Global Distribution and Genetic Heterogeneity of Border Disease Virus
Abstract
1. Introduction
2. Genomic Organization of BDV
3. Global Distribution of BDV Genotypes
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Valdazo-Gonzalez, B.; Alvarez-Martinez, M.; Greiser-Wilke, I. Genetic typing and prevalence of Border disease virus (BDV) in small ruminant flocks in Spain. Vet. Microbiol. 2006, 117, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Hughes, L.E.; Kershaw, G.F.; Shaw, I.G. Border Disease. An undescribed disease of sheep. Vet. Rec. 1959, 71, 313–317. [Google Scholar]
- Nettleton, P.F.; Gilray, J.A.; Russo, P.; Dlissi, E. Border disease of sheep and goats. Vet. Res. 1998, 29, 327–340. [Google Scholar] [PubMed]
- Chappuis, G.; Brun, A.; Kato, F.; Dauvergne, M.; Reynaud, G.; Duret, C. Études Sérologiques et Immunologiques Réalisées à la Suite de l’Isolement d’un Pestivirus dans un Foyer Ovin chez des Moutons de l’Aveyron. In Pestivirose des Ovinset des Bovins: Nouvelles Connaissances, Utilisation pour une Stratégie de Contrôle, Journées Nationales de la Société Française de Buiatrie et de son Groupe d’Étudesur la Pathologie des Ovins et des Caprins (GEPOC); Espinasse, J., Savey, M., Eds.; Societe Francaise de Buiatrie: Paris, France, 1986; pp. 55–65. [Google Scholar]
- Vilček, S.; Leskova, V.; Meyer, D.; Postel, A.; Becher, P. Molecular characterization of border disease virus strain Aveyron. Vet. Microbiol. 2014, 171, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Vega, S.; Rosell, R.; Orden, J.A.; Pérez, T.; Marín, C.; González, S.; Marco, I.; Cabezón, O.; de la Fuente, R. Antigenic and molecular characterisation of Border disease virus associated with high mortality in lambs in Spain. Vet. Rec. Open 2015, 2, e000048. [Google Scholar] [CrossRef]
- Arnal, M.; Fernandez-de-Luco, D.; Riba, L.; Maley, M.; Gilray, J.; Willoughby, K.; Vilček, S.; Nettleton, P.F. A novel pestivirus associated with deaths in Pyrenean chamois (Rupicapra pyrenaica pyrenaica). J. Gen. Virol. 2004, 85, 3653–3657. [Google Scholar] [CrossRef]
- Marco, I.; López-Olvera, J.R.; Rosell, R.; Vidal, E.; Hurtado, A.; Juste, R.; Pumarola, M.; Lavin, S. Severe outbreak of disease in the Southern chamois (Rupicapra pyrenaica) associated with border disease virus infection. Vet. Microbiol. 2007, 120, 33–41. [Google Scholar] [CrossRef]
- Marco, I.; Rosell, R.; Cabezón, O.; Mentaberre, G.; Casas, E.; Velarde, R.; Lavín, S. Border disease virus among chamois, Spain. Emerg. Infect. Dis. 2009, 15, 448–451. [Google Scholar] [CrossRef]
- Smith, D.B.; Meyers, G.; Bukh, J.; Gould, E.A.; Monath, T.; Muerhoff, A.S.; Pletnev, A.; Rico-Hesse, R.; Stapleton, J.T.; Simmonds, P.; et al. Proposed revision to the taxonomy of the genus Pestivirus, family Flaviviridae. J. Gen. Virol. 2017, 98, 2106–2112. [Google Scholar] [CrossRef]
- King, A.M.Q.; Lefkowitz, E.J.; Mushegian, A.R.; Adams, M.J.; Dutilh, B.E.; Gorbalenya, A.E.; Harrach, B.; Harrison, R.L.; Junglen, S.; Knowles, N.J.; et al. Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses. Arch. Virol. 2018, 163, 2601–2631. [Google Scholar] [CrossRef] [PubMed]
- Becher, P.; Avalos Ramirez, R.; Orlich, M.; Cedillo Rosales, S.; Kosmidou, A.; Konig, M.; Schweizer, M.; Stalder, H.; Schirmeier, H.; Thiel, H.J. Genetic and antigenic characterization of novel pestivirus genotypes: Implications for classification. Virology 2003, 311, 96–104. [Google Scholar] [CrossRef]
- Valdazo-Gonzalez, B.; Alvarez-Martinez, M.; Sandvik, T. Genetic and antigenic typing of border disease virus isolates in sheep from the Iberian Peninsula. Vet. J. 2007, 174, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Dubois, E.; Russo, P.; Prigent, M.; Thiery, R. Genetic characterization of ovine pestiviruses isolated in France, between 1985 and 2006. Vet. Microbiol. 2008, 130, 69–79. [Google Scholar] [CrossRef]
