Concepts to Reveal Parvovirus–Nucleus Interactions
Abstract
:1. Introduction
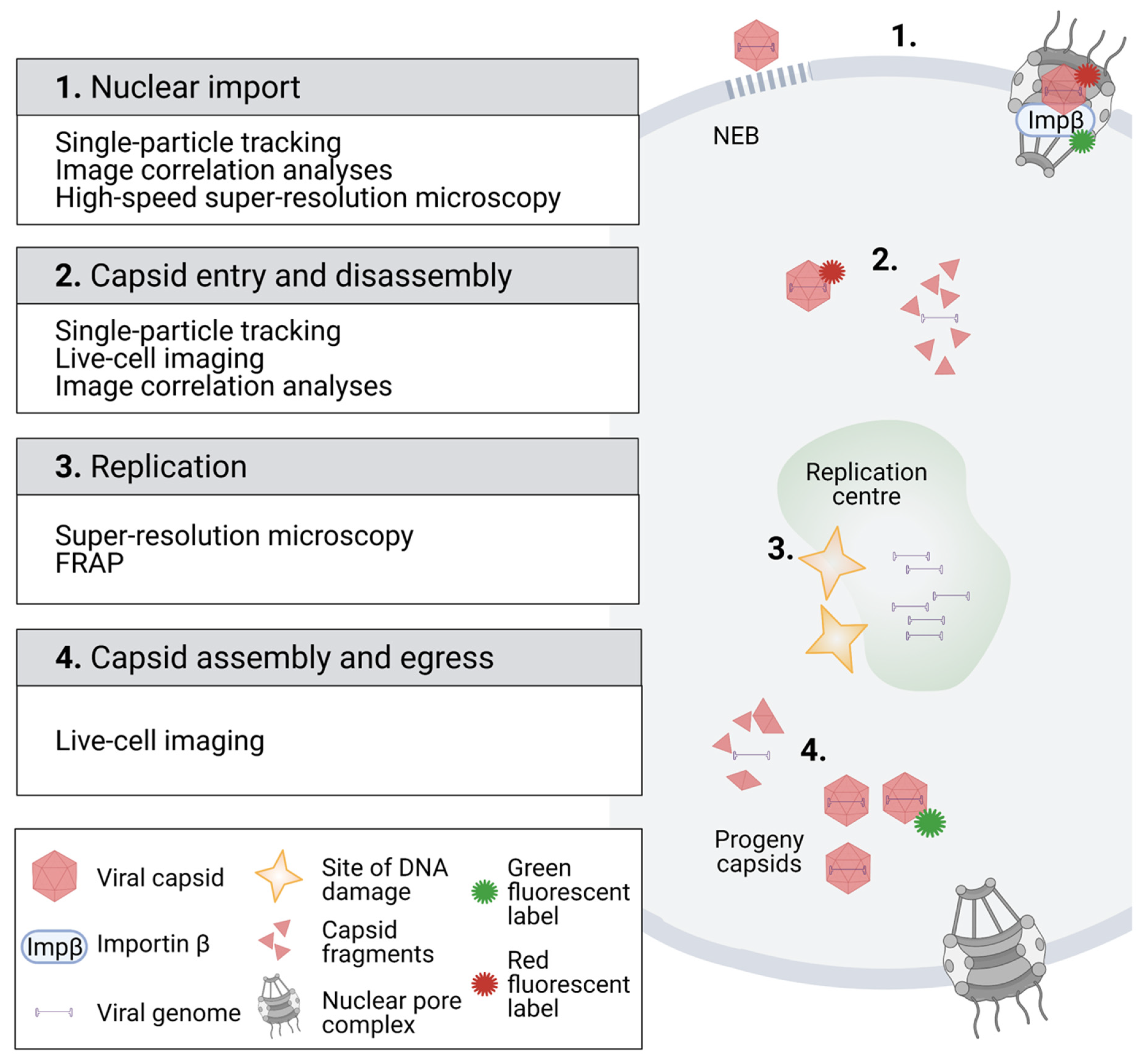
2. Imaging of Viral Interactions and Dynamics in the Cytoplasm and Nucleus
3. Screening and Validation of Protein–Protein Interactions
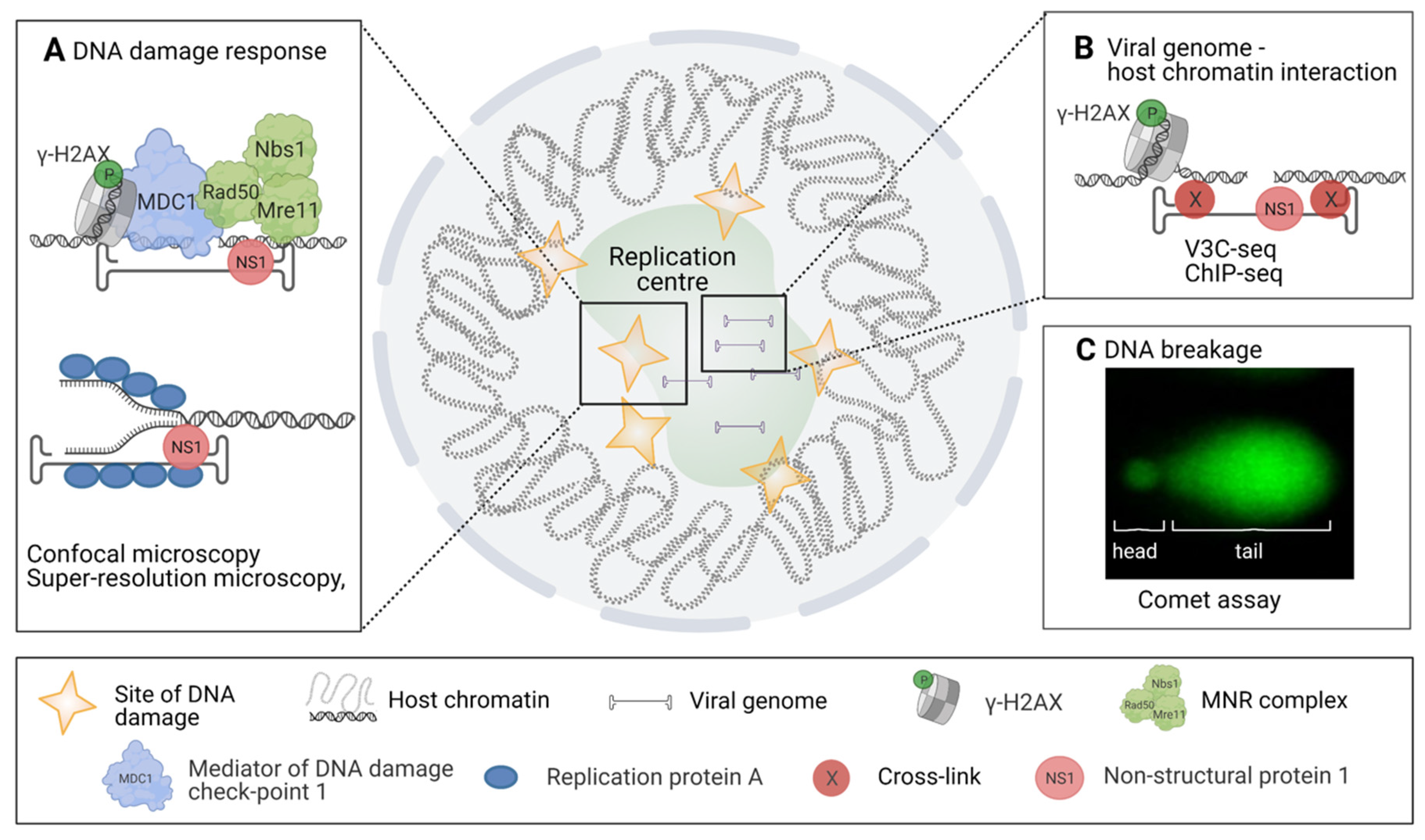
4. Detection of DNA Damage, DNA Repair, and Virus–DNA Interactions
5. Recent Methods for Future Studies of Parvovirus–Nucleus Interactions
6. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Heegaard, E.D.; Brown, K.E. Human Parvovirus B19. Clin. Microbiol. Rev. 2002, 15, 485–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nandi, S.; Kumar, M. Canine Parvovirus: Current Perspective. Indian J. Virol. 2010, 21, 31–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotterman, M.A.; Schaffer, D.V. Engineering Adeno-Associated Viruses for Clinical Gene Therapy. Nat. Rev. Genet. 2014, 15, 445–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Samulski, R.J. Engineering Adeno-Associated Virus Vectors for Gene Therapy. Nat. Rev. Genet. 2020, 21, 255–272. [Google Scholar] [CrossRef] [PubMed]
- Marchini, A.; Bonifati, S.; Scott, E.M.; Angelova, A.L.; Rommelaere, J. Oncolytic Parvoviruses: From Basic Virology to Clinical Applications. Virol. J. 2015, 12, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Zabaleta, N.; Dai, W.; Bhatt, U.; Chichester, J.A.; Estelien, R.; Sanmiguel, J.; Michalson, K.T.; Diop, C.; Maciorowski, D.; Qi, W.; et al. Immunogenicity of an AAV-Based, Room-Temperature Stable, Single Dose COVID-19 Vaccine in Mice and Non-Human Primates. bioRxiv 2021. [Google Scholar] [CrossRef]
- Tse, L.V.; Meganck, R.M.; Graham, R.L.; Baric, R.S. The Current and Future State of Vaccines, Antivirals and Gene Therapies Against Emerging Coronaviruses. Front. Microbiol. 2020, 11, 658. [Google Scholar] [CrossRef]
- Pénzes, J.J.; Söderlund-Venermo, M.; Canuti, M.; Eis-Hübinger, A.M.; Hughes, J.; Cotmore, S.F.; Harrach, B. Reorganizing the Family Parvoviridae: A Revised Taxonomy Independent of the Canonical Approach Based on Host Association. Arch. Virol. 2020, 165, 2133–2146. [Google Scholar] [CrossRef]
- Cotmore, S.F.; Tattersall, P. Dna Replication in the Autonomous Parvoviruses. Semin. Virol. 1995, 6, 271–281. [Google Scholar] [CrossRef]
- Li, X.; Rhode, S.L. Mutation of Lysine 405 to Serine in the Parvovirus H-1 NS1 Abolishes Its Functions for Viral DNA Replication, Late Promoter Trans Activation, and Cytotoxicity. J. Virol. 1990, 64, 4654–4660. [Google Scholar] [CrossRef] [Green Version]
- Christensen, J.; Cotmore, S.F.; Tattersall, P. Minute Virus of Mice Transcriptional Activator Protein NS1 Binds Directly to the Transactivation Region of the Viral P38 Promoter in a Strictly ATP-Dependent Manner. J. Virol. 1995, 69, 5422–5430. [Google Scholar] [CrossRef] [Green Version]
- Naeger, L.K.; Cater, J.; Pintel, D.J. The Small Nonstructural Protein (NS2) of the Parvovirus Minute Virus of Mice Is Required for Efficient DNA Replication and Infectious Virus Production in a Cell-Type-Specific Manner. J. Virol. 1990, 64, 6166–6175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tullis, G.E.; Labieniec-Pintel, L.; Clemens, K.E.; Pintel, D. Generation and Characterization of a Temperature-Sensitive Mutation in the NS-1 Gene of the Autonomous Parvovirus Minute Virus of Mice. J. Virol. 1988, 62, 2736–2744. [Google Scholar] [CrossRef] [Green Version]
- Cotmore, S.F.; Gottlieb, R.L.; Tattersall, P. Replication Initiator Protein NS1 of the Parvovirus Minute Virus of Mice Binds to Modular Divergent Sites Distributed throughout Duplex Viral DNA. J. Virol. 2007, 81, 13015–13027. [Google Scholar] [CrossRef] [Green Version]
- McCarty, D.M.; Pereira, D.J.; Zolotukhin, I.; Zhou, X.; Ryan, J.H.; Muzyczka, N. Identification of Linear DNA Sequences That Specifically Bind the Adeno-Associated Virus Rep Protein. J. Virol. 1994, 68, 4988–4997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niskanen, E.A.; Ihalainen, T.O.; Kalliolinna, O.; Häkkinen, M.M.; Vihinen-Ranta, M. Effect of ATP Binding and Hydrolysis on Dynamics of Canine Parvovirus NS1. J. Virol. 2010, 84, 5391–5403. [Google Scholar] [CrossRef] [Green Version]
- Niskanen, E.A.; Kalliolinna, O.; Ihalainen, T.O.; Hakkinen, M.; Vihinen-Ranta, M. Mutations in DNA Binding and Transactivation Domains Affect the Dynamics of Parvovirus NS1 Protein. J. Virol. 2013, 87, 11762–11774. [Google Scholar] [CrossRef] [Green Version]
