30, 1723–1729. [Google Scholar] [CrossRef][Green Version]
- Singh, K.K.; Wang, Y.; Gray, K.P.; Farhad, M.; Brummel, S.; Fenton, T.; Trout, R.; Spector, S.A. Genetic Variants in the Host Restriction Factor APOBEC3G are Associated With HIV-1–Related Disease Progression and Central Nervous System Impairment in Children. JAIDS J. Acquir. Immune Defic. Syndr. 2013, 62, 197–203. [Google Scholar] [CrossRef]
- Do, H.; Vasilescu, A.; Diop, G.; Hirtzig, T.; Heath, S.C.; Coulonges, C.; Rappaport, J.; Therwath, A.; Lathrop, M.; Matsuda, F.; et al. Exhaustive Genotyping of theCEM15 (APOBEC3G)Gene and Absence of Association with AIDS Progression in a French Cohort. J. Infect. Dis. 2005, 191, 159–163. [Google Scholar] [CrossRef]
- Valcke, H.S.; Bernard, N.F.; Bruneau, J.; Alary, M.; Tsoukas, C.M.; Roger, M. APOBEC3G genetic variants and their association with risk of HIV infection in highly exposed Caucasians. AIDS 2006, 20, 1984–1986. [Google Scholar] [CrossRef]
- De Maio, F.A.; Rocco, C.A.; Aulicino, P.C.; Bologna, R.; Mangano, A.; Sen, L. Effect of HIV-1 Vif variability on progression to pediatric AIDS and its association with APOBEC3G and CUL5 polymorphisms. Infect. Genet. Evol. 2011, 11, 1256–1262. [Google Scholar] [CrossRef]
- De Maio, F.A.; Rocco, C.A.; Aulicino, P.C.; Bologna, R.; Mangano, A.; Sen, L. APOBEC3-Mediated Editing in HIV Type 1 from Pediatric Patients and Its Association withAPOBEC3G/CUL5Polymorphisms and Vif Variability. AIDS Res. Hum. Retrovir. 2012, 28, 619–627. [Google Scholar] [CrossRef]
- Singh, H.; Marathe, S.; Nain, S.; Nema, V.; Angadi, M.; Bapat, S.; Pawar, J.; Ghate, M.; Sahay, S.; Gangakhedkar, R.R. Coding region variant 186H/R in Exon 4 of APOBEC3G among individuals of Western India. APMIS 2016, 124, 401–405. [Google Scholar] [CrossRef]
- Rathore, A.; Chatterjee, A.; Yamamoto, N.; Dhole, T.N.T.N. Absence of H186R Polymorphism in Exon 4 of the APOBEC3G Gene among North Indian Individuals. Genet. Test. 2008, 12, 453–456. [Google Scholar] [CrossRef]
- Reddy, K.; A Winkler, C.; Werner, L.; Mlisana, K.; Karim, S.A.; Ndung’U, T. APOBEC3G expression is dysregulated in primary HIV-1 infection and polymorphic variants influence CD4+ T-cell counts and plasma viral load. AIDS 2010, 24, 195–204. [Google Scholar] [CrossRef]
- Compaore, T.R.; Soubeiga, S.T.; Ouattara, A.K.; Obiri-Yeboah, D.; Tchelougou, D.; Maiga, M.; Assih, M.; Bisseye, C.; Bakouan, D.; Compaore, I.P.; et al. APOBEC3G Variants and Protection against HIV-1 Infection in Burkina Faso. PLoS ONE 2016, 11, e0146386. [Google Scholar] [CrossRef][Green Version]
- Compaore, T.R.; Diarra, B.; Assih, M.; Obiri-Yeboah, D.; Soubeiga, S.T.; Ouattara, A.K.; Tchelougou, D.; Bisseye, C.; Bakouan, D.R.; Compaore, I.P.; et al. HBV/HIV co-infection and APOBEC3G polymorphisms in a population from Burkina Faso. BMC Infect. Dis. 2016, 16, 336. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bizinoto, M.C.; Leal, É.; Diaz, R.S.; Janini, L.M. Loci Polymorphisms of the APOBEC3G Gene in HIV Type 1-Infected Brazilians. AIDS Res. Hum. Retrovir. 2011, 27, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.; Gangakhedkar, R. Occurrence of APOBEC3G variations in West Indian HIV patients. Microb. Pathog. 2018, 121, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Mhandire, K.; Duri, K.; Mhandire, D.; Musarurwa, C.; Stray-Pedersen, B.; Dandara, C. Evaluating the contribution of APOBEC3G haplotypes, on influencing HIV infection in a Zimbabwean paediatric population. South. Afr. Med. J. 2016, 106, 119–123. [Google Scholar] [CrossRef]
