Influenza Virus-Induced Novel miRNAs Regulate the STAT Pathway
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Viral Infections
2.3. Next-Generation Sequencing (NGS)
2.4. NGS Data Analysis
2.5. MicroRNA Analysis
2.6. Transfection Studies
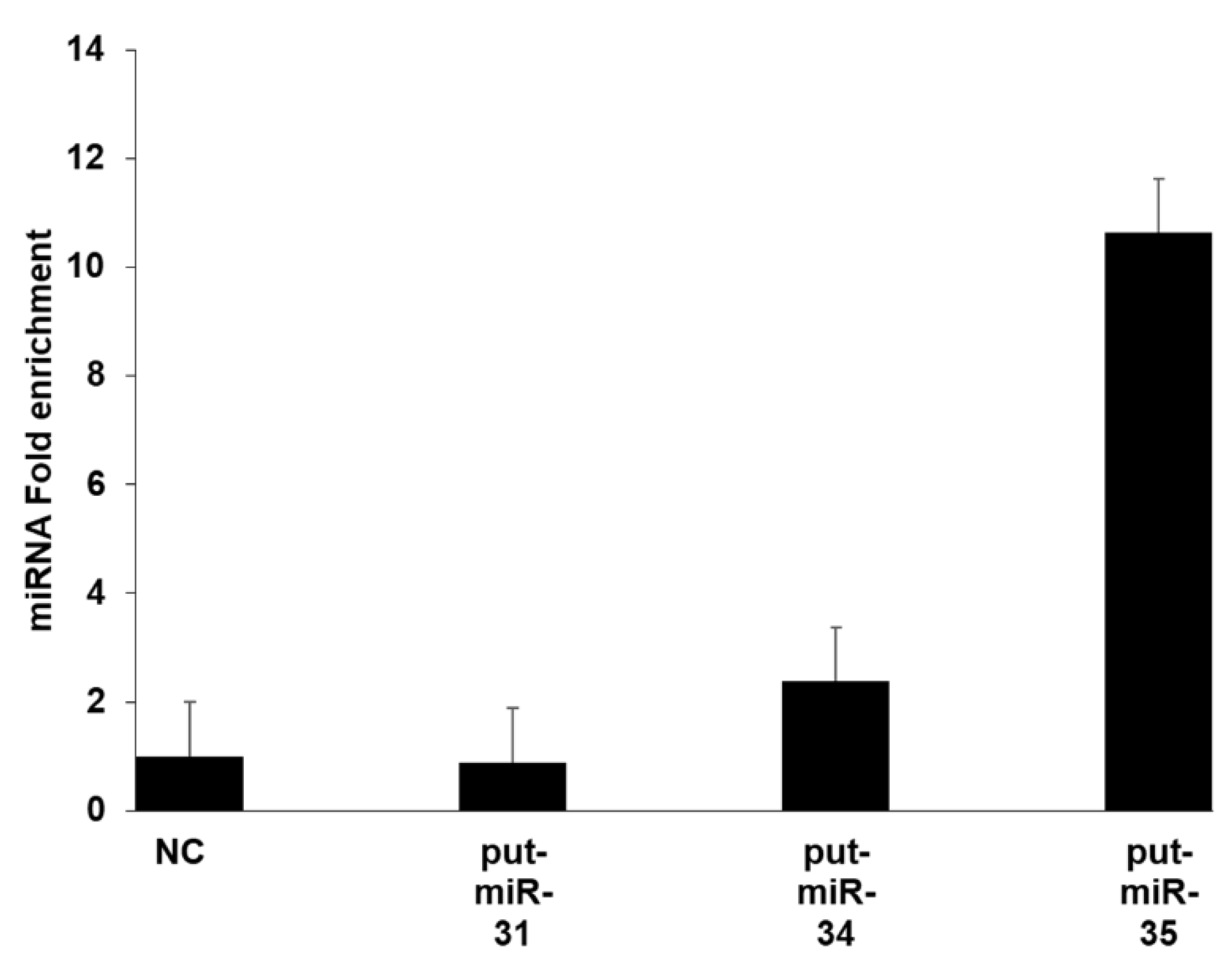
2.7. Argonaute (AGO) Immunoprecipitation
2.8. Western Immunoblotting
2.9. Statistical Analysis
3. Results
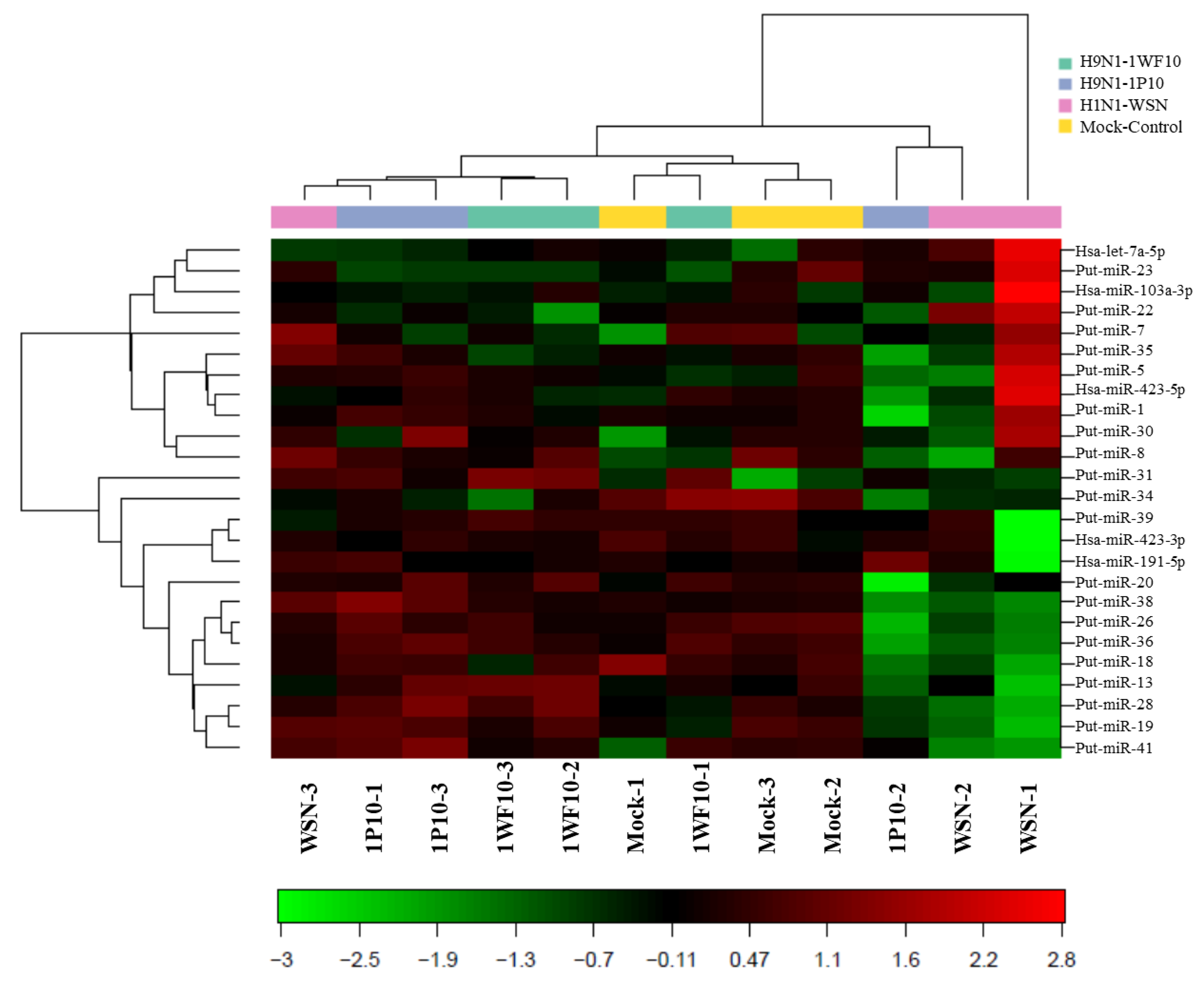
3.1. Influenza Virus Infection Induce Novel Putative miRNAs
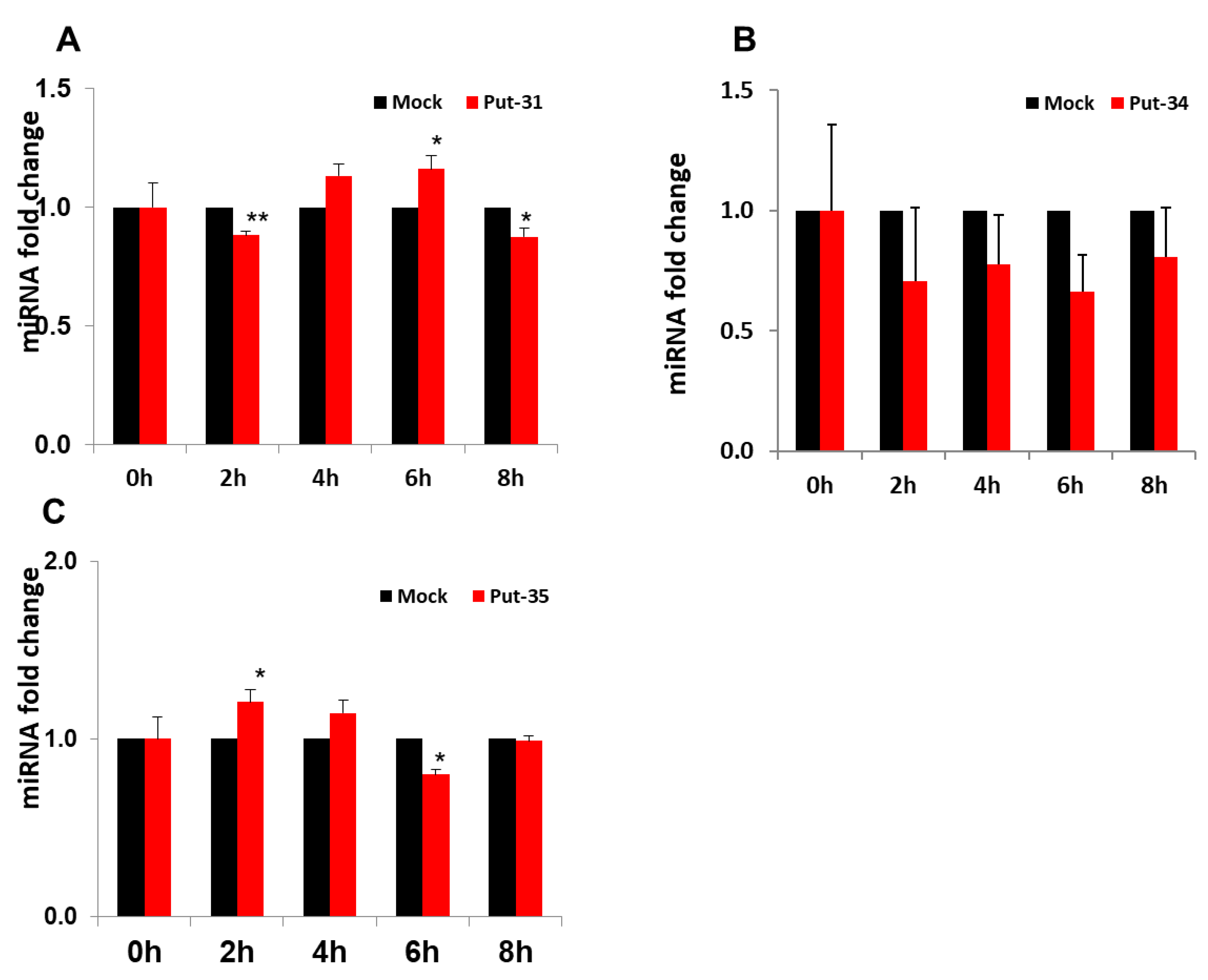
3.2. Confirmatory Studies of Selected Putative miRNAs
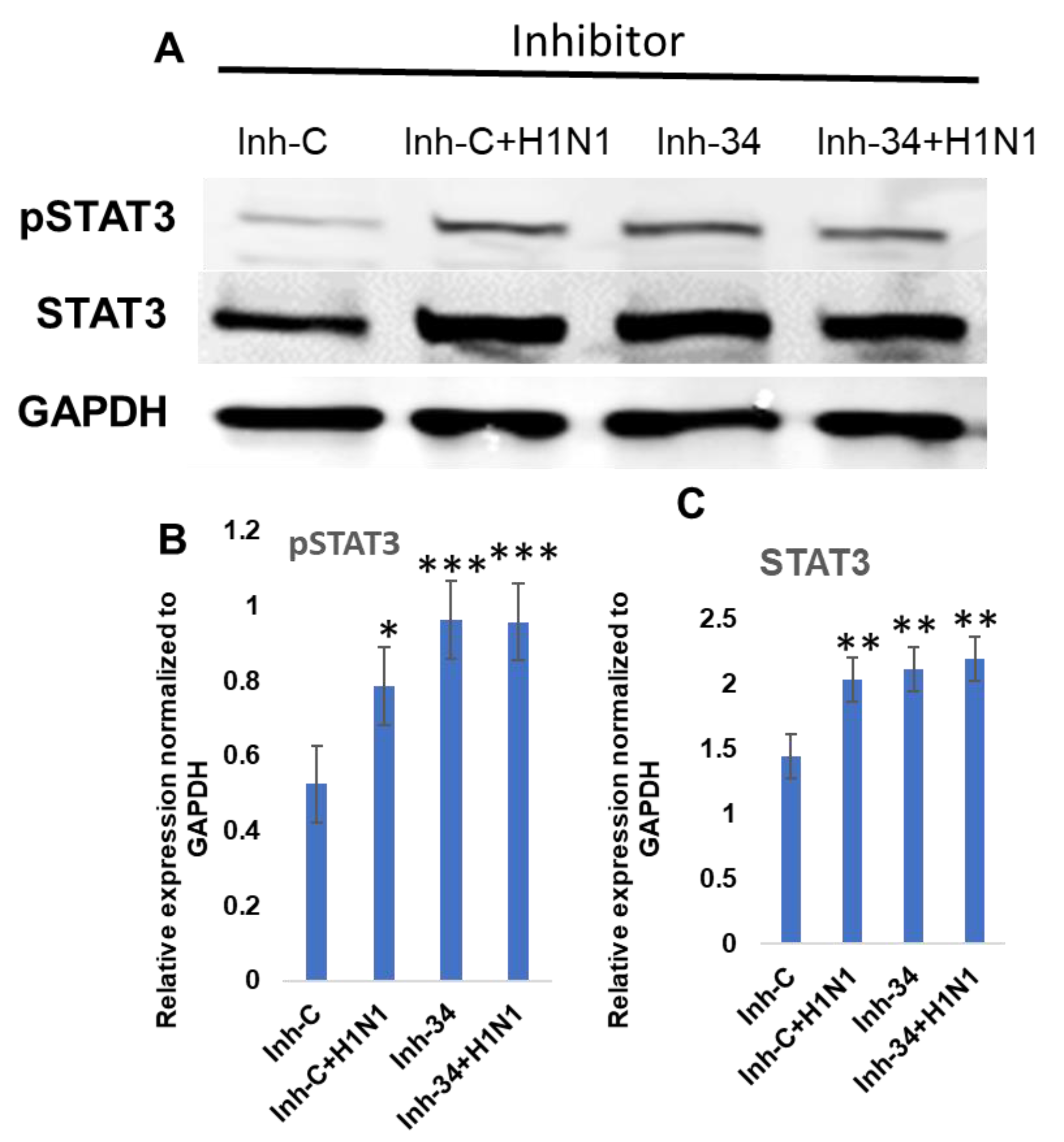
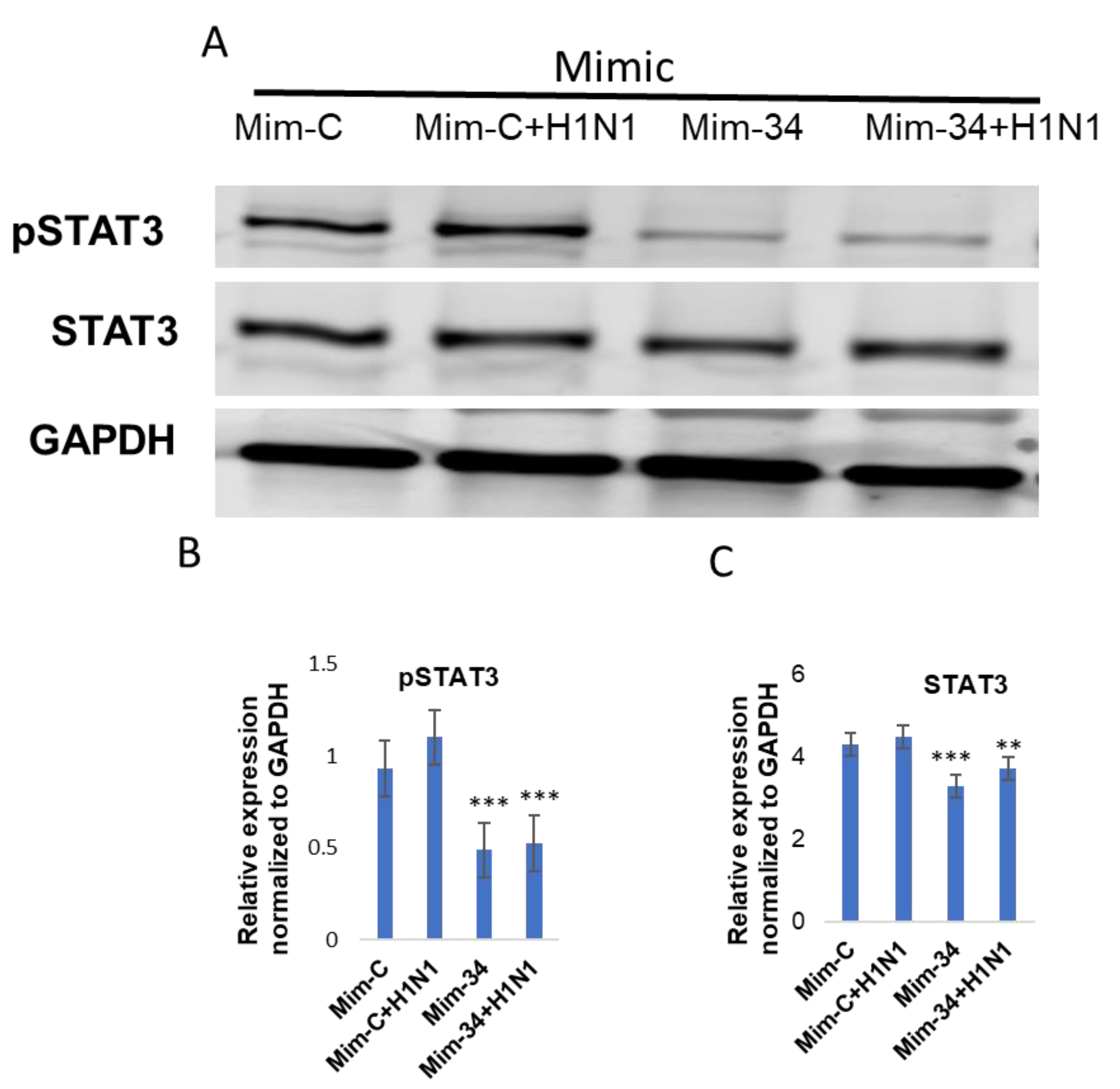
3.3. STAT3 Expression Is Regulated by Put-miR-34.
3.4. Functional Analysis of Put-miR-34
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Wang, Y.; Brahmakshatriya, V.; Lupiani, B.; Reddy, S.M.; Soibam, B.; Benham, A.L.; Gunaratne, P.; Liu, H.-C.; Trakooljul, N.; Ing, N.; et al. Integrated analysis of microRNA expression and mRNA transcriptome in lungs of avian influenza virus infected broilers. BMC Genom. 2012, 13, 278. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Feng, G.; Chen, H.; Wang, L.; Wang, Y. Identification of host encoded microRNAs interacting with novel swine-origin influenza A (H1N1) virus and swine influenza virus. Bioinformation 2009, 4, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Othumpangat, S.; Bryan, N.B.; Beezhold, D.H.; Noti, J.D. Upregulation of miRNA-4776 in Influenza Virus Infected Bronchial Epithelial Cells Is Associated with Downregulation of NFKBIB and Increased Viral Survival. Viruses 2017, 9, 94. [Google Scholar] [CrossRef] [PubMed]
- Othumpangat, S.; Noti, J.D.; Blachere, F.M.; Beezhold, D.H. Expression of non-structural-1A binding protein in lung epithelial cells is modulated by miRNA-548an on exposure to influenza A virus. Virology 2013, 447, 84–94. [Google Scholar] [CrossRef]
- Ghosh, S.; May, M.J.; Kopp, E.B. NF-kappa B and Rel proteins: Evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 1998, 16, 225–260. [Google Scholar] [CrossRef]