- Brownstein, D.G.; Smith, A.L.; Johnson, E.A.; Pintel, D.J.; Naeger, L.K.; Tattersall, P. The Pathogenesis of Infection with Minute Virus of Mice Depends on Expression of the Small Nonstructural Protein NS2 and on the Genotype of the Allotropic Determinants VP1 and VP2. J. Virol. 1992, 66, 3118–3124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz, Z.; Mihaylov, I.S.; Cotmore, S.F.; Tattersall, P. Recruitment of DNA Replication and Damage Response Proteins to Viral Replication Centers during Infection with NS2 Mutants of Minute Virus of Mice (MVM). Virology 2011, 410, 375–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naeger, L.K.; Salomé, N.; Pintel, D.J. NS2 Is Required for Efficient Translation of Viral MRNA in Minute Virus of Mice-Infected Murine Cells. J. Virol. 1993, 67, 1034–1043. [Google Scholar] [CrossRef] [Green Version]
- Cotmore, S.F.; D’Abramo, A.M.; Carbonell, L.F.; Bratton, J.; Tattersall, P. The NS2 Polypeptide of Parvovirus MVM Is Required for Capsid Assembly in Murine Cells. Virology 1997, 231, 267–280. [Google Scholar] [CrossRef] [Green Version]
- Engelsma, D.; Valle, N.; Fish, A.; Salomé, N.; Almendral, J.M.; Fornerod, M. A Supraphysiological Nuclear Export Signal Is Required for Parvovirus Nuclear Export. Mol. Biol. Cell 2008, 19, 2544–2552. [Google Scholar] [CrossRef] [Green Version]
- Eichwald, V.; Daeffler, L.; Klein, M.; Rommelaere, J.; Salomé, N. The NS2 Proteins of Parvovirus Minute Virus of Mice Are Required for Efficient Nuclear Egress of Progeny Virions in Mouse Cells. J. Virol. 2002, 76, 10307–10319. [Google Scholar] [CrossRef] [Green Version]
- Miller, C.L.; Pintel, D.J. Interaction between Parvovirus NS2 Protein and Nuclear Export Factor Crm1 Is Important for Viral Egress from the Nucleus of Murine Cells. J. Virol. 2002, 76, 3257–3266. [Google Scholar] [CrossRef] [Green Version]
- López-Bueno, A.; Valle, N.; Gallego, J.M.; Pérez, J.; Almendral, J.M. Enhanced Cytoplasmic Sequestration of the Nuclear Export Receptor CRM1 by NS2 Mutations Developed in the Host Regulates Parvovirus Fitness. J. Virol. 2004, 78, 10674–10684. [Google Scholar] [CrossRef] [Green Version]
- Hashemi, H.; Condurat, A.L.; Stroh-Dege, A.; Weiss, N.; Geiss, C.; Pilet, J.; Bartolomé, C.C.; Rommelaere, J.; Salomé, N.; Dinsart, C. Mutations in the Non-Structural Protein-Coding Sequence of Protoparvovirus h-1pv Enhance the Fitness of the Virus and Show Key Benefits Regarding the Transduction Efficiency of Derived Vectors. Viruses 2018, 10, 150. [Google Scholar] [CrossRef] [Green Version]
- Kelich, J.M.; Ma, J.; Dong, B.; Wang, Q.; Chin, M.; Magura, C.M.; Xiao, W.; Yang, W. Super-Resolution Imaging of Nuclear Import of Adeno-Associated Virus in Live Cells. Mol. Ther. Methods Clin. Dev. 2015, 2, 15047. [Google Scholar] [CrossRef]
- Junod, S.L.; Saredy, J.; Yang, W. Nuclear Import of Adeno-Associated Viruses Imaged by High-Speed Single-Molecule Microscopy. Viruses 2021, 13, 167. [Google Scholar] [CrossRef]
- Mäntylä, E.; Kann, M.; Vihinen-Ranta, M. Protoparvovirus Knocking at the Nuclear Door. Viruses 2017, 9, 286. [Google Scholar] [CrossRef] [Green Version]
- Mäntylä, E.; Chacko, J.V.; Aho, V.; Parrish, C.R.; Shahin, V.; Kann, M.; Digman, M.A.; Gratton, E.; Vihinen-Ranta, M. Viral Highway to Nucleus Exposed by Image Correlation Analyses. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Porwal, M.; Cohen, S.; Snoussi, K.; Popa-Wagner, R.; Anderson, F.; Dugot-Senant, N.; Wodrich, H.; Dinsart, C.; Kleinschmidt, J.A.; Panté, N.; et al. Parvoviruses Cause Nuclear Envelope Breakdown by Activating Key Enzymes of Mitosis. PLoS Pathog. 2013, 9, e1003671. [Google Scholar] [CrossRef] [Green Version]
- Cohen, S.; Panté, N. Pushing the Envelope: Microinjection of Minute Virus of Mice into Xenopus Oocytes Causes Damage to the Nuclear Envelope. J. Gen. Virol. 2005, 86, 3243–3252. [Google Scholar] [CrossRef]
- Cohen, S.; Behzad, A.R.; Carroll, J.B.; Panté, N. Parvoviral Nuclear Import: Bypassing the Host Nuclear-Transport Machinery. J. Gen. Virol. 2006, 87, 3209–3213. [Google Scholar] [CrossRef]
- Ros, C.; Bayat, N.; Wolfisberg, R.; Almendral, J.M. Protoparvovirus Cell Entry. Viruses 2017, 9, 313. [Google Scholar] [CrossRef]
- Lombardo, E.; Ramírez, J.C.; Garcia, J.; Almendral, J.M. Complementary Roles of Multiple Nuclear Targeting Signals in the Capsid Proteins of the Parvovirus Minute Virus of Mice during Assembly and Onset of Infection. J. Virol. 2002, 76, 7049–7059. [Google Scholar] [CrossRef] [Green Version]