- Pace, C.; Keller, J.; Nolan, D.; James, I.; Gaudieri, S.; Moore, C.; Mallal, S. Population Level Analysis of Human Immunodeficiency Virus Type 1 Hypermutation and Its Relationship with APOBEC3G and vif Genetic Variation. J. Virol. 2007, 81, 8843–8845. [Google Scholar] [CrossRef]
- Wang, X.; Abudu, A.; Son, S.; Dang, Y.; Venta, P.J.; Zheng, Y.-H. Analysis of Human APOBEC3H Haplotypes and Anti-Human Immunodeficiency Virus Type 1 Activity. J. Virol. 2011, 85, 3142–3152. [Google Scholar] [CrossRef]
- Oh Ainle, M.; Kerns, J.A.; Li, M.M.; Malik, H.S.; Emerman, M. Antiretroelement Activity of APOBEC3H Was Lost Twice in Recent Human Evolution. Cell Host Microbe 2008, 4, 249–259. [Google Scholar] [CrossRef]
- Chesarino, N.M.; Emerman, M. Polymorphisms in Human APOBEC3H Differentially Regulate Ubiquitination and Antiviral Activity. Viruses 2020, 12, 378. [Google Scholar] [CrossRef]
- Ooms, M.; Majdak, S.; Seibert, C.W.; Harari, A.; Simon, V. The Localization of APOBEC3H Variants in HIV-1 Virions Determines Their Antiviral Activity. J. Virol. 2010, 84, 7961–7969. [Google Scholar] [CrossRef]
- Gourraud, P.-A.; Karaouni, A.; Woo, J.; Schmidt, T.; Oksenberg, J.; Hecht, F.; Liegler, T.; Barbour, J. APOBEC3H haplotypes and HIV-1 pro-viral vif DNA sequence diversity in early untreated human immunodeficiency virus–1 infection. Hum. Immunol. 2011, 72, 207–212. [Google Scholar] [CrossRef]
- Binka, M.; Ooms, M.; Steward, M.; Simon, V. The Activity Spectrum of Vif from Multiple HIV-1 Subtypes against APOBEC3G, APOBEC3F, and APOBEC3H. J. Virol. 2011, 86, 49–59. [Google Scholar] [CrossRef]
- Ooms, M.; Letko, M.; Binka, M.; Simon, V. The Resistance of Human APOBEC3H to HIV-1 NL4-3 Molecular Clone Is Determined by a Single Amino Acid in Vif. PLoS ONE 2013, 8, e57744. [Google Scholar] [CrossRef]
- Benito, J.M.; Hillung, J.; Restrepo, C.; Cuevas, J.M.; Leon, A.; Ruiz-Mateos, E.; Palacios-Muñoz, R.; Górgolas, M.; Sanjuán, R.; Rallón, N. Role of APOBEC3H in the Viral Control of HIV Elite Controller Patients. Int. J. Med. Sci. 2018, 15, 95–100. [Google Scholar] [CrossRef]
- Sakurai, D.; Iwatani, Y.; Ohtani, H.; Naruse, T.K.; Terunuma, H.; Sugiura, W.; Kimura, A. APOBEC3H polymorphisms associated with the susceptibility to HIV-1 infection and AIDS progression in Japanese. Immunogenetics 2015, 67, 253–257. [Google Scholar] [CrossRef]
- Naruse, T.K.; Sakurai, D.; Ohtani, H.; Sharma, G.; Sharma, S.K.; Vajpayee, M.; Mehra, N.K.; Kaur, G.; Kimura, A. APOBEC3H polymorphisms and susceptibility to HIV-1 infection in an Indian population. J. Hum. Genet. 2015, 61, 263–265. [Google Scholar] [CrossRef]
| Description | Allelic Frequency | Studied Populations and References | Effect | |||
|---|---|---|---|---|---|---|
| African | European | Asian | ||||
| APOBEC3A/B | ||||||