- Othumpangat, S.; Noti, J.D.; McMillen, C.M.; Beezhold, D.H. ICAM-1 regulates the survival of influenza virus in lung epithelial cells during the early stages of infection. Virology 2016, 487, 85–94. [Google Scholar] [CrossRef]
- Kuchipudi, S.V. The Complex Role of STAT3 in Viral Infections. J. Immunol Res. 2015, 2015, 272359. [Google Scholar] [CrossRef]
- Jones, M.; Davidson, A.; Hibbert, L.; Gruenwald, P.; Schlaak, J.; Ball, S.; Foster, G.R.; Jacobs, M. Dengue virus inhibits alpha interferon signaling by reducing STAT2 expression. J. Virol. 2005, 79, 5414–5420. [Google Scholar] [CrossRef] [PubMed]
- Richardson, C.; Fielding, C.; Rowe, M.; Brennan, P. Epstein-Barr virus regulates STAT1 through latent membrane protein 1. J. Virol. 2003, 77, 4439–4443. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Antunes, K.H.; Becker, A.; Franceschina, C.; de Freitas, D.d.N.; Lape, I.; da Cunha, M.D.Á.; Leitão, L.; Rigo, M.M.; Pinto, L.A.; Stein, R.T.; et al. Respiratory syncytial virus reduces STAT3 phosphorylation in human memory CD8 T cells stimulated with IL-21. Sci. Rep. 2019, 9, 17766. [Google Scholar] [CrossRef]
- Zhang, H.; Hu, H.; Greeley, N.; Jin, J.; Matthews, A.J.; Ohashi, E.; Caetano, M.S.; Li, H.S.; Wu, X.; Mandal, P.K.; et al. STAT3 restrains RANK- and TLR4-mediated signalling by suppressing expression of the E2 ubiquitin-conjugating enzyme Ubc13. Nat. Commun. 2014, 5, 5798. [Google Scholar] [CrossRef] [PubMed]
- Jia, D.; Rahbar, R.; Chan, R.W.; Lee, S.M.; Chan, M.C.; Wang, B.X.; Baker, D.P.; Sun, B.; Peiris, J.S.; Nicholls, J.M.; et al. Influenza virus non-structural protein 1 (NS1) disrupts interferon signaling. PLoS ONE 2010, 5, e13927. [Google Scholar] [CrossRef]
- Debnath, B.; Xu, S.; Neamati, N. Small molecule inhibitors of signal transducer and activator of transcription 3 (Stat3) protein. J. Med. Chem. 2012, 55, 6645–6668. [Google Scholar] [CrossRef]
