Evaluation of ELISA-Based Multiplex Peptides for the Detection of Human Serum Antibodies Induced by Zika Virus Infection across Various Countries
Abstract
:1. Introduction
2. Materials and Methods
2.1. Serum Sources
2.2. High-Throughput ELISA Plate Preparation, Quantification, and Normalization
2.3. Computational Processing of Peptide ELISA Data
2.4. Validation Using Mikrogen Commercial Assay
2.5. Validation Using PRNT Assay
2.6. Validation Using Microneutralization Assay
3. Results
3.1. Peptide-Based ELISA vs. Mikrogen Commercial Assay
3.2. Comparison of Peptide-Based ELISA against DENV qRT-PCR
3.3. Peptide-Based ELISA vs. IgG in Longitudinal Samples from Chilean Patients
3.4. Peptide-Based ELISA vs. qRT-PCR and PRNT Assays in Characterized Brazilian Serum Samples
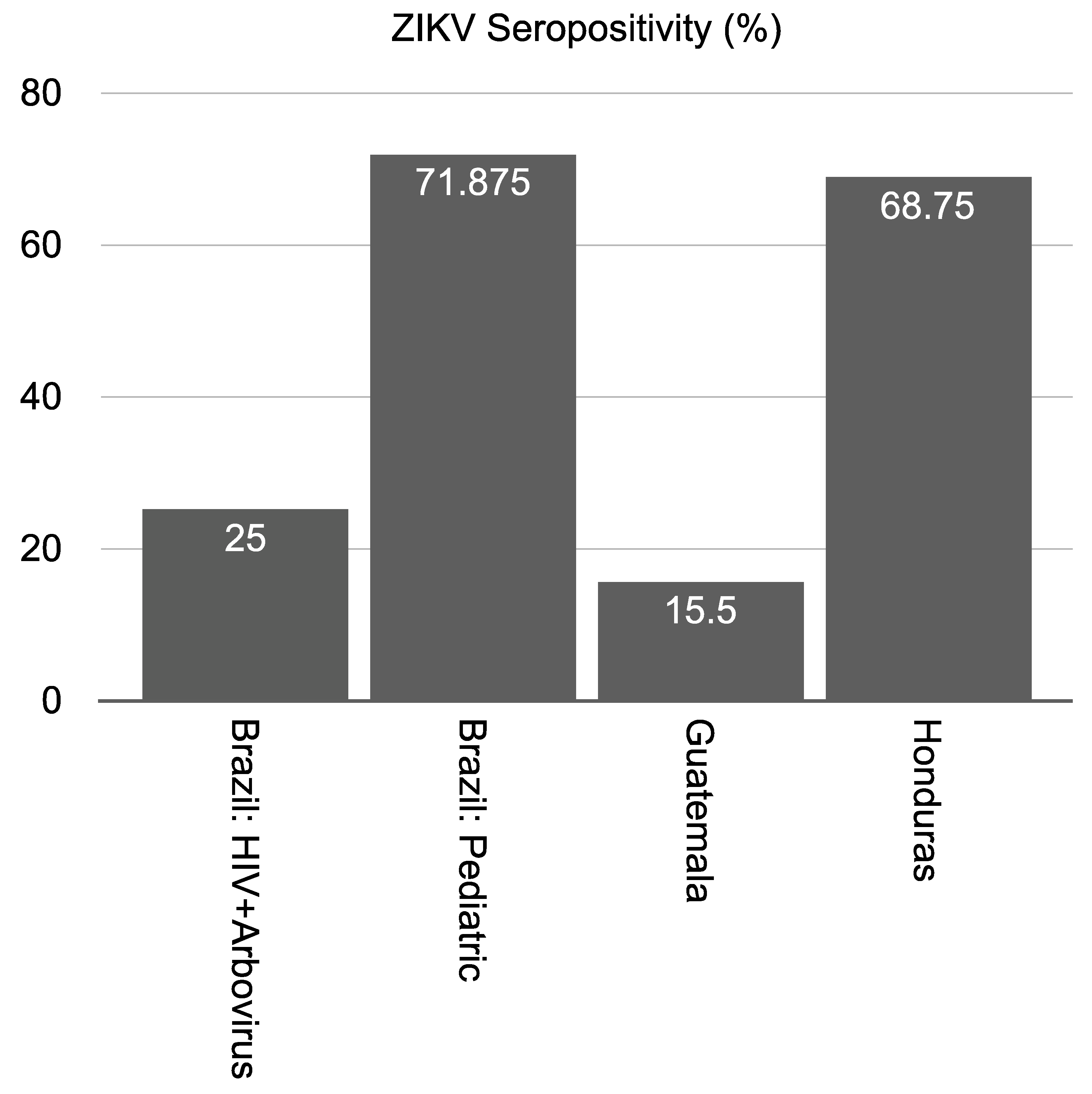
3.5. Peptide-Based ELISA to Estimate Seropositivity among Individuals in ZIKV-Endemic Regions
3.6. Comparison of Peptide-Based ELISA with Microneutralization Assays in Uncharacterized Guatemalan Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- D’Ortenzio, E.; Matheron, S.; Yazdanpanah, Y.; de Lamballerie, X.; Hubert, B.; Piorkowski, G.; Maquart, M.; Descamps, D.; Damond, F.; Leparc-Goffart, I. Evidence of Sexual Transmission of Zika Virus. N. Engl. J. Med. 2016, 374, 2195–2198. [Google Scholar] [CrossRef]
- McDonald, E.M.; Duggal, N.K.; Brault, A.C. Pathogenesis and Sexual Transmission of Spondweni and Zika Viruses. PLoS Negl. Trop. Dis. 2017, 11, e0005990. [Google Scholar] [CrossRef]
- Moreira, J.; Lamas, C.C.; Siqueira, A. Sexual Transmission of Zika Virus: Implications for Clinical Care and Public Health Policy. Clin. Infect. Dis. 2016, 63, 141–142. [Google Scholar] [CrossRef] [Green Version]
- Musso, D.; Richard, V.; Teissier, A.; Stone, M.; Lanteri, M.C.; Latoni, G.; Alsina, J.; Reik, R.; Busch, M.P. Recipient Epidemiology and Donor Evaluation Study (REDS-III) ZIKV Study Group. Detection of Zika Virus RNA in Semen of Asymptomatic Blood Donors. Clin. Microbiol. Infect. 2017, 23, 1001.e1–1001.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duffy, M.R.; Chen, T.H.; Hancock, W.T.; Powers, A.M.; Kool, J.L.; Lanciotti, R.S.; Pretrick, M.; Marfel, M.; Holzbauer, S.; Dubray, C.; et al. Zika Virus Outbreak on Yap Island, Federated States of Micronesia. N. Engl. J. Med. 2009, 360, 2536–2543. [Google Scholar] [CrossRef] [PubMed]
- Galindo-Fraga, A.; Ochoa-Hein, E.; Sifuentes-Osornio, J.; Ruiz-Palacios, G. Zika Virus: A New Epidemic on our Doorstep. Rev. Investig. Clin. 2015, 67, 329–332. [Google Scholar]