- Vihinen-Ranta, M.; Kakkola, L.; Kalela, A.; Vilja, P.; Vuento, M. Characterization of a Nuclear Localization Signal of Canine Parvovirus Capsid Proteins. Eur. J. Biochem. 1997, 250, 389–394. [Google Scholar] [CrossRef]
- Vihinen-Ranta, M.; Wang, D.; Weichert, W.S.; Parrish, C.R. The VP1 N-Terminal Sequence of Canine Parvovirus Affects Nuclear Transport of Capsids and Efficient Cell Infection. J. Virol. 2002, 76, 1884–1891. [Google Scholar] [CrossRef] [Green Version]
- Popa-Wagner, R.; Sonntag, F.; Schmidt, K.; King, J.; Kleinschmidt, J.A. Nuclear Translocation of Adeno-Associated Virus Type 2 Capsid Proteins for Virion Assembly. J. Gen. Virol. 2012, 93, 1887–1898. [Google Scholar] [CrossRef]
- Liu, P.; Chen, S.; Wang, M.; Cheng, A. The Role of Nuclear Localization Signal in Parvovirus Life Cycle. Virol. J. 2017, 14, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Grieger, J.C.; Snowdy, S.; Samulski, R.J. Separate Basic Region Motifs within the Adeno-Associated Virus Capsid Proteins Are Essential for Infectivity and Assembly. J. Virol. 2006, 80, 5199–5210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sonntag, F.; Bleker, S.; Leuchs, B.; Fischer, R.; Kleinschmidt, J.A. Adeno-Associated Virus Type 2 Capsids with Externalized VP1/VP2 Trafficking Domains Are Generated Prior to Passage through the Cytoplasm and Are Maintained until Uncoating Occurs in the Nucleus. J. Virol. 2006, 80, 11040–11054. [Google Scholar] [CrossRef] [Green Version]
- Mäntylä, E.; Aho, V.; Kann, M.; Vihinen-Ranta, M. Cytoplasmic Parvovirus Capsids Recruit Importin Beta for Nuclear Delivery. J. Virol. 2019, 94. [Google Scholar] [CrossRef]
- Fay, N.; Panté, N. Old Foes, New Understandings: Nuclear Entry of Small Non-Enveloped DNA Viruses. Curr. Opin. Virol. 2015, 12, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Tattersall, P.; Ward, D.C. Rolling Hairpin Model for Replication of Parvovirus and Linear Chromosomal DNA. Nature 1976, 263, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Adeyemi, R.O.; Landry, S.; Davis, M.E.; Weitzman, M.D.; Pintel, D.J. Parvovirus Minute Virus of Mice Induces a DNA Damage Response That Facilitates Viral Replication. PLoS Pathog. 2010, 6, 1001141. [Google Scholar] [CrossRef] [PubMed]
- Bashir, T.; Rommelaere, J.; Cziepluch, C. In Vivo Accumulation of Cyclin A and Cellular Replication Factors in Autonomous Parvovirus Minute Virus of Mice-Associated Replication Bodies. J. Virol. 2001, 75, 4394–4398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ihalainen, T.O.; Niskanen, E.A.; Jylhävä, J.; Paloheimo, O.; Dross, N.; Smolander, H.; Langowski, J.; Timonen, J.; Vihinen-Ranta, M. Parvovirus Induced Alterations in Nuclear Architecture and Dynamics. PLoS ONE 2009, 4. [Google Scholar] [CrossRef]
- Adeyemi, R.O.; Pintel, D.J. Replication of Minute Virus of Mice in Murine Cells Is Facilitated by Virally Induced Depletion of P21. J. Virol. 2012, 86, 8328–8332. [Google Scholar] [CrossRef] [Green Version]
- Adeyemi, R.O.; Pintel, D.J. Parvovirus-Induced Depletion of Cyclin B1 Prevents Mitotic Entry of Infected Cells. PLoS Pathog. 2014, 10, 1003891. [Google Scholar] [CrossRef] [Green Version]
- Lou, S.; Luo, Y.; Cheng, F.; Huang, Q.; Shen, W.; Kleiboeker, S.; Tisdale, J.F.; Liu, Z.; Qiu, J. Human Parvovirus B19 DNA Replication Induces a DNA Damage Response That Is Dispensable for Cell Cycle Arrest at Phase G2/M. J. Virol. 2012, 86, 10748–10758. [Google Scholar] [CrossRef] [Green Version]
- Majumder, K.; Etingov, I.; Pintel, D.J. Protoparvovirus Interactions with the Cellular DNA Damage Response. Viruses 2017, 9, 323. [Google Scholar] [CrossRef] [Green Version]
- Majumder, K.; Wang, J.; Boftsi, M.; Fuller, M.S.; Rede, J.E.; Joshi, T.; Pintel, D.J. Parvovirus Minute Virus of Mice Interacts with Sites of Cellular DNA Damage to Establish and Amplify Its Lytic Infection. eLife 2018, 7. [Google Scholar] [CrossRef]
- Cotmore, S.F.; Tattersall, P. Parvoviruses: Small Does Not Mean Simple. Annu. Rev. Virol. 2014, 1, 517–537. [Google Scholar] [CrossRef] [Green Version]
- Op De Beeck, A.; Caillet-Fauquet, P. The NS1 Protein of the Autonomous Parvovirus Minute Virus of Mice Blocks Cellular DNA Replication: A Consequence of Lesions to the Chromatin? J. Virol. 1997, 71, 5323–5329. [Google Scholar] [CrossRef] [Green Version]
- Moffatt, S.; Yaegashi, N.; Tada, K.; Tanaka, N.; Sugamura, K. Human Parvovirus B19 Nonstructural (NS1) Protein Induces Apoptosis in Erythroid Lineage Cells. J. Virol. 1998, 72, 3018–3028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, A.Y.; Qiu, J. Parvovirus Infection-Induced Cell Death and Cell Cycle Arrest. Future Virol. 2010, 5, 731–743. [Google Scholar] [CrossRef] [Green Version]