| rs12628403 (surrogate for ~30 kb APOBEC3B deletion) | ~30 kb deletion (Δ) of all APOBEC3B exons and introns except for exon 8; In-frame fusion to APOBEC3A. | n = 3394 Δ = 0.03 | n = 20,404 Δ = 0.08 | n = 116 Δ = 0.4 | Indonesian PMID: 26305823 | HIV-1 co-infections: Δ > reference |
| Moroccan PMID: 24010642 | %Δ: HBV patients ~ healthy donors Progression to liver diseases: Δ > reference | |||||
| Western Indian PMID: 27522954 | Risk of HIV-1 acquisition: Δ > reference | |||||
| European Americans and African Americans PMID: 19698078 | HIV-1 acquisition, progression to AIDS, and viral set point: Δ/Δ > reference | |||||
| Japanese and Indian PMID: 20684727 | %Δ: HIV-1 patients ~ healthy donors | |||||
| Japanese PMID: 19788695 | %Δ: HBV patients ~ healthy donors Hypermutation context: Δ ~ reference | |||||
| Japanese PMID: 24667791 | %Δ: HIV-1 patients ~ healthy donors HIV-1 co-infections: Δ ~ reference HIV-1 progression: Δ ~ reference | |||||
| APOBEC3C | ||||||
| rs112120857 | Missense (G > C / G > T) S188I | n = 3574 G = 0.94 C = 0.00 T = 0.06 | n = 37,290 G = 0.99962 C = 0.00003 T = 0.00035 | n = 168 G = 1.00 C = 0.00 T = 0.00 | PMIDs: 27732658, 30558640 | In vitro anti-HIV-1 activity: I > S |
| APOBEC3D | ||||||
| rs201709403 | Missense (A > G) T238A | n = 2898 A = 1.00 G = 0.00 | n = 9690 A = 1.00 G = 0.00 | n = 112 A = 1.00 G = 0.00 | Northern South African PMID: 30660178 | %G (238A): HIV-1 patients > 1000 Genomes Africans |
| rs75858538 | Missense (C > T) R97C | n = 5146 C = 0.98 T = 0.02 | n = 180,258 C = 0.9999 T = 0.0001 | n = 6364 C = 0.9998 T = 0.0002 | Northern South African PMID: 30660178 | %T (97C): HIV-1 patients > 1000 Genomes Africans |
| PMID: 23755966 | Protein expression: R ~ C Anti-HIV-1 activity: R > C Sensitivity to HIV-1 Vif: R ~ C Anti-Alu activity: R ~ C | |||||
| rs61748819 | Missense (G > A) R248K | n = 3574 G = 0.91 A = 0.09 | n = 32,834 G = 0.9997 A = 0.00034 | n = 168 G = 1.00 A = 0.000 | PMID: 23755966 | Protein expression: R > K Anti-HIV-1 activity: R > K Sensitivity to HIV-1 Vif: R ~ K Anti-Alu activity: R > K |
| APOBEC3F | ||||||
| rs2020390 | Missense (G > T) A108S | n = 3512 G = 0.60 T = 0.40 | n = 37,024 G = 0.49 T = 0.51 | n = 168 G = 0.29 T = 0.71 | Northern South African PMID: 30660178 | %T (108S): HIV-1 patients > 1000 Genomes African populations |
| rs12157816 | Missense (A > G) Y307C | n = 3574 A = 0.97 G = 0.03 | n = 37,264 A = 0.9881 G = 0.0119 | n = 168 A = 1.0 G = 0.0 | Northern South African PMID: 30660178 | %G (307C): HIV-1 patients > 1000 Genomes African populations |
| rs2076101 | Missense (G > A) V231I | n = 8220 G = 0.73 A = 0.27 | n = 256,418 G = 0.48 A = 0.52 | n = 3812 G = 0.3 A = 0.7 | European American and African American PMID: 26942578 | Set-point viral load: I > V Progression to AIDS: I > V Pneumocystis pneumonia: I > V |
| PMID: 30448640 | HIV-1 restriction: V > I Protection against HIV-1 Vif: V > I Level of viral encapsidation: V > I | |||||
| APOBEC3FΔ2 | Isoform lacking exon 2 | PMID: 20624919 | Sensitivity to Vif: main isoform > APOBEC3FΔ2 Viral packaging: main isoform > APOBEC3FΔ2 Antiviral activity: main isoform > APOBEC3FΔ2 Deamination: main isoform > APOBEC3FΔ2 | |||