- Giammarioli, M.; La Rocca, S.A.; Steinbach, F.; Casciari, C.; De Mia, G.M. Genetic and antigenic typing of border disease virus (BDV) isolates from Italy reveals the existence of a novel BDV group. Vet. Microbiol. 2011, 147, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Peletto, S.; Caruso, C.; Cerutti, F.; Modesto, P.; Zoppi, S.; Dondo, A.; Acutis, P.L.; Masoero, L. A new genotype of border disease virus with implications for molecular diagnostics. Arch. Virol. 2016, 161, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Thabti, F.; Letellier, C.; Hammami, S.; Pepin, M.; Ribiere, M.; Mesplede, A.; Kerkhofs, P.; Russo, P. Detection of a novel border disease virus subgroup in Tunisian sheep. Arch. Virol. 2005, 150, 215–229. [Google Scholar] [CrossRef]
- Ciulli, S.; Purpari, G.; Agnello, S.; Di Marco, P.; Di Bella, S.; Volpe, E.; Mira, F.; De Aguiar Saldanha Pinheiro, A.C.; Vullo, S.; Guercio, A. Evidence for Tunisian-like pestiviruses presence in small ruminants in Italy since 2007. Transbound. Emerg. Dis. 2017, 64, 1243–1253. [Google Scholar] [CrossRef]
- Oguzoglu, T.C.; Tan, M.T.; Toplu, N.; Demir, A.B.; Bilge-Dagalp, S.; Karaoglu, T.; Ozkul, A.; Alkan, F.; Haas, L.; Greiser-Wilke, I. Border disease virus (BDV) infections of small ruminants in Turkey: A new BDV subgroup. Vet. Microbiol. 2009, 135, 374–379. [Google Scholar] [CrossRef]
- Becher, P.; Schmeiser, S.; Cigdem, T.; Postel, A. Complete genome sequence of a novel pestivirus from sheep. J. Virol. 2012, 86, 11412. [Google Scholar] [CrossRef]
- Postel, A.; Schmeiser, S.; Oguzoglu, T.C.; Indenbirken, D.; Alawi, M.; Fischer, N.; Grundhoff, A.; Becher, P. Close Relationship of Ruminant Pestiviruses and Classical Swine Fever Virus. Emerg. Infect. Dis. 2015, 21, 668. Available online: www.cdc.gov/eid (accessed on 20 May 2020). [CrossRef]
- Sozzi, E.; Lavazza, A.; Gaffuri, A.; Bencetti, F.C.; Prosperi, A.; Lelli, D.; Chiapponi, C.; Moreno, A. Isolation and full-length sequence analysis of a Pestivirus from aborted lamb foetuses in Italy. Viruses 2019, 11, 744. [Google Scholar] [CrossRef]
- Casciari, C.; Sozzi, E.; Bazzucchi, M.; Martin, A.M.M.; Gaffuri, A.; Giammarioli, M.; Lavazza, A.; De Mia, G.M. Serological relationship between a novel ovine pestivirus and classical swine fever virus. Transbound. Emerg. Dis. 2020, 67, 1406–1410. [Google Scholar] [CrossRef] [PubMed]
- Vilček, S.; Belak, S. Genetic identification of pestivirus strain Frijters as a border disease virus from pigs. J. Virol. Methods 1996, 60, 103–108. [Google Scholar] [CrossRef]
- Nagai, M.; Aoki, H.; Sakoda, Y.; Kozasa, T.; Tominaga-Teshima, K.; Mine, J.; Abe, Y.; Tamura, T.; Kobayashi, T.; Nishine, K.; et al. Molecular, biological, and antigenic characterization of a Border disease virus isolated from a pig during classical swine fever surveillance in Japan. J. Vet. Diagn. Investig. 2014, 26, 547–552. [Google Scholar] [CrossRef] [PubMed]
- Kawanishi, N.; Tsuduku, S.; Shimizu, H.; Ohtani, Y.; Kameyama, K.; Yamakawa, M.; Tsutsui, T.; Matsuura, K.; Ohashi, S.; Isobe, T.; et al. First isolation of border disease virus in Japan is from a pig farm with no ruminants. Vet. Microbiol. 2014, 171, 210–214. [Google Scholar] [CrossRef]
- Rosell, R.; Cabezón, O.; Pujols, J.; Domingo, M.; Muñoz, I.; Núñez, J.I.; Ganges, L. Identification of a porcine pestivirus as a border disease virus from naturally infected pigs in Spain. Vet. Rec. 2014, 174, 18. [Google Scholar] [CrossRef] [PubMed]