- Othumpangat, S.; Noti, J.D.; Beezhold, D.H. Lung epithelial cells resist influenza A infection by inducing the expression of cytochrome c oxidase VIc which is modulated by miRNA 4276. Virology 2014, 468–470, 256–264. [Google Scholar] [CrossRef]
- Ramakers, C.; Ruijter, J.M.; Deprez, R.H.; Moorman, A.F. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.; Karlen, Y.; Bakker, O.; van den Hoff, M.J.; Moorman, A.F. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Benjamini, Y.; Drai, D.; Elmer, G.; Kafkafi, N.; Golani, I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001, 125, 279–284. [Google Scholar] [CrossRef]
- Kuhn, D.E.; Martin, M.M.; Feldman, D.S.; Terry, A.V., Jr.; Nuovo, G.J.; Elton, T.S. Experimental validation of miRNA targets. Methods 2008, 44, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Doench, J.G.; Petersen, C.P.; Sharp, P.A. siRNAs can function as miRNAs. Genes Dev. 2003, 17, 438–442. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Wen, Z.; Darnell, J.E., Jr. Stat3: A STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science 1994, 264, 95–98. [Google Scholar] [CrossRef]
- Ruff-Jamison, S.; Zhong, Z.; Wen, Z.; Chen, K.; Darnell, J.E., Jr.; Cohen, S. Epidermal growth factor and lipopolysaccharide activate Stat3 transcription factor in mouse liver. J. Biol. Chem. 1994, 269, 21933–21935. [Google Scholar] [CrossRef]
- Tian, S.S.; Lamb, P.; Seidel, H.M.; Stein, R.B.; Rosen, J. Rapid activation of the STAT3 transcription factor by granulocyte colony-stimulating factor. Blood 1994, 84, 1760–1764. [Google Scholar] [CrossRef]
- Zhong, Z.; Wen, Z.; Darnell, J.E., Jr. Stat3 and Stat4: Members of the family of signal transducers and activators of transcription. Proc. Natl. Acad. Sci. USA 1994, 91, 4806–4810. [Google Scholar] [CrossRef]
- Yu, Z.; Kone, B.C. The STAT3 DNA-binding domain mediates interaction with NF-kappaB p65 and inducible nitric oxide synthase transrepression in mesangial cells. J. Am. Soc. Nephrol. 2004, 15, 585–591. [Google Scholar] [CrossRef]