- Cotmore, S.F.; Tattersall, P. Parvovirus Diversity and DNA Damage Responses. Cold Spring Harb. Perspect. Biol. 2013, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lombardo, E.; Ramírez, J.C.; Agbandje-McKenna, M.; Almendral, J.M. A Beta-Stranded Motif Drives Capsid Protein Oligomers of the Parvovirus Minute Virus of Mice into the Nucleus for Viral Assembly. J. Virol. 2000, 74, 3804–3814. [Google Scholar] [CrossRef] [Green Version]
- Riolobos, L.; Reguera, J.; Mateu, M.G.; Almendral, J.M. Nuclear Transport of Trimeric Assembly Intermediates Exerts a Morphogenetic Control on the Icosahedral Parvovirus Capsid. J. Mol. Biol. 2006, 357, 1026–1038. [Google Scholar] [CrossRef]
- Gil-Ranedo, J.; Hernando, E.; Riolobos, L.; Domínguez, C.; Kann, M.; Almendral, J.M. The Mammalian Cell Cycle Regulates Parvovirus Nuclear Capsid Assembly. PLoS Pathog. 2015, 11, e1004920. [Google Scholar] [CrossRef] [PubMed]
- Russell, D.W.; Miller, A.D.; Alexander, I.E. Adeno-Associated Virus Vectors Preferentially Transduce Cells in S Phase. Proc. Natl. Acad. Sci. USA 1994, 91, 8915–8919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartlett, J.S.; Wilcher, R.; Samulski, R.J. Infectious Entry Pathway of Adeno-Associated Virus and Adeno-Associated Virus Vectors. J. Virol. 2000, 74, 2777–2785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vihinen-Ranta, M.; Yuan, W.; Parrish, C.R. Cytoplasmic Trafficking of the Canine Parvovirus Capsid and Its Role in Infection and Nuclear Transport. J. Virol. 2000, 74, 4853–4859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seisenberger, G.; Ried, M.U.; Endreß, T.; Büning, H.; Hallek, M.; Bräuchle, C. Real-Time Single-Molecule Imaging of the Infection Pathway of Anadeno-Associated Virus. Science 2001, 294, 1929–1932. [Google Scholar] [CrossRef]
- Lee, D.W.; Allison, A.B.; Bacon, K.B.; Parrish, C.R.; Daniel, S. Single-Particle Tracking Shows That a Point Mutation in the Carnivore Parvovirus Capsid Switches Binding between Host-Specific Transferrin Receptors. J. Virol. 2016, 90, 4849–4853. [Google Scholar] [CrossRef] [Green Version]
- Cureton, D.K.; Harbison, C.E.; Cocucci, E.; Parrish, C.R.; Kirchhausen, T. Limited Transferrin Receptor Clustering Allows Rapid Diffusion of Canine Parvovirus into Clathrin Endocytic Structures. J. Virol. 2012, 86, 5330–5340. [Google Scholar] [CrossRef] [Green Version]
- Xiao, P.-J.; Samulski, R.J. Cytoplasmic Trafficking, Endosomal Escape, and Perinuclear Accumulation of Adeno-Associated Virus Type 2 Particles Are Facilitated by Microtubule Network. J. Virol. 2012, 86, 10462–10473. [Google Scholar] [CrossRef] [Green Version]
- Saxton, M.J. A Biological Interpretation of Transient Anomalous Subdiffusion. I. Qualitative Model. Biophys. J. 2007, 92, 1178–1191. [Google Scholar] [CrossRef] [Green Version]
- Warrington, K.H.; Gorbatyuk, O.S.; Harrison, J.K.; Opie, S.R.; Zolotukhin, S.; Muzyczka, N. Adeno-Associated Virus Type 2 VP2 Capsid Protein Is Nonessential and Can Tolerate Large Peptide Insertions at Its N Terminus. J. Virol. 2004, 78, 6595–6609. [Google Scholar] [CrossRef] [Green Version]
- Lux, K.; Goerlitz, N.; Schlemminger, S.; Perabo, L.; Goldnau, D.; Endell, J.; Leike, K.; Kofler, D.M.; Finke, S.; Hallek, M.; et al. Green Fluorescent Protein-Tagged Adeno-Associated Virus Particles Allow the Study of Cytosolic and Nuclear Trafficking. J. Virol. 2005, 79, 11776–11787. [Google Scholar] [CrossRef] [Green Version]
- Chandran, J.S.; Sharp, P.S.; Karyka, E.; Aves-Cruzeiro, J.M.D.C.; Coldicott, I.; Castelli, L.; Hautbergue, G.; Collins, M.O.; Azzouz, M. Site Specific Modification of Adeno-Associated Virus Enables Both Fluorescent Imaging of Viral Particles and Characterization of the Capsid Interactome. Sci. Rep. 2017, 7, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Michelfelder, S.; Varadi, K.; Raupp, C.; Hunger, A.; Körbelin, J.; Pahrmann, C.; Schrepfer, S.; Müller, O.J.; Kleinschmidt, J.A.; Trepel, M. Peptide Ligands Incorporated into the Threefold Spike Capsid Domain to Re-Direct Gene Transduction of AAV8 and AAV9 in Vivo. PLoS ONE 2011, 6, e23101. [Google Scholar] [CrossRef] [Green Version]
- Digman, M.A.; Gratton, E. Lessons in Fluctuation Correlation Spectroscopy. Annu. Rev. Phys. Chem. 2011, 62, 645–668. [Google Scholar] [CrossRef] [Green Version]