- Becher, P.; Orlich, M.; Shannon, A.D.; Horner, G.; König, M.; Thiel, H.J. Phylogenetic analysis of pestiviruses from domestic and wild ruminants. J. Gen. Virol. 1997, 78, 1357–1366. [Google Scholar] [CrossRef]
- Cranwell, M.P.; Otter, A.; Errington, J.; Hogg, R.A.; Wakeley, P.; Sandvik, T. Detection of border disease virus in cattle. Vet. Rec. 2007, 161, 211–212. [Google Scholar] [CrossRef]
- Krametter-Frötscher, R.; Benetka, V.; Möstl, K.; Baumgartner, W. Transmission of border disease virus from sheep to calves—a possible risk factor for the Austrian BVD eradication programme in cattle? Wien. Tierarztl. Monatsschr. 2008, 95, 200–203. [Google Scholar]
- Schirrmeier, H.; Strebelow, G.; Tavella, A.; Stifter, E. Border disease virus infection in cattle—Epidemiolocical and diagnostic impact. In Proceedings of the 7th ESVV Pestivirus Symposium, Uppsala, Sweden, 16–19 September 2008. [Google Scholar]
- Hornberg, A.; Fernández, S.R.; Vogl, C.; Vilček, S.; Matt, M.; Fink, M.; Köfer, J.; Schöpf, K. Genetic diversity of pestivirus isolates in cattle from Western Austria. Vet. Microbiol. 2009, 135, 205–213. [Google Scholar] [CrossRef]
- Strong, R.; La Rocca, S.A.; Ibata, G.; Sandvik, T. Antigenic and genetic characterisation of border disease viruses isolated from UK cattle. Vet. Microbiol. 2010, 141, 208–215. [Google Scholar] [CrossRef] [PubMed]
- McFadden, A.M.; Tisdall, D.J.; Hill, F.I.; Otterson, P.; Pulford, D.; Peake, J.; Finnegan, C.J.; La Rocca, S.A.; Kok-Mun, T.; Weir, A.M. The first case of a bull persistently infected with border disease virus in New Zealand. N. Z. Vet. J. 2012, 60, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Braun, U.; Reichle, S.F.; Reichert, C.; Hässig, M.; Stalder, H.P.; Bachofen, C.; Peterhans, E. Sheep persistently infected with Border disease readily transmit virus to calves seronegative to BVD virus. Vet. Microbiol. 2014, 168, 98–104. [Google Scholar] [CrossRef]
- Schoepf, K.; Revilla-Fernández, S.; Steinrigl, A.; Fuchs, R.; Sailer, A.; Weikel, J.; Schmoll, F. Retrospective epidemiological evaluation of molecular and animal husbandry data within the bovine viral diarrhoea virus (BVDV) control programme in Western Austria during 2009–2014. Berl. Münch. Tierärztl. Wschr. 2016, 129, 196–201. [Google Scholar]
- Gómez-Romero, N.; Basurto-Alcántara, F.J.; Verdugo-Rodríguez, A.; Lagunes-Quintanilla, R.; Bauermann, F.V.; Ridpath, J.F. Detection of border disease virus in Mexican cattle. Transbound. Emerg. Dis. 2018, 65, 267–271. [Google Scholar] [CrossRef]
- Braun, U.; Hilbe, M.; Peterhans, E.; Schweizer, M. Border disease in cattle. Vet. J. 2019, 246, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Becher, P.; Orlich, M.; Kosmidou, A.; Konig, M.; Baroth, M.; Thiel, H.J. Genetic diversity of pestiviruses: Identification of novel groups and implications for classification. Virology 1999, 262, 64–67. [Google Scholar] [CrossRef] [PubMed]
- Danuser, R.; Vogt, H.-R.; Kaufmann, T.; Peterhans, E.; Zanoni, R. Seroprevalence and characterization of pestivirus infections in small ruminants and new world camelids in Switzerland. Schweizer. Archiv. für Tierheilkunde 2013, 151, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.; Duquesne, V.; Adam, G.; Belleau, E.; Gauthier, D.; Champion, J.L.; Saegerman, C.; Thiéry, R.; Dubois, E. Pestiviruses infections at the wild and domestic ruminants interface in the French Southern Alps. Vet. Microbiol. 2015, 175, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Vilček, S.; Bjorklund, H.V.; Horner, G.W.; Meers, J.; Belak, S. Genetic typing of pestiviruses from New Zealand. N. Z. Vet. J. 1998, 46, 35–37. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, D.G.; Chang, D.J.; Akkina, R.K. Genetic characterization of ruminant pestiviruses: Sequence analysis of viral genotypes isolated from sheep. Virus Res. 1997, 47, 19–29. [Google Scholar] [CrossRef]