- Yan, H.; Wu, J.; Liu, W.; Zuo, Y.; Chen, S.; Zhang, S.; Zeng, M.; Huang, W. MicroRNA-20a overexpression inhibited proliferation and metastasis of pancreatic carcinoma cells. Hum. Gene. 2010, 21, 1723–1734. [Google Scholar] [CrossRef] [PubMed]
- Othumpangat, S.; Lindsley, W.G.; Beezhold, D.H.; Kashon, M.L.; Burrell, C.N.; Mubareka, S.; Noti, J.D. Differential Expression of Serum Exosome microRNAs and Cytokines in Influenza A and B Patients Collected in the 2016 and 2017 Influenza Seasons. Pathogens 2021, 10, 149. [Google Scholar] [CrossRef]
- Hale, B.G.; Randall, R.E.; Ortin, J.; Jackson, D. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 2008, 89, 2359–2376. [Google Scholar] [CrossRef] [PubMed]
- Hui, K.P.; Li, H.S.; Cheung, M.C.; Chan, R.W.; Yuen, K.M.; Mok, C.K.; Nicholls, J.M.; Peiris, J.S.; Chan, M.C. Highly pathogenic avian influenza H5N1 virus delays apoptotic responses via activation of STAT3. Sci Rep. 2016, 6, 28593. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Yan, R.; Chen, B.; Pan, Q.; Chen, Y.; Hong, J.; Zhang, L.; Liu, W.; Wang, S.; Chen, J.L. Influenza Virus-Induced Robust Expression of SOCS3 Contributes to Excessive Production of IL-6. Front. Immunol. 2019, 10, 1843. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Li, J.; Li, J.; Yang, Y.; Kang, X.; Li, Y.; Wu, X.; Zhu, Q.; Zhou, Y.; Hu, Y. Up-regulation of microRNA-203 in influenza A virus infection inhibits viral replication by targeting DR1. Sci. Rep. 2018, 8, 6797. [Google Scholar] [CrossRef]
- Song, L.; Liu, H.; Gao, S.; Jiang, W.; Huang, W. Cellular microRNAs inhibit replication of the H1N1 influenza A virus in infected cells. J. Virol. 2010, 84, 8849–8860. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Wang, J.; Wei, S.; Li, C.; Zhou, K.; Hu, J.; Ye, X.; Yan, J.; Liu, W.; Gao, G.F.; et al. Endogenous Cellular MicroRNAs Mediate Antiviral Defense against Influenza A Virus. Mol. Nucleic Acids 2018, 10, 361–375. [Google Scholar] [CrossRef]




| Putative miRNA | Chromosome | Strand | Start | Stop | Sequence |
|---|---|---|---|---|---|
| put-miR-1 | 10 | + | 64,075,172 | 64,075,189 | GAGTGTGAGTCTGAAACT |