- Hinde, E.; Cardarelli, F.; Digman, M.A.; Gratton, E. In Vivo Pair Correlation Analysis of EGFP Intranuclear Diffusion Reveals DNA-Dependent Molecular Flow. Proc. Natl. Acad. Sci. USA 2010, 107, 16560–16565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Digman, M.A.; Gratton, E. Imaging Barriers to Diffusion by Pair Correlation Functions. Biophys. J. 2009, 97, 665–673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, L.; Wang, X.; Xing, D.; Chen, T.; Chen, W.R. Noninvasive Determination of Cell Nucleoplasmic Viscosity by Fluorescence Correlation Spectroscopy. J. Biomed. Opt. 2009, 14, 024013. [Google Scholar] [CrossRef]
- Seksek, O.; Biwersi, J.; Verkman, A.S. Translational Diffusion of Macromolecule-Sized Solutes in Cytoplasm and Nucleus. J. Cell Biol. 1997, 138, 131–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ros, C.; Baltzer, C.; Mani, B.; Kempf, C. Parvovirus Uncoating in Vitro Reveals a Mechanism of DNA Release without Capsid Disassembly and Striking Differences in Encapsidated DNA Stability. Virology 2006, 345, 137–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mani, B.; Baltzer, C.; Valle, N.; Almendral, J.M.; Kempf, C.; Ros, C. Low PH-Dependent Endosomal Processing of the Incoming Parvovirus Minute Virus of Mice Virion Leads to Externalization of the VP1 N-Terminal Sequence (N-VP1), N-VP2 Cleavage, and Uncoating of the Full-Length Genome. J. Virol. 2006, 80, 1015–1024. [Google Scholar] [CrossRef] [Green Version]
- Suikkanen, S.; Antila, M.; Jaatinen, A.; Vihinen-Ranta, M.; Vuento, M. Release of Canine Parvovirus from Endocytic Vesicles. Virology 2003, 316, 267–280. [Google Scholar] [CrossRef] [Green Version]
- Caliaro, O.; Marti, A.; Ruprecht, N.; Leisi, R.; Subramanian, S.; Hafenstein, S.; Ros, C. Parvovirus B19 Uncoating Occurs in the Cytoplasm without Capsid Disassembly and It Is Facilitated by Depletion of Capsid-Associated Divalent Cations. Viruses 2019, 11, 430. [Google Scholar] [CrossRef] [Green Version]
- Cotmore, S.F.; D’Abramo, A.M.; Ticknor, C.M.; Tattersall, P. Controlled Conformational Transitions in the MVM Virion Expose the VP1 N-Terminus and Viral Genome without Particle Disassembly. Virology 1999, 254, 169–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cotmore, S.F.; Hafenstein, S.; Tattersall, P. Depletion of Virion-Associated Divalent Cations Induces Parvovirus Minute Virus of Mice to Eject Its Genome in a 3′-to-5′ Direction from an Otherwise Intact Viral Particle. J. Virol. 2010, 84, 1945–1956. [Google Scholar] [CrossRef] [Green Version]
- Cotmore, S.F.; Tattersall, P. Mutations at the Base of the Icosahedral Five-Fold Cylinders of Minute Virus of Mice Induce 3’-to-5’ Genome Uncoating and Critically Impair Entry Functions. J. Virol. 2012, 86, 69–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ros, C.; Kempf, C. The Ubiquitin-Proteasome Machinery Is Essential for Nuclear Translocation of Incoming Minute Virus of Mice. Virology 2004, 324, 350–360. [Google Scholar] [CrossRef] [Green Version]
- Bernaud, J.; Rossi, A.; Fis, A.; Gardette, L.; Aillot, L.; Büning, H.; Castelnovo, M.; Salvetti, A.; Faivre-Moskalenko, C. Characterization of AAV Vector Particle Stability at the Single-Capsid Level. J. Biol. Phys. 2018, 44, 181–194. [Google Scholar] [CrossRef]
- Cziepluch, C.; Lampel, S.; Grewenig, A.; Grund, C.; Lichter, P.; Rommelaere, J. H-1 Parvovirus-Associated Replication Bodies: A Distinct Virus-Induced Nuclear Structure. J. Virol. 2000, 74, 4807–4815. [Google Scholar] [CrossRef] [Green Version]
- Mäntylä, E.; Salokas, K.; Oittinen, M.; Aho, V.; Mäntysaari, P.; Palmujoki, L.; Kalliolinna, O.; Ihalainen, T.O.; Niskanen, E.A.; Timonen, J.; et al. Promoter-Targeted Histone Acetylation of Chromatinized Parvoviral Genome Is Essential for the Progress of Infection. J. Virol. 2016, 90, 4059–4066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ihalainen, T.O.; Willman, S.F.; Niskanen, E.A.; Paloheimo, O.; Smolander, H.; Laurila, J.P.; Kaikkonen, M.U.; Vihinen-Ranta, M. Distribution and Dynamics of Transcription-Associated Proteins during Parvovirus Infection. J. Virol. 2012, 86, 13779–13784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ihalainen, T.O.; Niskanen, E.A.; Jylhävä, J.; Turpeinen, T.; Rinne, J.; Timonen, J.; Vihinen-Ranta, M. Dynamics and Interactions of Parvoviral NS1 Protein in the Nucleus. Cell. Microbiol. 2007, 9, 1946–1959. [Google Scholar] [CrossRef] [PubMed]
- Riolobos, L.; Valle, N.; Hernando, E.; Maroto, B.; Kann, M.; Almendral, J.M. Viral Oncolysis That Targets Raf-1 Signaling Control of Nuclear Transport. J. Virol. 2010, 84, 2090–2099. [Google Scholar] [CrossRef] [Green Version]