- Giangaspero, M.; Harasawa, R.; Muschko, K.; Bǘttner, M. Characteristics of the 5′ untranslated region of wisent (Bison bonasus) and reindeer (Rangifer tarandus) Pestivirus isolates. Vet. Ital. 2006, 42, 165–172. [Google Scholar]
- Giammarioli, M.; Rossi, E.; Casciari, C.; Bazzucchi, M.; Torresi, C.; De Mia, G.M. Genetic characterization of border disease virus (BDV) isolates from small ruminants in Italy. Virus Genes 2015, 50, 321–324. [Google Scholar] [CrossRef]
- Krametter-Froetscher, R.; Kohler, H.; Benetka, V.; Moestl, K.; Golja, F.; Vilček, S.; Baumgartner, W. Influence of communal alpine pasturing on the spread of pestiviruses among sheep and goats in Austria: First identification of border disease virus in Austria. Zoonoses Public. Health 2007, 54, 209–213. [Google Scholar] [CrossRef]
- Rosamilia, A.; Grattarola, C.; Caruso, C.; Peletto, S.; Gobbi, E.; Tarello, V.; Caroggio, P.; Dondo, A.; Masoero, L.; Acutis, P.L. Detection of border disease virus (BDV) genotype 3 in Italian goat herds. Vet. J. 2014, 199, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Luzzago, C.; Ebranati, E.; Cabezón, O.; Fernández-Sirera, L.; Lavín, S.; Rosell, R.; Veo, C.; Rossi, L.; Cavallero, S.; Lanfranchi, P.; et al. Spatial and temporal phylogeny of border disease virus in Pyrenean chamois (Rupicapra pyrenaica). PLoS ONE 2016, 11, e0168232. [Google Scholar] [CrossRef]
- De Mia, G.M.; Greiser-Wilke, I.; Feliziani, F.; Giammarioli, M.; De Giuseppe, A. Genetic characterization of a caprine pestivirus as the first member of a novel pestivirus subgroup. J. Vet. Med. 2005, 52, 206–210. [Google Scholar] [CrossRef]
- Piras, I.M.; Dei Giudici, S.; Fadda, M.; Anfossi, A.G.; Oggiano, A.; Pittau, M.; Chessa, B. Distribution and Genetic Characterization of Border Disease Virus Circulating in Sardinian Ovine Flocks. Pathogens 2020, 9, 360. [Google Scholar] [CrossRef] [PubMed]
- Peterhans, E.; Bachofen, C.; Stalder, H.; Schweizer, M. Cytopathic bovine viral diarrhea viruses (BVDV): Emerging pestiviruses doomed to extinction. Vet. Res. 2010, 41, 44. [Google Scholar] [CrossRef]
- Caruso, C.; Peletto, S.; Cerutti, F.; Modesto, P.; Robetto, S.; Domenis, L.; Masoero, L.; Acuti, P. Evidence of circulation of the novel border disease virus genotype 8 in chamois. Arch. Virol. 2017, 162, 511–515. [Google Scholar] [CrossRef]
- Stalder, H.P.; Marti, S.; Flückiger, F.; Renevey, N.; Hofmann, M.A.; Schweizer, M. Complete genome sequences of three border disease virus strains of the same subgenotype, BD Swiss, isolated from sheep, cattle, and pigs in Switzerland. Genome Announc. 2017, 5, e01238-17. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, M.; Peterhans, E. Pestiviruses. Annu. Rev. Anim. Biosci. 2014, 2, 141–163. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.H.; He, B.; Li, K.R.; Li, F.; Zhang, L.Y.; Li, X.Q.; Zhao, L. First report of border disease virus in Melophagus ovinus (sheep ked) collected in Xinjiang, China. PLoS ONE 2019, 14, e0221435. [Google Scholar] [CrossRef]
- Colom-Cadena, A.; Cabezóna, O.; Rosell, R.; Fernández-Aguilar, X.; Blanch-Lázaro, B.; Tetas, E.; Lavín, S.; Marco, I. The European hare (Lepus europaeus) as a potential wild reservoir forruminant pestiviruses. Prev. Vet. Med. 2016, 131, 60–63. [Google Scholar] [CrossRef] [PubMed]
- Oğuzoğlu, T.C. A Review of Border Disease Virus Infection in Ruminants: Molecular Characterization, Pathogenesis, Diagnosis and Control. Anim. Health Prod. Hyg. 2012, 1, 1–9. [Google Scholar]
- Yesilbag, K.; Alpay, G.; Becher, P. Variability and Global Distribution of Subgenotypes of Bovine Viral Diarrhea Virus. Viruses 2017, 9, 128. [Google Scholar] [CrossRef] [PubMed]