| put-miR-2 | 12 | + | 12,227,225 | 12,227,242 | GCCCGGATAGCTCAGTCA |
| put-miR-3 | 14 | + | 103,804,182 | 103,804,209 | TTGCAAGCAACACTCTGTGGCAGATGAT |
| put-miR-4 | 1 | + | 28,880,952 | 28,880,969 | ATTACGATCTGCTGAGTA |
| put-miR-5 | 1 | + | 90,118,586 | 90,118,605 | GAGAGGGTGCTGTAGGCTCA |
| put-miR-6 | 1 | + | 145,951,004 | 145,951,032 | AGTTCAGTGGTAGAATTCTCGCCTCCCAC |
| put-miR-7 | 1 | + | 147,492,721 | 147,492,750 | CAGTTCAGTGGTAGAATTCTCGCCTCCCAC |
| put-miR-8 | 1 | + | 161,582,560 | 161,582,588 | TTCGATTCCCGGGTAACGAAACGTTTTTG |
| put-miR-9 | 1 | + | 183,212,789 | 183,212,813 | GAATAGACTGGATGGAAAGACAAAC |
| put-miR-10 | 1 | + | 183,212,907 | 183,212,930 | AATGTGACTAAAGGAAAAAACTTT |
| put-miR-11 | 1 | + | 183,213,617 | 183,213,638 | TGACATAGTCTCTGCCCTCATA |
| put-miR-12 | 20 | + | 3,194,760 | 3,194,784 | GCAAAATGATGAGGTACCTGATACT |
| put-miR-13 | 20 | + | 18,309,660 | 18,309,685 | ATGGTAGTGGGTTATCAGAACTTATT |
| put-miR-14 | 20 | + | 32,082,877 | 32,082,893 | TTGCTCTGATGAAATCT |
| put-miR-15 | 2 | + | 164,382,863 | 164,382,879 | ACAGTGACTGAGAGACT |
| put-miR-16 | 3 | + | 164,059,153 | 164,059,174 | TATCTCGCTGGGGCCTCCAAAA |
| put-miR-17 | 4 | + | 83,354,324 | 83,354,340 | ACTACCGTTTTCTGAAG |
| put-miR-18 | 5 | + | 105,889,151 | 105,889,173 | GTTCTTGTAGTTGAAATACAACG |
| put-miR-19 | 5 | + | 105,889,191 | 105,889,208 | TGGTCGTGGTTGTAGTCC |
| put-miR-20 | 5 | + | 180,528,918 | 180,528,945 | GCCAGCTTGTTGTGATTCCTCCATTTTT |
| put-miR-21 | 6 | + | 11,976,724 | 11,976,740 | TTTCCTTCTGAGAACAA |
| put-miR-22 | 6 | + | 27,656,041 | 27,656,059 | AGTATTCTCTGTGGCTTTT |
| put-miR-23 | 6 | + | 28,574,986 | 28,575,007 | TCAATCCCCGGCACCTCCACCA |
| put-miR-24 | 7 | + | 18,159,276 | 18,159,293 | AAATCTGACTGTCTAATT |
| put-miR-25 | 7 | + | 123,501,242 | 123,501,257 | AGTTTCTGTCTGATAA |
| put-miR-26 | 7 | + | 139,025,502 | 139,025,531 | GAGTCCCATCTGGGGTGGCCTGTGACTTTT |
| put-miR-27 | 9 | + | 122,841,421 | 122,841,437 | TAGCAAGACTGAGGCTT |
| put-miR-28 | 9 | + | 133,282,156 | 133,282,176 | AGCCTGTCTGAGCGCCGCTCT |