- Wistuba, A.; Kern, A.; Weger, S.; Grimm, D.; Kleinschmidt, J.A. Subcellular Compartmentalization of Adeno-Associated Virus Type 2 Assembly. J. Virol. 1997, 71, 1341–1352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naumer, M.; Sonntag, F.; Schmidt, K.; Nieto, K.; Panke, C.; Davey, N.E.; Popa-Wagner, R.; Kleinschmidt, J.A. Properties of the Adeno-Associated Virus Assembly-Activating Protein. J. Virol. 2012, 86, 13038–13048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plevka, P.; Hafenstein, S.; Li, L.; D’Abrgamo, A.; Cotmore, S.F.; Rossmann, M.G.; Tattersall, P. Structure of a Packaging-Defective Mutant of Minute Virus of Mice Indicates That the Genome Is Packaged via a Pore at a 5-Fold Axis. J. Virol. 2011, 85, 4822–4827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramanian, S.; Organtini, L.J.; Grossman, A.; Domeier, P.P.; Cifuente, J.O.; Makhov, A.M.; Conway, J.F.; D’Abramo, A.; Cotmore, S.F.; Tattersall, P.; et al. Cryo-EM Maps Reveal Five-Fold Channel Structures and Their Modification by Gatekeeper Mutations in the Parvovirus Minute Virus of Mice (MVM) Capsid. Virology 2017, 510, 216–223. [Google Scholar] [CrossRef]
- Maroto, B.; Valle, N.; Saffrich, R.; Almendral, J.M. Nuclear Export of the Nonenveloped Parvovirus Virion Is Directed by an Unordered Protein Signal Exposed on the Capsid Surface. J. Virol. 2004, 78, 10685–10694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Köhler, A.; Hurt, E. Exporting RNA from the Nucleus to the Cytoplasm. Nat. Rev. Mol. Cell Biol. 2007, 8, 761–773. [Google Scholar] [CrossRef] [PubMed]
- Panté, N.; Kann, M. Nuclear Pore Complex Is Able to Transport Macromolecules with Diameters of 39 Nm. Mol. Biol. Cell 2002, 13, 425–434. [Google Scholar] [CrossRef] [Green Version]
- Nicolson, S.C.; Samulski, R.J. Recombinant Adeno-Associated Virus Utilizes Host Cell Nuclear Import Machinery to Enter the Nucleus. J. Virol. 2014, 88, 4132–4144. [Google Scholar] [CrossRef] [Green Version]
- Söderberg, O.; Leuchowius, K.J.; Gullberg, M.; Jarvius, M.; Weibrecht, I.; Larsson, L.G.; Landegren, U. Characterizing Proteins and Their Interactions in Cells and Tissues Using the in Situ Proximity Ligation Assay. Methods 2008, 45, 227–232. [Google Scholar] [CrossRef]
- Gerold, G.; Bruening, J.; Pietschmann, T. Decoding Protein Networks during Virus Entry by Quantitative Proteomics. Virus Res. 2016, 218, 25–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richards, A.L.; Eckhardt, M.; Krogan, N.J. Mass Spectrometry-based Protein–Protein Interaction Networks for the Study of Human Diseases. Mol. Syst. Biol. 2021, 17, e8792. [Google Scholar] [CrossRef] [PubMed]
- Pankow, S.; Bamberger, C.; Calzolari, D.; Bamberger, A.; Yates, J.R. Deep Interactome Profiling of Membrane Proteins by Co-Interacting Protein Identification Technology. Nat. Protoc. 2016, 11, 2515–2528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kovács, I.A.; Luck, K.; Spirohn, K.; Wang, Y.; Pollis, C.; Schlabach, S.; Bian, W.; Kim, D.K.; Kishore, N.; Hao, T.; et al. Network-Based Prediction of Protein Interactions. Nat. Commun. 2019, 10, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Silverbush, D.; Sharan, R. A Systematic Approach to Orient the Human Protein–Protein Interaction Network. Nat. Commun. 2019, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zheng, J.; Sun, W.; Huo, Y.; Zhang, L.; Hao, P.; Wang, H.; Zhuang, M. A Proximity-Tagging System to Identify Membrane Protein–Protein Interactions. Nat. Methods 2018, 15, 715–722. [Google Scholar] [CrossRef] [PubMed]
- Roux, K.J.; Kim, D.I.; Raida, M.; Burke, B. A Promiscuous Biotin Ligase Fusion Protein Identifies Proximal and Interacting Proteins in Mammalian Cells. J. Cell Biol. 2012, 196, 801–810. [Google Scholar] [CrossRef] [Green Version]
- Sears, R.M.; May, D.G.; Roux, K.J. BioID as a tool for protein-proximity labeling in living cells. Methods Mol. Biol. 2019. [Google Scholar] [CrossRef]
- Liu, X.; Salokas, K.; Tamene, F.; Jiu, Y.; Weldatsadik, R.G.; Öhman, T.; Varjosalo, M. An AP-MS- and BioID-Compatible MAC-Tag Enables Comprehensive Mapping of Protein Interactions and Subcellular Localizations. Nat. Commun. 2018, 9, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Xu, P.; Cheng, F.; Li, Y.; Wang, Z.; Hao, S.; Wang, J.; Ning, K.; Ganaie, S.S.; Engelhardt, J.F.; et al. Cellular Cleavage and Polyadenylation Specificity Factor 6 (CPSF6) Mediates Nuclear Import of Human Bocavirus 1 NP1 Protein and Modulates Viral Capsid Protein Expression. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Smith-Moore, S.; Neil, S.J.D.; Fraefel, C.; Michael Linden, R.; Bollen, M.; Rowe, H.M.; Henckaerts, E. Adeno-Associated Virus Rep Proteins Antagonize Phosphatase PP1 to Counteract KAP1 Repression of the Latent Viral Genome. Proc. Natl. Acad. Sci. USA 2018, 115, E3529–E3538. [Google Scholar] [CrossRef] [Green Version]