- Menzies, P.I. Abortion in Sheep: Diagnosis and Control. In Current Therapy in Large Animal Theriogenology; Saunders: Philadelphia, PA, USA, 2007. [Google Scholar]
- Gillespie, J.H.; Baker, J.A.; McEntee, K. A cytopathogenic strain of virus diarrhea virus. Cornell Vet. 1960, 50, 73–79. [Google Scholar] [PubMed]
- Becher, P.; Meyers, G.; Shannon, A.D.; Thiel, H.J. Cytopathogenicity of border disease virus is correlated with integration of cellular sequences into the viral genome. J. Virol. 1996, 70, 2992–2998. [Google Scholar] [CrossRef]
- Nettleton, P.F.; Entrican, G. Ruminant pestiviruses. Br. Vet. J. 1995, 151, 615–642. [Google Scholar] [CrossRef]
- Monies, R.J.; Paton, D.J.; Vilcek, S. Mucosal disease-like lesions in sheep infected with Border disease virus. Vet. Rec. 2004, 155, 765–769. [Google Scholar]
- Hilbe, M.; Camenisch, U.; Braun, U.; Peterhans, E.; Stalder, H.P.; Zlinszky, K.; Ehrensperger, F. Mucosal lesions in a sheep infected with the Border Disease Virus (BDV). Schweiz. Arch. Tierheilk. 2009, 151, 391–396. [Google Scholar] [CrossRef]
- Giannitti, F.; Barr, B.; Sverlow, K.; Thompson, M.; Crossley, B. Border disease resembling mucosal disease in a lamb (Ovis aries). In Proceedings of the ACVP Annual Meeting, Seattle, WA, USA, 1–5 December 2012. [Google Scholar]
- Brun, A.; Lacoste, F.; Reynaud, G.; Kato, F.; Saint-Marc, B. Evaluation of the potency of an inactivated vaccine against border disease pestivirus infection in sheep. In Proceedings of the Second Symposium on Pestiviruses, Annecy, France, 1–3 October 1992; Edwards, S., Ed.; Fonda on Marcel Merieux: Annecy, France, 1992; pp. 257–259. [Google Scholar]
- Newcomer, B.W.; Givens, M.D. Approved and experimental countermeasures against pestiviral diseases: Bovine viral diarrhea, classical swine fever and border disease. Antivir. Res. 2013, 100, 133–150. [Google Scholar] [CrossRef] [PubMed]
- Firquet, S.; Beaujard, S.; Lobert, P.E.; Sane, F.; Caloone, D.; Izard, D.; Hober, D. Survival of enveloped and non-enveloped viruses on inanimate surfaces. Microbes Environ. 2015, 30, 140–144. [Google Scholar] [CrossRef] [PubMed]
- Collett, M.S.; Larson, R.; Gold, C.; Strick, D.; Anderson, D.K.; Purchio, A.F. Molecular cloning and nucleotide sequence of the pestivirus bovine viral diarrhea virus. Virology 1988, 165, 191–199. [Google Scholar] [CrossRef]
- Becher, P.; Orlich, M.; Thiel, H.J. Complete genomic sequence of border disease virus, a pestivirus from sheep. J. Virol. 1998, 72, 5165–5173. [Google Scholar] [CrossRef] [PubMed]
- Thiel, H.J.; Colle, M.S.; Gould, E.A.; Heinz, F.X.; Houghton, M.; Meyers, G.; Purcell, R.H.; Rice, C.M. Family Flaviviridae. In Virus Taxonomy; Eighth Report of the International Commitiee on Taxonomy of Viruses; Fauquet, C.M., Mayo, M.A., Manilo, J., Desselberger, U., Ball, L.A., Eds.; Elsevier Academic Press: San Diego, CA, USA, 2005; pp. 981–998. [Google Scholar]
- Vilček, S.; Nettleton, P.F.; Paton, D.J.; Belák, S. Molecular characterization of ovine pestiviruses. J. Gen. Virol. 1997, 78, 725–735. [Google Scholar] [CrossRef]
- Simmonds, P.; Becher, P.; Bukh, J.; Gould, E.A.; Meyers, G.; Monath, T.; Muerhoff, S.; Pletnev, A.; Rico-Hesse, R.; Smith, D.B.; et al. ICTV virus taxonomy profile: Flaviviridae. J. Gen. Virol. 2017, 98, 2–3. [Google Scholar] [CrossRef] [PubMed]