| put-miR-29 | 18 | + | 19,645,301 | 19,645,318 | AAAGAAGTTCTGAGCTTG |
| put-miR-30 | 18 | + | 68,180,033 | 68,180,063 | CAGGAGTTCTGGGCTGTAGTGCGCTATGCT |
| put-miR-31 | 22 | + | 35,126,604 | 35,126,623 | CCAACTTCCTTCTGAGAACA |
| put-miR-32 | 15 | + | 45,009,923 | 45,009,945 | ACATGGACATGATCTTCTTTATA |
| put-miR-33 | MT | − | 16,525 | 16,551 | AAGGGGAACGTGTGGGCTATTTAGGCT |
| put-miR-34 | X | + | 23,803,909 | 23,803,934 | AAGAGGGTTGGAGACTGTTCAAGATC |
| put-miR-35 | X | + | 34,234,100 | 34,234,123 | GAGCAGTAACAGGTCTGTGATGCT |
| put-miR-36 | 8 | + | 100,305,233 | 100,305,261 | GTTACTAGAGAAGTTTCTCTGAACGTGTA |
| put-miR-37 | 16 | + | 2,737,223 | 2,737,253 | CTCTGGTGATGAAATGGAACGTTTCTGATG |
| put-miR-38 | 16 | + | 33,963,846 | 33,963,873 | TTGGTGGAGTGATTTGTCTGGTTAATTC |
| put-miR-39 | 16 | + | 33,963,882 | 33,963,908 | AACGAGACTCTGGCATGCTAACTAGTT |
| put-miR-40 | 11 | + | 49,764,393 | 49,764,415 | TATCCCGGACGAGCCCCCATTAT |
| put-miR-41 | 11 | + | 62,126,505 | 62,126,531 | TGGTGTAATGGTTAGCACTCTGGACTC |
| put-miR-42 | 11 | + | 65,271,895 | 65,271,916 | GTGAAACGACTGGAGTATGATT |
| Putative miRNA | Fold Change H1N1 /Mock | t-Test p-Value | Benjamini– Hochberg FDR |
|---|---|---|---|
| put-miR-34 | −2.725 | 0.013 | 0.315 |
| put-miR-18 | −1.823 | 0.098 | 0.675 |
| put-miR-26 | −1.652 | 0.124 | 0.675 |
| put-miR-36 | −2.338 | 0.143 | 0.675 |
| put-miR-22 | 1.494 | 0.172 | 0.675 |
| put-miR-31 | 1.636 | 0.213 | 0.675 |
| put-miR-28 | −1.741 | 0.227 | 0.675 |
| put-miR-7 | 1.744 | 0.241 | 0.675 |
| put-miR-19 | −2.057 | 0.272 | 0.675 |
| put-miR-13 | −1.501 | 0.283 | 0.675 |
| put-miR-39 | −1.490 | 0.338 | 0.675 |
| put-miR-20 | −1.184 | 0.366 | 0.675 |
| put-miR-38 | −1.447 | 0.397 | 0.675 |
| put-miR-41 | −1.699 | 0.465 | 0.675 |
| put-miR-23 | 1.163 | 0.471 | 0.675 |
| put-miR-30 | 1.256 | 0.540 | 0.675 |
| put-miR-35 | 1.230 | 0.625 | 0.744 |
| put-miR-5 | 1.247 | 0.738 | 0.802 |
| put-miR-8 | −1.173 | 0.826 | 0.860 |
| put-miR-1 | 1.027 | 0.958 | 0.958 |
| put-miR-10 | 3.130 | ND | ND |
| put-miR-11 | −1.080 | ND | ND |