- White, M.K.; Wollebo, H.S.; David Beckham, J.; Tyler, K.L.; Khalili, K. Zika Virus: An Emergent Neuropathological Agent. Ann. Neurol. 2016, 80, 479–489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayes, E.B. Zika Virus Outside Africa. Emerg. Infect. Dis. 2009, 15, 1347–1350. [Google Scholar] [CrossRef]
- Lanciotti, R.S.; Kosoy, O.L.; Laven, J.J.; Velez, J.O.; Lambert, A.J.; Johnson, A.J.; Stanfield, S.M.; Duffy, M.R. Genetic and Serologic Properties of Zika Virus Associated with an Epidemic, Yap State, Micronesia, 2007. Emerg. Infect. Dis. 2008, 14, 1232–1239. [Google Scholar] [CrossRef] [PubMed]
- Beltramello, M.; Williams, K.L.; Simmons, C.P.; Macagno, A.; Simonelli, L.; Quyen, N.T.; Sukupolvi-Petty, S.; Navarro-Sanchez, E.; Young, P.R.; de Silva, A.M.; et al. The Human Immune Response to Dengue Virus is Dominated by Highly Cross-Reactive Antibodies Endowed with Neutralizing and Enhancing Activity. Cell. Host Microbe 2010, 8, 271–283. [Google Scholar] [CrossRef] [Green Version]
- Deng, Y.Q.; Dai, J.X.; Ji, G.H.; Jiang, T.; Wang, H.J.; Yang, H.O.; Tan, W.L.; Liu, R.; Yu, M.; Ge, B.X.; et al. A Broadly Flavivirus Cross-Neutralizing Monoclonal Antibody that Recognizes a Novel Epitope within the Fusion Loop of E Protein. PLoS ONE 2011, 6, e16059. [Google Scholar] [CrossRef] [Green Version]
- Stiasny, K.; Kiermayr, S.; Holzmann, H.; Heinz, F.X. Cryptic Properties of a Cluster of Dominant Flavivirus Cross-Reactive Antigenic Sites. J. Virol. 2006, 80, 9557–9568. [Google Scholar] [CrossRef] [Green Version]
- Stettler, K.; Beltramello, M.; Espinosa, D.A.; Graham, V.; Cassotta, A.; Bianchi, S.; Vanzetta, F.; Minola, A.; Jaconi, S.; Mele, F.; et al. Specificity, Cross-Reactivity, and Function of Antibodies Elicited by Zika Virus Infection. Science 2016, 353, 823–826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Priyamvada, L.; Quicke, K.M.; Hudson, W.H.; Onlamoon, N.; Sewatanon, J.; Edupuganti, S.; Pattanapanyasat, K.; Chokephaibulkit, K.; Mulligan, M.J.; Wilson, P.C.; et al. Human Antibody Responses after Dengue Virus Infection are Highly Cross-Reactive to Zika Virus. Proc. Natl. Acad. Sci. USA 2016, 113, 7852–7857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kam, Y.W.; Lum, F.M.; Teo, T.H.; Lee, W.W.; Simarmata, D.; Harjanto, S.; Chua, C.L.; Chan, Y.F.; Wee, J.K.; Chow, A.; et al. Early Neutralizing IgG Response to Chikungunya Virus in Infected Patients Targets a Dominant Linear Epitope on the E2 Glycoprotein. EMBO Mol. Med. 2012, 4, 330–343. [Google Scholar] [CrossRef] [PubMed]