| APOBEC3FΔ2-4 | Isoform lacking exons 2, 3 and 4 | PMID: 20624919 | Sensitivity to Vif: APOBEC3FΔ2–4 > main isoform Viral packaging: main isoform > APOBEC3FΔ2–4 Antiviral activity: Main isoform > APOBEC3FΔ2–4 deamination: main isoform > APOBEC3FΔ2–4 | |||
| APOBEC3G | ||||||
| rs5757463 | -571G/C upstream (G > C) | n = 106 G = 0.29 C = 0.71 | n = 6564 G = 0.06 C = 0.94 | n = 4 G = 0.0 C = 1.0 | African Americans and European Americans PMID: 15452227 | Susceptibility to HIV-1 infection: G ~ C |
| Brazilian PMID: 20874421 | CD4 T-cell count: CC > CG and GG | |||||
| West Indian PMID: 29864532 | %CG: HIV patients > healthy donors %(GC + CC): HIV patients > healthy donors | |||||
| Zimbabwean PMID: 27245545 | Susceptibility to HIV-1 infection: G ~ C | |||||
| rs34550797 | -199G/A upstream (G > A) | n = 2816 G = 0.995 A = 0.005 | n = 7618 G = 0.9992 A = 0.0008 | n = 108 G = 1.0 A = 0.0 | African Americans and European Americans PMID: 15452227 | Susceptibility to HIV-1 infection: G ~ A |
| rs5750743 | -90C/G upstream/5′UTR (C > G) | n = 276 C = 1.00 G = 0.00 | n = 5330 C = 0.54 G = 0.46 | n = 8 C = 1.0 G = 0.0 | African Americans and European Americans PMID: 15452227 | Susceptibility to HIV-1 infection: C ~ G |
| West Indian PMID: 29864532 | %CG: HIV-1 patients > healthy donors %GG: healthy donors > HIV patients | |||||
| Zimbabwean PMID: 27245545 | Susceptibility to HIV-1 infection: C ~ G | |||||
| rs5757465 | Synonymous (T > C) F119F | n = 11,214 T = 0.90 C = 0.10 | n = 248,594 T = 0.56 C = 0.44 | n = 3908 T = 0.76 C = 0.24 | African Americans and European Americans PMID: 15452227 | Susceptibility to HIV-1 infection: T ~ C |
| Diverse populations PMID: 23138837 | HIV-1 disease progression: T > C CNS impairment: T > C | |||||
| rs3736685 | 197193T/C intron (T > C) | n = 7982 T = 0.63 C = 0.37 | n = 223,498 T = 0.97 C = 0.03 | n = 3870 T = 0.92 C = 0.08 | African Americans and European Americans PMID: 15452227 | Susceptibility to HIV-1 infection: T ~ C |
| Zimbabwean PMID: 27245545 | Susceptibility to HIV-1 infection: T ~ C | |||||
| rs2294367 | 199376G/C intron (C > G) | n = 2036 C = 0.99 G = 0.01 | n = 8254 C = 0.57 G = 0.43 | n = 16 C = 0.75 G = 0.25 | African Americans and European Americans PMID: 15452227 | Susceptibility to HIV-1 infection: C ~ G Progression to AIDS: G > C |
| Zimbabwean PMID: 27245545 | Susceptibility to HIV-1 infection: C ~ G | |||||
| rs8177832 | Missense (A > G) H186R | n = 11,678 A = 0.66 G = 0.34 | n = 252,598 A = 0.97 G = 0.03 | n = 6820 A = 0.92 G = 0.08 | African Americans and European Americans PMID: 15452227 | Susceptibility to HIV-1 infection: H ~ R Rate of CD4 T cell loss: R > H Progression to AIDS: R > H |
| PMID: 27064995 | Antiviral activity: H > R Counteraction by Vif: H ~ R | |||||
| Diverse populations PMID: 23138837 | HIV-1 disease progression: R > H CNS impairment: R > H | |||||
| South Africans PMID: 19996938 | Viral load: R > H CD4 T cell count: H > R | |||||
| French PMID: 15609224 | Progression to AIDS: H ~ R | |||||
| Caucasians PMID: 16988524 | Progression to AIDS: H ~ R | |||||
| Argentinian PMID: 22145963 | Progression to AIDS: H ~ R Viral G-to-A mutation: H ~ R | |||||
| Argentinian PMID: 21571098 | HIV-1 transmission: H ~ R Progression to AIDS: H ~ R Viral G-to-A mutation: H ~ R | |||||
| Zimbabwean PMID: 27245545 | Susceptibility to HIV-1 infection: H ~ R | |||||