| put-miR-27 | 1.207 | ND | ND |
| put-miR-29 | 2.776 | ND | ND |
| put-miR-37 | −1.147 | ND | ND |
| put-miR-40 | −1.079 | ND | ND |
| put-miR-6 | 1.524 | ND | ND |
| Putative miRNA | Fold Change H9N1-IP10/Mock | t-Test p-Value | Benjamini–Hochberg FDR |
|---|---|---|---|
| put-miR-31 | 2.213 | 0.050 | 0.675 |
| put-miR-34 | −3.009 | 0.067 | 0.866 |
| put-miR-23 | −1.211 | 0.192 | 0.989 |
| put-miR-22 | −1.237 | 0.205 | 0.989 |
| put-miR-41 | 1.718 | 0.293 | 0.989 |
| put-miR-18 | −1.377 | 0.323 | 0.989 |
| put-miR-26 | −1.413 | 0.454 | 0.989 |
| put-miR-39 | −1.056 | 0.503 | 0.989 |
| put-miR-35 | −1.312 | 0.525 | 0.989 |
| put-miR-20 | −1.452 | 0.590 | 0.989 |
| put-miR-1 | −1.471 | 0.593 | 0.989 |
| put-miR-30 | 1.149 | 0.656 | 0.989 |
| put-miR-36 | −1.378 | 0.673 | 0.989 |
| put-miR-8 | −1.229 | 0.686 | 0.989 |
| put-miR-7 | 1.146 | 0.710 | 0.989 |
| put-miR-28 | 1.101 | 0.768 | 0.989 |
| put-miR-19 | −1.087 | 0.794 | 0.989 |
| put-miR-6 | 1.221 | 0.802 | 0.989 |
| put-miR-5 | −1.024 | 0.948 | 0.989 |
| put-miR-13 | 1.006 | 0.985 | 0.989 |
| put-miR-38 | −1.007 | 0.989 | 0.989 |
| put-miR-10 | 1.105 | ND | ND |
| put-miR-11 | −1.016 | ND | ND |
| put-miR-27 | 1.691 | ND | ND |
| put-miR-29 | 1.090 | ND | ND |
| put-miR-37 | −1.086 | ND | ND |
| put-miR-40 | −1.286 | ND | ND |
| Raw Data Counts | ||||
|---|---|---|---|---|
| Ago-IP | Put-miR-31 | Put-miR-34 | Put-miR-35 | Control-SCR |
| Put-miR-31 | 29,275 | 1949 | 681 | 3866 |
| Put-miR-34 | 221 | 117,229 | 901 | 422 |
| Put-miR-35 | 115 | 138 | 263,227 | 61 |
| GO_ID | Term | Annotated | Significant | Expected | p-Value |
|---|---|---|---|---|---|
| GO:0006281 | DNA repair | 427 | 5 | 5.07 | 5.1 × 10−14 |
| GO:0019048 | modulation by virus of host morphology or physiology | 336 | 8 | 3.99 | 2.1 × 10−11 |
| GO:0007067 | mitotic nuclear division | 359 | 4 | 4.26 | 6.9 × 10−10 |
| GO:0051301 | cell division | 577 | 6 | 6.85 | 1.4 × 10−09 |
| GO:0006886 | intracellular protein transport | 821 | 8 | 9.75 | 6.3 × 10−09 |