- Schirrmeier, H.; Strebelow, G.; Depner, K.; Hoffmann, B.; Beer, M. Genetic and antigenic characterization of an atypical pestivirus isolate, a putative member of a novel pestivirus species. J. Gen. Virol. 2004, 85, 3647–3652. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, P.; Becher, P.; Collett, M.S.; Gould, E.A.; Heinz, F.X.; Meyers, G.; Monath, T.; Pletnev, A.; Rice, C.M.; Stiasny, K.; et al. Family Flaviviridae. In Virus Taxonomy; Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Lefkowitz, E., Adams, M.J., Carstens, E.B., Eds.; Elsevier Academic Press: San Diego, CA, USA, 2012; pp. 1003–1020. [Google Scholar]
- Giangaspero, M.; Harasawa, R. Characterization of genotypes among bovine viral diarrhea virus type 1 strains according to palindromic nucleotide substitutions in the genomic 50-untranslated region. J. Virol. Methods 2014, 195, 34–53. [Google Scholar] [CrossRef]
- Vilček, S.; Herring, A.J.; Herring, J.A.; Nettleton, P.F.; Lowings, J.P.; Paton, D.J. Pestiviruses isolated from pigs, cattle and sheep can be allocated into at least three genogroups using polymerase chain reaction and restriction endonuclease analysis. Arch. Virol. 1994, 136, 309–323. [Google Scholar] [CrossRef]
- Stalder, H.; Hug, C.; Zanoni, R.; Vogt, H.R.; Peterhans, E.; Schweizer, M.; Bachofen, C. A nationwide database linking information on the hosts with sequence data of their virus strains: A useful tool for the eradication of bovine viral diarrhea (BVD) in Switzerland. Virus Res. 2016, 218, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Krametter-Froetscher, R.V.; Benetka, K.; Rasser, F.; Tockner, G.; Moesslacher, K.; Moestl, B.W. BVDV control program in Austria—Is a monitoring of BDV status in sheep in Austria necessary? Vet. Med. 2009, 54, 517–524. [Google Scholar] [CrossRef]
- Krametter-Froetscher, R.; Duenser, M.; Preyler, B.; Theiner, A.; Benetka, V.; Moestl, K.; Baumgartner, W. Pestivirus infection in sheep and goats in West Austria. Vet. J. 2010, 186, 342–346. [Google Scholar] [CrossRef]
- Pioz, M.; Loison, A.; Gibert, P.; Dubray, D.; Menaut, P.; Le Tallec, B.; Artois, M.; Gilot-Fromont, E. Transmission of a pestivirus infection in a population of Pyrenean chamois. Vet. Microbiol. 2007, 119, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Avalos-Ramirez, R.; Orlich, M.; Thiel, H.J.; Becher, P. Evidence for the Presence of Two Novel Pestivirus Species. Virology 2001, 286, 456–465. [Google Scholar] [CrossRef] [PubMed]
- Becher, P.; Shannon, A.D.; Tautz, N.; Thiel, H.J. Molecular characterization of border disease virus, a pestivirus from sheep. Virology 1994, 198, 542–551. [Google Scholar] [CrossRef]
- Orsel, K.; Antonis, A.F.; Oosterloo, J.C.; Vellema, P. Van der Meer, F.J. Seroprevalence of antibodies against pestiviruses in small ruminants in the Netherlands. Tijdschr. Diergeneeskd. 2009, 134, 380–384. [Google Scholar]
- Leskova, V.; Jackova, A.; Vlásákova, M.; Vilček, S. Genetic characterization of a border disease virus isolate originating from Slovakia. Acta Virol. 2013, 57, 17–25. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hurtado, A.; García-Pérez, A.L.; Aduriz, G.; Juste, R.A. Genetic diversity of ruminant pestiviruses from Spain. Virus Res. 2003, 92, 67–73. [Google Scholar] [CrossRef]
- Hurtado, A.; Aduriz, G.; Gómez, N.; Oporto, B.; Juste, R.A.; Lavin, S.; Lopez-Olvera, J.R.; Marco, I. Molecular Identification of a New Pestivirus Associated with Increased Mortality in the Pyrenean Chamois (Rupicapra pyrenaica pyrenaica) in Spain. J. Wildl. Dis. 2004, 40, 796–800. [Google Scholar] [CrossRef]