- Kam, Y.W.; Pok, K.Y.; Eng, K.E.; Tan, L.K.; Kaur, S.; Lee, W.W.; Leo, Y.S.; Ng, L.C.; Ng, L.F. Sero-Prevalence and Cross-Reactivity of Chikungunya Virus Specific Anti-E2EP3 Antibodies in Arbovirus-Infected Patients. PLoS Negl. Trop. Dis. 2015, 9, e3445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martins, K.A.; Gregory, M.K.; Valdez, S.M.; Sprague, T.R.; Encinales, L.; Pacheco, N.; Cure, C.; Porras-Ramirez, A.; Rico-Mendoza, A.; Chang, A.; et al. Neutralizing Antibodies from Convalescent Chikungunya Virus Patients can Cross-Neutralize Mayaro and Una Viruses. Am. J. Trop. Med. Hyg. 2019, 100, 1541–1544. [Google Scholar] [CrossRef]
- Wikan, N.; Suputtamongkol, Y.; Yoksan, S.; Smith, D.R.; Auewarakul, P. Immunological Evidence of Zika Virus Transmission in Thailand. Asian Pac. J. Trop. Med. 2016, 9, 141–144. [Google Scholar] [CrossRef]
- Balmaseda, A.; Zambrana, J.V.; Collado, D.; Garcia, N.; Saborio, S.; Elizondo, D.; Mercado, J.C.; Gonzalez, K.; Cerpas, C.; Nunez, A.; et al. Comparison of Four Serological Methods and Two Reverse Transcription-PCR Assays for Diagnosis and Surveillance of Zika Virus Infection. J. Clin. Microbiol. 2018, 56, e01785-17. [Google Scholar] [CrossRef] [Green Version]
- Mishra, N.; Caciula, A.; Price, A.; Thakkar, R.; Ng, J.; Chauhan, L.V.; Jain, K.; Che, X.; Espinosa, D.A.; Montoya Cruz, M.; et al. Diagnosis of Zika Virus Infection by Peptide Array and Enzyme-Linked Immunosorbent Assay. MBio 2018, 9, e00095-18. [Google Scholar] [CrossRef] [Green Version]
- Bosch, I.; de Puig, H.; Hiley, M.; Carre-Camps, M.; Perdomo-Celis, F.; Narvaez, C.F.; Salgado, D.M.; Senthoor, D.; O’Grady, M.; Phillips, E.; et al. Rapid Antigen Tests for Dengue Virus Serotypes and Zika Virus in Patient Serum. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef] [Green Version]
- Nascimento, E.J.M.; George, J.K.; Velasco, M.; Bonaparte, M.I.; Zheng, L.; DiazGranados, C.A.; Marques, E.T.A.; Huleatt, J.W. Development of an Anti-Dengue NS1 IgG ELISA to Evaluate Exposure to Dengue Virus. J. Virol. Methods 2018, 257, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.J.; Bhattacharya, R.; Scheuermann, R.H.; Pickett, B.E. Identification of Diagnostic Peptide Regions that Distinguish Zika Virus from Related Mosquito-Borne Flaviviruses. PLoS ONE 2017, 12, e0178199. [Google Scholar] [CrossRef] [PubMed]
- Martinez Viedma, M.D.P.; Kose, N.; Parham, L.; Balmaseda, A.; Kuan, G.; Lorenzana, I.; Harris, E.; Crowe, J.E.; Pickett, B.E. Peptide Arrays Incubated with Three Collections of Human Sera from Patients Infected with Mosquito-Borne Viruses. F1000Research 2019, 8, 1875. [Google Scholar] [CrossRef] [PubMed]
- Ramanakumar, A.V.; Thomann, P.; Candeias, J.M.; Ferreira, S.; Villa, L.L.; Franco, E.L. Use of the Normalized Absorbance Ratio as an Internal Standardization Approach to Minimize Measurement Error in Enzyme-Linked Immunosorbent Assays for Diagnosis of Human Papillomavirus Infection. J. Clin. Microbiol. 2010, 48, 791–796. [Google Scholar] [CrossRef] [Green Version]