- Mattola, S.; Salokas, K.; Aho, V.; Hakanen, S.; Salminen, S.; Mäntylä, E.; Niskanen, E.A.; Svirskaite, J.; Ihalainen, T.O.; Parrish, C.R.; et al. Parvovirus Nonstructural Protein 2 Interacts with Proteins of Cellular Machinery Regulating Chromatin Functions (Manuscript).
- Lilley, C.E.; Schwartz, R.A.; Weitzman, M.D. Using or Abusing: Viruses and the Cellular DNA Damage Response. Trends Microbiol. 2007, 15, 119–126. [Google Scholar] [CrossRef]
- Trigg, B.J.; Ferguson, B.J. Functions of DNA Damage Machinery in the Innate Immune Response to DNA virus Infection. Curr. Opin. Virol. 2015, 15, 56–62. [Google Scholar] [CrossRef]
- Kastan, M.B.; Bartek, J. Cell-Cycle Checkpoints and Cancer. Nature 2004, 432, 316–323. [Google Scholar] [CrossRef]
- Shiotani, B.; Zou, L. Single-Stranded DNA Orchestrates an ATM-to-ATR Switch at DNA Breaks. Mol. Cell 2009, 33, 547–558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, X.; Yan, Z.; Cheng, F.; Engelhardt, J.F.; Qiu, J. Replication of an Autonomous Human Parvovirus in Non-Dividing Human Airway Epithelium Is Facilitated through the DNA Damage and Repair Pathways. PLoS Pathog. 2016, 12. [Google Scholar] [CrossRef]
- Luo, Y.; Lou, S.; Deng, X.; Liu, Z.; Li, Y.; Kleiboeker, S.; Qiu, J. Parvovirus B19 Infection of Human Primary Erythroid Progenitor Cells Triggers ATR-Chk1 Signaling, Which Promotes B19 Virus Replication. J. Virol. 2011, 85, 8046–8055. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, R.A.; Carson, C.T.; Schuberth, C.; Weitzman, M.D. Adeno-Associated Virus Replication Induces a DNA Damage Response Coordinated by DNA-Dependent Protein Kinase. J. Virol. 2009, 83, 6269–6278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vassin, V.M.; Anantha, R.W.; Sokolova, E.; Kanner, S.; Borowiec, J.A. Human RPA Phosphorylation by ATR Stimulates DNA Synthesis and Prevents SsDNA Accumulation during DNA-Replication Stress. J. Cell Sci. 2009, 122, 4070–4080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majumder, K.; Boftsi, M.; Whittle, F.B.; Wang, J.; Fuller, M.S.; Joshi, T.; Pintel, D.J. The NS1 Protein of the Parvovirus MVM Aids in the Localization of the Viral Genome to Cellular Sites of DNA Damage. PLoS Pathog. 2020, 16, e1009002. [Google Scholar] [CrossRef] [PubMed]
- Dixon, J.R.; Gorkin, D.U.; Ren, B. Chromatin Domains: The Unit of Chromosome Organization. Mol. Cell 2016, 62, 668–680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ostling, O.; Johanson, K.J. Microelectrophoretic Study of Radiation-Induced DNA Damages in Individual Mammalian Cells. Biochem. Biophys. Res. Commun. 1984, 123, 291–298. [Google Scholar] [CrossRef]
- Olive, P.L.; Banáth, J.P. The Comet Assay: A Method to Measure DNA Damage in Individual Cells. Nat. Protoc. 2006, 1, 23–29. [Google Scholar] [CrossRef]
- Collins, A.R. The Comet Assay: A Heavenly Method! Mutagenesis 2015, 30, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Alekseev, O.; Donegan, W.E.; Donovan, K.R.; Limonnik, V.; Azizkhan-Clifford, J. HSV-1 Hijacks the Host Dna Damage Response in Corneal Epithelial Cells through ICP4-Mediated Activation of ATM. Invest. Ophthalmol. Vis. Sci. 2020, 61, 39. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Kleiboeker, S.; Deng, X.; Qiu, J. Human Parvovirus B19 Infection Causes Cell Cycle Arrest of Human Erythroid Progenitors at Late S Phase That Favors Viral DNA Replication. J. Virol. 2013, 87, 12766–12775. [Google Scholar] [CrossRef] [Green Version]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of Age: Ten Years of next-Generation Sequencing Technologies. Nat. Rev. Genet. 2016, 17, 333–351. [Google Scholar] [CrossRef]
- Metzker, M.L. Sequencing Technologies the next Generation. Nat. Rev. Genet. 2010, 11, 31–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marx, V. Method of the Year: Spatially Resolved Transcriptomics. Nat. Methods 2021, 18, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Stickels, R.R.; Murray, E.; Kumar, P.; Li, J.; Marshall, J.L.; Di Bella, D.J.; Arlotta, P.; Macosko, E.Z.; Chen, F. Highly Sensitive Spatial Transcriptomics at Near-Cellular Resolution with Slide-SeqV2. Nat. Biotechnol. 2021, 39, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Larsson, L.; Frisén, J.; Lundeberg, J. Spatially Resolved Transcriptomics Adds a New Dimension to Genomics. Nat. Methods 2021, 18, 15–18. [Google Scholar] [CrossRef]