- Berriatua, E.; Barandika, J.; Aduriz, G.; Atxaerandio, R.; Garrido, J.; Garcıa-Perez, A.L. Age-specific seroprevalence of Border disease virus and presence of persistently infected sheep in Basque dairy-sheep flocks. Vet. J. 2004, 168, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Berriatua, E.; Barandika, J.; Aduriz, G.; Hurtado, A.; Estevez, L.; Atxaerandio, R.; García-Pérez, A. Flock-prevalence of border disease virus infection in Basque dairy-sheep estimated by bulk-tank milk analysis. Vet. Microbiol. 2006, 118, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Marco, I.; Rosell, R.; Cabezon, O.; Mentaberre, G.; Casas, E.; Velarde, R.; Lopez-Olvera, J.R.; Hurtado, A.; Lavín, S. Epidemiological study of border disease virus infection in southern chamois (Rupicapra pyrenaica) after an outbreak of disease in the pyrenees (NE Spain). Vet. Microbiol. 2008, 127, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Cabezon, O.; Rosell, R.; Velarde, R.; Mentaberre, G.; Casas-Diaz, E.; Lavin, S.; Marco, I. Border disease virus shedding and detection in naturally infected Pyrenean chamois (Rupicapra pyrenaica). J. Vet. Diagn. Invest. 2010, 22, 744–747. [Google Scholar] [CrossRef] [PubMed]
- Giangaspero, M. Genetic variation of Border disease virus species strains. Vet. Ital. 2011, 47, 415–435. [Google Scholar]
- Marco, I.; Cabezón, O.; Velarde, R.; Fernández-Sirera, L.; Colom-Cadena, A.; Serrano, E.; Rosell, R.; Casas-Díaz, E.; Lavín, S. The two sides of border disease in Pyrenean chamois (Rupicapra pyrenaica): Silent persistence and population collapse. Anim. Health Res. Rev. 2015, 16, 70–77. [Google Scholar] [CrossRef]
- Marco, I.; Cabezon, O.; Rosell, R.; Fernandez-Sirera, L.; Allepuz, A.; Lavin, S. Retrospective study of pestivirus infection in Pyrenean chamois (Rupicapra pyrenaica) and other ungulates in the Pyrenees (NE Spain). J. Vet. Microbiol. 2011, 149, 17–22. [Google Scholar] [CrossRef]
- Fernandez-Sirera, L.; Cabezon, O.; Dematteis, A.; Rossi, L.; Meneguz, P.G.; Gennero, M.S.; Allepuz, A.; Rosell, R.; Lavın, S.; Marco, I. Survey of Pestivirus infection in wild and domestic ungulates from south-western Italian Alps. Eur. J. Wild. Res. 2012, 58, 425–431. [Google Scholar] [CrossRef]
- Schaller, P.; Vogt, H.R.; Strasser, M.; Nettleton, P.F.; Peterhans, E.; Zanoni, R. Seroprevalence of maedi-visna and border disease in Switzerland. Schweiz. Arch. Tierheilkd. 2000, 142, 145–153. [Google Scholar]
- Stalder, H.P.; Meier, P.; Pfaffen, G.; Wageck-Canal, C.; Rufenacht, J.; Schaller, P.; Bachofen, C.; Marti, S.; Vogt, H.R.; Peterhans, E. Genetic heterogeneity of pestiviruses of ruminants in Switzerland. Prev. Vet. Med. 2005, 72, 37–41. [Google Scholar] [CrossRef]
- Braun, U.; Bachofen, C.; Büchi, R.; Hässig, M.; Peterhans, E. Infection of cattle with Border disease virus by sheep on communal alpine pastures. Schweizer Archiv. für Tierheilkunde 2013, 155, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Braun, U.; Hilbe, M.; Janett, F.; Hässig, M.; Zanoni, R.; Frei, S.; Schweizer, M. Transmission of border disease virus from a persistently infected calf to seronegative heifers in early pregnancy. BMC Vet. 2015, 11, 43. [Google Scholar] [CrossRef]
- Barlow, R.M.; Patterson, D.S.P. Border Disease of sheep: A virus-induced teratogenic disorder. Adv. Vet. Med. Suppl. J. Vet. Med. 1982, 90, 87–90. [Google Scholar]
- Li, W.L.; Mao, L.; Zhao, Y.Q.; Sun, Y.H.; He, K.W.; Jiang, J.Y. Detection of border disease virus (BDV) in goat herds suffering diarrhea in eastern China. Virol. J. 2013, 10, 80. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Liu, X.; Li, W.; Yang, L.; Zhang, W.; Jiang, J. Characterization of one sheep border disease virus in China. Virol. J. 2015, 12, 15. [Google Scholar] [CrossRef] [PubMed]
- Mishra, N.; Rajukumar, K.; Vilček, S.; Kalaiyarasu, S.; Behera, S.P.; Dubey, P.; Nema, R.K.; Gavade, V.B.; Dubey, S.C.; Kulkarni, D.D. Identification and molecular characterization of border disease virus (BDV) from sheep in India. Comp. Immunol. Microbiol. Infect. Dis. 2016, 44, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Anoyatbekova, A.M.; Alexeyenkova, S.V.; Yurov, K.P. Molecular-evolutionary genetic analysis of the Border disease virus detected in sheep in Tajikistan. Veterinariya 2017, 1, 23–26. [Google Scholar]