- Baer, A.; Kehn-Hall, K. Viral Concentration Determination through Plaque Assays: Using Traditional and Novel Overlay Systems. J. Vis. Exp. 2014, 93, e52065. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, E.J.M.; Bonaparte, M.I.; Luo, P.; Vincent, T.S.; Hu, B.; George, J.K.; Anez, G.; Noriega, F.; Zheng, L.; Huleatt, J.W. Use of a Blockade-of-Binding ELISA and Microneutralization Assay to Evaluate Zika Virus Serostatus in Dengue-Endemic Areas. Am. J. Trop. Med. Hyg. 2019, 101, 708–715. [Google Scholar] [CrossRef] [Green Version]
- Sabalza, M.; Barber, C.A.; Abrams, W.R.; Montagna, R.; Malamud, D. Zika Virus Specific Diagnostic Epitope Discovery. J. Vis. Exp. 2017, 130. [Google Scholar] [CrossRef]
- Hoen, B.; Carpentier, M.; Gaete, S.; Tressieres, B.; Herrmann-Storck, C.; Vingadassalom, I.; Huc-Anais, P.; Funk, A.L.; Fontanet, A.; de Lamballerie, X. Kinetics of Anti-Zika Virus Antibodies After Acute Infection in Pregnant Women. J. Clin. Microbiol. 2019, 57. [Google Scholar] [CrossRef]
- Miller, E.; Becker, Z.; Shalev, D.; Lee, C.T.; Cioroiu, C.; Thakur, K. Probable Zika Virus-Associated Guillain-Barre Syndrome: Challenges with Clinico-Laboratory Diagnosis. J. Neurol. Sci. 2017, 375, 367–370. [Google Scholar] [CrossRef]
- Shan, C.; Xie, X.; Ren, P.; Loeffelholz, M.J.; Yang, Y.; Furuya, A.; Dupuis, A.P.; Kramer, L.D.; Wong, S.J.; Shi, P.Y. A Rapid Zika Diagnostic Assay to Measure Neutralizing Antibodies in Patients. EBioMedicine 2017, 17, 157–162. [Google Scholar] [CrossRef] [Green Version]
- Balmaseda, A.; Stettler, K.; Medialdea-Carrera, R.; Collado, D.; Jin, X.; Zambrana, J.V.; Jaconi, S.; Cameroni, E.; Saborio, S.; Rovida, F.; et al. Antibody-Based Assay Discriminates Zika Virus Infection from other Flaviviruses. Proc. Natl. Acad. Sci. USA 2017, 114, 8384–8389. [Google Scholar] [CrossRef] [Green Version]
- Gake, B.; Vernet, M.A.; Leparc-Goffart, I.; Drexler, J.F.; Gould, E.A.; Gallian, P.; de Lamballerie, X. Low Seroprevalence of Zika Virus in Cameroonian Blood Donors. Braz. J. Infect. Dis. 2017, 21, 481–483. [Google Scholar] [CrossRef] [PubMed]
- Gallian, P.; Cabie, A.; Richard, P.; Paturel, L.; Charrel, R.N.; Pastorino, B.; Leparc-Goffart, I.; Tiberghien, P.; de Lamballerie, X. Zika Virus in Asymptomatic Blood Donors in Martinique. Blood 2017, 129, 263–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saba Villarroel, P.M.; Nurtop, E.; Pastorino, B.; Roca, Y.; Drexler, J.F.; Gallian, P.; Jaenisch, T.; Leparc-Goffart, I.; Priet, S.; Ninove, L.; et al. Zika Virus Epidemiology in Bolivia: A Seroprevalence Study in Volunteer Blood Donors. PLoS Negl. Trop. Dis. 2018, 12, e0006239. [Google Scholar] [CrossRef]