- Zhuang, X. Spatially Resolved Single-Cell Genomics and Transcriptomics by Imaging. Nat. Methods 2021, 18, 18–22. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A Revolutionary Tool for Transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Buenrostro, J.D.; Giresi, P.G.; Zaba, L.C.; Chang, H.Y.; Greenleaf, W.J. Transposition of Native Chromatin for Fast and Sensitive Epigenomic Profiling of Open Chromatin, DNA-Binding Proteins and Nucleosome Position. Nat. Methods 2013, 10, 1213–1218. [Google Scholar] [CrossRef]
- Aughey, G.N.; Cheetham, S.W.; Southall, T.D. DamID as a Versatile Tool for Understanding Gene Regulation. Dev. Camb. 2019, 146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaya-Okur, H.S.; Wu, S.J.; Codomo, C.A.; Pledger, E.S.; Bryson, T.D.; Henikoff, J.G.; Ahmad, K.; Henikoff, S. CUT&Tag for Efficient Epigenomic Profiling of Small Samples and Single Cells. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Furey, T.S. ChIP-Seq and beyond: New and Improved Methodologies to Detect and Characterize Protein-DNA Interactions. Nat. Rev. Genet. 2012, 13, 840–852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Wit, E.; de Laat, W. A Decade of 3C Technologies: Insights into Nuclear Organization. Genes Dev. 2012, 26, 11–24. [Google Scholar] [CrossRef] [Green Version]
- Kong, X.; Wei, G.; Chen, N.; Zhao, S.; Shen, Y.; Zhang, J.; Li, Y.; Zeng, X.; Wu, X. Dynamic Chromatin Accessibility Profiling Reveals Changes in Host Genome Organization in Response to Baculovirus Infection. PLoS Pathog. 2020, 16, e1008633. [Google Scholar] [CrossRef]
- Lamontagne, R.J.; Soldan, S.S.; Su, C.; Wiedmer, A.; Won, K.J.; Lu, F.; Goldman, A.R.; Wickramasinghe, J.; Tang, H.Y.; Speicher, D.W.; et al. A Multi-Omics Approach to Epstein-Barr Virus Immortalization of B-Cells Reveals EBNA1 Chromatin Pioneering Activities Targeting Nucleotide Metabolism. PLoS Pathog. 2021, 17, e1009208. [Google Scholar] [CrossRef]
- Van Steensel, B.; Henikoff, S. Identification of in Vivo DNA Targets of Chromatin Proteins Using Tethered Dam Methyltransferase. Nat. Biotechnol. 2000, 18, 424–428. [Google Scholar] [CrossRef]
- Kind, J.; Pagie, L.; De Vries, S.S.; Nahidiazar, L.; Dey, S.S.; Bienko, M.; Zhan, Y.; Lajoie, B.; De Graaf, C.A.; Amendola, M.; et al. Genome-Wide Maps of Nuclear Lamina Interactions in Single Human Cells. Cell 2015, 163, 134–147. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Shen, Y.; Draper, W.; Buenrostro, J.D.; Litzenburger, U.; Cho, S.W.; Satpathy, A.T.; Carter, A.C.; Ghosh, R.P.; East-Seletsky, A.; et al. ATAC-See Reveals the Accessible Genome by Transposase-Mediated Imaging and Sequencing. Nat. Methods 2016, 13, 1013–1020. [Google Scholar] [CrossRef] [PubMed]
- Kind, J.; Pagie, L.; Ortabozkoyun, H.; Boyle, S.; De Vries, S.S.; Janssen, H.; Amendola, M.; Nolen, L.D.; Bickmore, W.A.; Van Steensel, B. Single-Cell Dynamics of Genome-Nuclear Lamina Interactions. Cell 2013, 153, 178–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takei, Y.; Yun, J.; Zheng, S.; Ollikainen, N.; Pierson, N.; White, J.; Shah, S.; Thomassie, J.; Suo, S.; Eng, C.H.L.; et al. Integrated Spatial Genomics Reveals Global Architecture of Single Nuclei. Nature 2021, 590, 344–350. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mattola, S.; Hakanen, S.; Salminen, S.; Aho, V.; Mäntylä, E.; Ihalainen, T.O.; Kann, M.; Vihinen-Ranta, M. Concepts to Reveal Parvovirus–Nucleus Interactions. Viruses 2021, 13, 1306. https://doi.org/10.3390/v13071306
Mattola S, Hakanen S, Salminen S, Aho V, Mäntylä E, Ihalainen TO, Kann M, Vihinen-Ranta M. Concepts to Reveal Parvovirus–Nucleus Interactions. Viruses. 2021; 13(7):1306. https://doi.org/10.3390/v13071306
Chicago/Turabian StyleMattola, Salla, Satu Hakanen, Sami Salminen, Vesa Aho, Elina Mäntylä, Teemu O. Ihalainen, Michael Kann, and Maija Vihinen-Ranta. 2021. "Concepts to Reveal Parvovirus–Nucleus Interactions" Viruses 13, no. 7: 1306. https://doi.org/10.3390/v13071306
APA StyleMattola, S., Hakanen, S., Salminen, S., Aho, V., Mäntylä, E., Ihalainen, T. O., Kann, M., & Vihinen-Ranta, M. (2021). Concepts to Reveal Parvovirus–Nucleus Interactions. Viruses, 13(7), 1306. https://doi.org/10.3390/v13071306