| Morocco PMID: 24010642 | Risk of HBV acquisition: H ~ R | |||||
| Burkina Fasoian PMID: 26741797 | Protection against HIV-1 infection: GGT [rs6001417, rs8177832 (H186R), rs35228531] > Other haplotypes Risk of HIV-1 infection: GGC and CGC > Other haplotypes | |||||
| Burkina Fasoian PMID: 27449138 | HIV-1/HBV co-infection rate: other haplotypes > GGT [rs6001417, rs8177832 (H186R), rs35228531] | |||||
| Western Indian PMID: 26853443 | Risk of HIV-1 acquisition: H ~ R Progression to AIDS: H ~ R | |||||
| North Indian PMID: 18652534 | Absence of 186R variant | |||||
| rs17496018 | C40693T Intron (C > T) | n = 4296 C = 0.94 T = 0.06 | n = 26,952 C = 0.93 T = 0.07 | n = 128 C = 0.97 T = 0.03 | Caucasians PMID: 16988524 | Risk of HIV-1 infection: T > C |
| Argentinian PMID 22145963 | Progression to AIDS: H ~ R Viral G > A mutation: H ~ R | |||||
| Argentinian PMID: 21571098 | HIV-1 transmission: H ~ R Progression to AIDS: H ~ R Viral G-to-A mutation: H ~ R C > T polymorphism was associated with Vif variation. | |||||
| rs17496046 | Missense (C > G) Q275E | n = 4374 C = 0.87 G = 0.13 | n = 96,948 C = 0.93 G = 0.07 | n = 3326 C = 0.97 G = 0.03 | Caucasians PMID: 16988524 | Risk of HIV-1 infection: C ~ G |
| Northern South African PMID: 30660178 | % G (275E): HIV-1 patients > 1000 Genomes Africans | |||||
| rs6001417 | Intron (C > G) | n = 2960 C = 0.63 G = 0.37 | n = 16,474 C = 0.97 G = 0.03 | n = 112 C = 0.94 G = 0.06 | Burkina Fasoian PMID: 26741797 | Protection against HIV-1 infection: GGT [rs6001417, rs8177832 (H186R), rs35228531] > Other haplotypes Risk of HIV-1 infection: GGC and CGC > Other haplotypes |
| Burkina Fasoian PMID: 27449138 | HIV-1/HBV co-infection rate: other haplotypes > GGT [rs6001417, rs8177832 (H186R), rs35228531] | |||||
| rs35228531 | Downstream (C > T) | n = 1564 C = 0.994 T = 0.006 | n = 9814 C = 1.00 T = 0.00 | n = 112 C = 1.00 T = 0.00 | South African PMID: 19996938 | Viral load: T > C CD4 T-cell count: C > T |
| Burkina Fasoian PMID: 26741797 | Protection against HIV-1 infection: GGT [rs6001417, rs8177832 (H186R), rs35228531] > Other haplotypes Risk of HIV-1 infection: GGC and CGC > Other haplotypes | |||||
| Burkina Fasoian PMID: 27449138 | HIV-1/HBV co-infection rate: C > T HIV-1/HBV co-infection rate: Other haplotypes > GGT [rs6001417, rs8177832 (H186R), rs35228531] | |||||
| rs5757467 | Intron (C > T) | n = 2946 C = 0.31 T = 0.69 | n = 15,414 C = 0.36 T = 0.64 | n = 112 C = 0.21 T = 0.79 | West Australian PMID: 16940537 | HIV-1 hypermutation: C > T |
| APOBEC3H | ||||||
| rs139292 (Previously rs140936762) | Indel (CAA/Δ) N15Δ | n = 3506 Δ = 0.32 | n = 20,264 Δ = 0.34 | n = 168 Δ = 0.23 | PMID: 32235597 | Antiviral activity: Reference > Δ |
| Northern South African PMID: 30660178 | %Δ: HIV-1 patients >1000 Genomes Africans | |||||
| Japanese PMID: 25721876 | Susceptibility to HIV-1 infection: Δ > reference | |||||
| Indian PMID: 26559750 | Susceptibility to HIV-1 infection: Δ > reference | |||||
| rs139293 | Missense (G > T) R18L | n = 3158 G = 0.96 T = 0.04 | n = 36,146 G = 0.75 T = 0.25 | n = 166 G = 0.92 T = 0.08 | Northern South Africa PMID: 30660178 | %T(18L): HIV-1 patients > 1000 Genomes Africans |