- L’Huillier, A.G.; Hamid-Allie, A.; Kristjanson, E.; Papageorgiou, L.; Hung, S.; Wong, C.F.; Stein, D.R.; Olsha, R.; Goneau, L.W.; Dimitrova, K.; et al. Evaluation of Euroimmun Anti-Zika Virus IgM and IgG Enzyme-Linked Immunosorbent Assays for Zika Virus Serologic Testing. J. Clin. Microbiol. 2017, 55, 2462–2471. [Google Scholar] [CrossRef] [Green Version]
- Dhar-Chowdhury, P.; Paul, K.K.; Haque, C.E.; Hossain, S.; Lindsay, L.R.; Dibernardo, A.; Brooks, W.A.; Drebot, M.A. Dengue Seroprevalence, Seroconversion and Risk Factors in Dhaka, Bangladesh. PLoS Negl. Trop. Dis. 2017, 11, e0005475. [Google Scholar] [CrossRef]
- Petitdemange, C.; Becquart, P.; Wauquier, N.; Beziat, V.; Debre, P.; Leroy, E.M.; Vieillard, V. Unconventional Repertoire Profile is Imprinted during Acute Chikungunya Infection for Natural Killer Cells Polarization toward Cytotoxicity. PLoS Pathog. 2011, 7, e1002268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Collection | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Control | Sri Lanka | Chile | Brazil (HIV) | Brazil (Pediatric) | Brazil (Arbovirus) | Honduras | Guatemala | ||
| Number Starting Samples | 6 | 9 | 27 | 108 | 32 | 200 | |||
| Number Samples (after QC) | 6 | 9 | 19 | 4 | 22 | 32 | 32 | 175 | |
| Validation Assays | Mikrogen IgG | ✓ * | |||||||
| qRT-PCR | ✓ * | ✓ | ✓ * | ||||||
| IgM (ELISA) | ✓ | ✓ | ✓ | ||||||
| IgG (ELISA) | ✓ * | ✓ * | ✓ | ||||||
| Computed Tomography | ✓ * | ||||||||
| CDC MAC ELISA | ✓ | ||||||||
| PRNT | Y * | ||||||||
| Micro-neutralization | ✓ * | ||||||||
| BOB-ELISA | ✓ | ||||||||
| Peptide ELISA | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Patient Number | Clinical Diagnosis | 1 Week ** | 2 Weeks ** | 2 Months ** | 5 Months ** | Notes |
|---|---|---|---|---|---|---|
| 1 | ZIKV | ND | ND | ND | * | |
| 2 | ZIKV | * | Neg | Pos | * | |
| 3 | ZIKV | * | Pos | Pos | * | |
| 4 | ZIKV | Pos | Pos | ND | * | |
| 5 | ZIKV | * | * | * | ND | |
| 6 | ZIKV | Neg,ND,Pos + | Pos | Pos | * | Asymptomatic |
| 7 | ZIKV | ND | Pos | Neg | * | False positive |
| Sample ID | qRT-PCR (Plasma) | qRT-PCR (Urine) | qRT-PCR (Oral) | qRT-PCR (Blood) | CDC MAC-ELISA | Euroimmun (IgM) | PRNT | Peptide ELISA Results |
|---|---|---|---|---|---|---|---|---|
| 1 | Neg | 38.42 | Neg | 34.82 | Pos | Neg | 1:64 | Pos |
| 2 | 29.33 | Neg | Neg | 38.98 | Neg | Neg | <1:16 | Pos |
| 3 | 20.21 | 36 | 38.27 | 25.64 | Neg | Neg | <1:8 | Pos |
| 4 | Neg | 35.77 | Neg | Neg | inconclusive | Neg | 1:16 | Pos |
| 5 | Neg | 34.9 | Neg | 36.88 | Pos | Pos | ND | Pos |
| 6 | Neg | 29.01 | Neg | 36.31 | Pos | Neg | 1:2048 | Pos |
| 7 | Neg | 21.26 | Neg | 34.94 | Pos | Neg | 1:256 | Pos |