| GO:0000398 | mRNA splicing, via spliceosome | 210 | 3 | 2.49 | 6.4 × 10−08 |
| GO:0006355 | regulation of transcription, DNA templated | 3012 | 37 | 35.78 | 6.6 × 10−08 |
| GO:0043123 | positive regulation of I-kB kinase/NF-κB signaling | 153 | 6 | 1.82 | 8.6 × 10−08 |
| GO:0016197 | endosomal transport | 152 | 2 | 1.81 | 2.0 × 10−07 |
| GO:0000209 | protein polyubiquitination | 163 | 0 | 1.94 | 2.7 × 10−07 |
| GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 109 | 0 | 1.29 | 2.8 × 10−07 |
| GO:0006364 | rRNA processing | 114 | 1 | 1.35 | 3.0 × 10−07 |
| GO:0000086 | G2/M transition of mitotic cell cycle | 158 | 2 | 1.88 | 4.2 × 10−07 |
| GO:0000082 | G1/S transition of mitotic cell cycle | 223 | 0 | 2.65 | 1.5 × 10−06 |
| GO:0006614 | SRP-dependent co-translational protein targeting to membrane | 101 | 1 | 1.2 | 1.5 × 10−06 |
| GO:0010467 | gene expression | 4410 | 49 | 52.38 | 2.5 × 10−06 |
| GO:0006413 | translational initiation | 156 | 3 | 1.85 | 2.6 × 10−06 |
| GO:0006415 | translational termination | 86 | 0 | 1.02 | 3.3 × 10−06 |
| GO:0034138 | toll-like receptor 3 signaling pathway | 80 | 1 | 0.95 | 3.5 × 10−06 |
| GO:0051726 | regulation of cell cycle | 701 | 7 | 8.33 | 4.1 × 10−06 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Othumpangat, S.; Beezhold, D.H.; Umbright, C.M.; Noti, J.D. Influenza Virus-Induced Novel miRNAs Regulate the STAT Pathway. Viruses 2021, 13, 967. https://doi.org/10.3390/v13060967
Othumpangat S, Beezhold DH, Umbright CM, Noti JD. Influenza Virus-Induced Novel miRNAs Regulate the STAT Pathway. Viruses. 2021; 13(6):967. https://doi.org/10.3390/v13060967
Chicago/Turabian StyleOthumpangat, Sreekumar, Donald H. Beezhold, Christina M. Umbright, and John D. Noti. 2021. "Influenza Virus-Induced Novel miRNAs Regulate the STAT Pathway" Viruses 13, no. 6: 967. https://doi.org/10.3390/v13060967
APA StyleOthumpangat, S., Beezhold, D. H., Umbright, C. M., & Noti, J. D. (2021). Influenza Virus-Induced Novel miRNAs Regulate the STAT Pathway. Viruses, 13(6), 967. https://doi.org/10.3390/v13060967