| rs139297 | Missense (G > C) G105R | n = 1308 G = 0.66 C = 0.34 | n = 35,022 G = 0.56 C = 0.44 | n = 148 G = 0.89 C = 0.11 | PMID: 32235597 | Ubiquitination: G (HapI) > R (HapII) |
| PMID: 20519396 | Antiviral activity: R (HapII) > G (HapI) HIV-1 encapsidation: R (HapII) ~ G (HapI) Protein expression: R (HapII) > G (HapI) Interaction with Gag: R (HapII) with nucleocapsid; G (HapI) with C-terminal of matrix and N-terminal of capsid | |||||
| Japanese PMID: 25721876 | Susceptibility to HIV-1 infection: G (HapI) > R (HapII) Progression to AIDS: G (HapI) > R (HapII) | |||||
| Indian PMID: 26559750 | %15Δ-105R: HIV-1 patients > healthy donors | |||||
| Diverse populations PMID: 21167246 | Rate of HIV-1 GA-to-AA mutation: R (HapII) > G (HapI) | |||||
| rs139299 | Missense (G > C) K121D | n = 3574 G = 0.24 C = 0.76 | n = 37,158 G = 0.52 C = 0.48 | n = 168 G = 0.72 C = 0.28 | Diverse populations PMID: 21167246 | Rate of HIV-1 GA-to-AA mutation: D (HapII) > K (HapI) |
| rs139298 | Missense (A > G) K121D | n = 11,188 A = 0.21 G = 0.79 | n = 252,052 A = 0.53 G = 0.47 | n = 6744 A = 0.69 G = 0.31 | Diverse populations PMID: 21167246 | Rate of HIV-1 GA-to-AA mutation: D (HapII) > K (HapI) |
| rs139302 | Missense (G > C) E178D | n = 3882 G = 0.26 C = 0.74 | n = 97,472 G = 0.54 C = 0.46 | n = 3326 G = 0.67 C = 0.33 | PMID: 21167246 | Rate of HIV-1 GA-to-AA mutation: D (HapII) > E (HapI) |
| Northern South African PMID: 30660178 | %C (178D): HIV-1 patients > 1000 Genomes Africans | |||||
| PMID: 27534815 | Cytosine deamination: E (HapV) > D (other haplotypes) Methyl cytosine deamination: D (HapII) >> E (other haplotypes) DNA binding: all haplotypes are the same. | |||||
| rs149229175 | Indel (CTC/Δ) intron | n = 3410 Δ = 0.32 | n = 18,142 Δ = 0.13 | n = 164 Δ = 0.08 | Diverse populations PMID: 30297863 | %SV200: Δ > CTC |
| SV200, SV183, SV182, and SV154 | Splice variants with different C-terminals | Diverse populations PMID: 18945781 | Viral restriction: SV200 (HapII) > other variants | |||
| Diverse populations PMID: 30297863 | Antiviral activity: HapII-SV200 > SV182/183 Viral encapsidation: HapII-SV182/183 > SV200 Protease processing: only HapII SV200 L1 fragment in transcript: only HapII SV200 | |||||
| PMID: 27534815 | Cytosine and methyl cytosine deamination: HapI-SV182/183 > SV200 >> SV154 | |||||
| PMID: 31400856 | HBV restriction: HapII SV183 > other variants | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sadeghpour, S.; Khodaee, S.; Rahnama, M.; Rahimi, H.; Ebrahimi, D. Human APOBEC3 Variations and Viral Infection. Viruses 2021, 13, 1366. https://doi.org/10.3390/v13071366
Sadeghpour S, Khodaee S, Rahnama M, Rahimi H, Ebrahimi D. Human APOBEC3 Variations and Viral Infection. Viruses. 2021; 13(7):1366. https://doi.org/10.3390/v13071366
Chicago/Turabian StyleSadeghpour, Shiva, Saeideh Khodaee, Mostafa Rahnama, Hamzeh Rahimi, and Diako Ebrahimi. 2021. "Human APOBEC3 Variations and Viral Infection" Viruses 13, no. 7: 1366. https://doi.org/10.3390/v13071366
APA StyleSadeghpour, S., Khodaee, S., Rahnama, M., Rahimi, H., & Ebrahimi, D. (2021). Human APOBEC3 Variations and Viral Infection. Viruses, 13(7), 1366. https://doi.org/10.3390/v13071366