| Sample ID | ZIKV | HIV Status * |
|---|---|---|
| 1 | Neg | Not detected |
| 2 | Neg | 79 |
| 3 | ND | <LOD |
| 4 | Pos | Not detected |
| Sample ID | Age (Months) | IgM+ | IgG+ & CT+ | Peptide ELISA ZIKV |
|---|---|---|---|---|
| 1 | 32 | Pos | Neg | Pos |
| 2 | 34 | Neg | Pos | Pos |
| 3 | 32 | Neg | Pos | Pos |
| 4 | 21 | Neg | Pos | Pos |
| 5 | 34 | Neg | Pos | Pos |
| 6 | 32 | Neg | Pos | Pos |
| 7 | 34 | Neg | Pos | Pos |
| 8 | 30 | Pos | Neg | Pos |
| 9 | 29 | Neg | Pos | Pos |
| 10 | 32 | Pos | Neg | Pos |
| 11 | 32 | Neg | Pos | Pos |
| 12 | 35 | Neg | Pos | Pos |
| 13 | 30 | Pos | Neg | Pos |
| 14 | 33 | Neg | Pos | Pos |
| 15 | 30 | Neg | Pos | Pos |
| 16 | 32 | Neg | Pos | Pos |
| 17 | 35 | Neg | Pos | Pos |
| 18 | 32 | Neg | Pos | Pos |
| 19 | 32 | Pos | Neg | Pos |
| 20 | 33 | Neg | Pos | Pos |
| 21 | 32 | Neg | Pos | Pos |
| 22 | 16 | Neg | Pos | Pos |
| ZIKV (n) | |
|---|---|
| Sensitivity | 30.19% (175) |
| Specificity | 83.07% (175) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martinez Viedma, M.d.P.; Panossian, S.; Gifford, K.; García, K.; Figueroa, I.; Parham, L.; de Moraes, L.; Nunes Gomes, L.; García-Salum, T.; Perret, C.; et al. Evaluation of ELISA-Based Multiplex Peptides for the Detection of Human Serum Antibodies Induced by Zika Virus Infection across Various Countries. Viruses 2021, 13, 1319. https://doi.org/10.3390/v13071319
Martinez Viedma MdP, Panossian S, Gifford K, García K, Figueroa I, Parham L, de Moraes L, Nunes Gomes L, García-Salum T, Perret C, et al. Evaluation of ELISA-Based Multiplex Peptides for the Detection of Human Serum Antibodies Induced by Zika Virus Infection across Various Countries. Viruses. 2021; 13(7):1319. https://doi.org/10.3390/v13071319
Chicago/Turabian StyleMartinez Viedma, Maria del Pilar, Stephen Panossian, Kennedy Gifford, Kimberly García, Isis Figueroa, Leda Parham, Laise de Moraes, Lillian Nunes Gomes, Tamara García-Salum, Cecilia Perret, and et al. 2021. "Evaluation of ELISA-Based Multiplex Peptides for the Detection of Human Serum Antibodies Induced by Zika Virus Infection across Various Countries" Viruses 13, no. 7: 1319. https://doi.org/10.3390/v13071319
APA StyleMartinez Viedma, M. d. P., Panossian, S., Gifford, K., García, K., Figueroa, I., Parham, L., de Moraes, L., Nunes Gomes, L., García-Salum, T., Perret, C., Weiskopf, D., Tan, G. S., Augusto Silva, A., Boaventura, V., Ruiz-Palacios, G. M., Sette, A., De Silva, A. D., Medina, R. A., Lorenzana, I., ... Pickett, B. E. (2021). Evaluation of ELISA-Based Multiplex Peptides for the Detection of Human Serum Antibodies Induced by Zika Virus Infection across Various Countries. Viruses, 13(7), 1319. https://doi.org/10.3390/v13071319