- Ridpath, J.F.; Bolin, S.R. Comparison of the complete genomic sequence of the border disease virus, BD31, to other pestiviruses. Virus Res. 1997, 50, 237–243. [Google Scholar] [CrossRef]
- Osburn, B.I.; Crenshaw, G.L.; Jackson, T.A. Unthriftiness, hairy fleece and tremors in newborn lambs. J. Am. Vet. Med. Assoc. 1972, 160, 442–445. [Google Scholar]
- Tadich, N.; Nettleton, P.F.; Morgan, K.L.; Hodgson, A.; Macaulay, R.; Reinhardt, G.; Riedemann, S. Seroprevalencia de Border disease en ovejerías del sur de Chile. Arch. Med. Vet. 1998, 30, 191–196. [Google Scholar] [CrossRef]
- Tegtmeier, C.; Stryhn, H.; Uttenthal, A.; Kjeldsen, A.M.; Nielsen, T.K. Seroprevalence of border disease in Danish sheep and goat herds. Acta Vet. Scand. 2000, 41, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Hemmatzadeh, F.; Boardman, W.; Alinejad, A.; Hematzade, A.; Moghadam, M.K. Molecular and Serological Survey of Selected Viruses in Free-Ranging Wild Ruminants in Iran. PLoS ONE 2016, 11, e0168756. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.D. Prevalence of border disease virus in sheep and goats in Mosul, Iraq. Iraqi J. V. Sci. 2021, 35, 257–262. [Google Scholar] [CrossRef]
- Graham, D.A.; Calvert, V.; German, A.; Mc Cullough, S.J. Pestiviral infections in sheep and pigs in Northern Ireland. Vet. Rec. 2001, 148, 69–72. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, R.G.; O’Connor, M.; O’Reilly, P.J. A survey of antibodies to pestivirus in sheep in the Republic of Ireland. Ir. Vet. J. 2004, 57, 525–530. [Google Scholar] [CrossRef]
- Rivera, H.; Pezo, D.; García, W. Detección de anticuerpos contra pestivirus en rumiantes de una comunidad campesina de la provincia de Canchis, Cusco. Rev. Inv. Vet. Perú 2002, 13, 46–51. [Google Scholar]
- Llancares, N.; Rivera, H.; Arainga, M.; Falcón, N. Seroprevalencia de pestivirus de rumiantes en ovinos reproductores de una empresa de la sierra central del Perú. Rev. Inv. Vet. Perú 2012, 23, 504–509. [Google Scholar] [CrossRef]
- Kautto, A.H.; Alenius, S.; Mossing, T.; Becher, P.; Belak, S.; Larska, M. Pestivirus and alphaherpesvirus infections in Swedish reindeer (Rangifer tarandus tarandus L.). Vet. Microb. 2012, 156, 64–71. [Google Scholar] [CrossRef]
- Paton, D.J.; Christiansen, K.; Alenius, S. Prevalence of antibodies to bovine virus diarrhoea virus and other viruses in bulk tank milk in England and Wales. Vet. Rec. 1998, 142, 385–391. [Google Scholar] [CrossRef]
- Feknous, N.; Hanon, J.B.; Tignon, M.; Khaled, H.; Bouyoucef, A.; Cay, B. Seroprevalence of border disease virus and other pestiviruses in sheep in Algeria and associated risk factors. BMC Vet. Res. 2018, 14, 339. [Google Scholar] [CrossRef]
- Fihri, O.F.; Jammar, N.; Amrani, N.; El Berbri, I.; Alali, S. Sheep pestivirus in Morocco: Sero-epidemiological and molecular study. Vet. Rec. Open 2019, 6, e000324. [Google Scholar]
- Guidoum, K.A.; Benallou, B.; Pailler, L.; Espunyes, J.; Napp, S.; Cabezón, O. Ruminant pestiviruses in North Africa. Prev. Vet. Med. 2020, 184, 105156. [Google Scholar] [CrossRef] [PubMed]
- Ataseven, V.S.; Ataseven, L.; Tan, T.; Babür, C.; Oguzoglu, T.C. Seropositivity of agents causing abortion in local goat breeds in Eastern and South-eastern Anatolia, Turkey. Revue Méd. Vét. 2006, 157, 545–550. [Google Scholar]
- Bazzucchi, M.; Pierini, I.; Giammarioli, M.; De Mia, G.M. Near-complete nucleotide sequence of a border disease virus genotype BDV-7 isolate from Italy. Arch. Virol. 2020, 165, 3007–3009. [Google Scholar] [CrossRef] [PubMed]
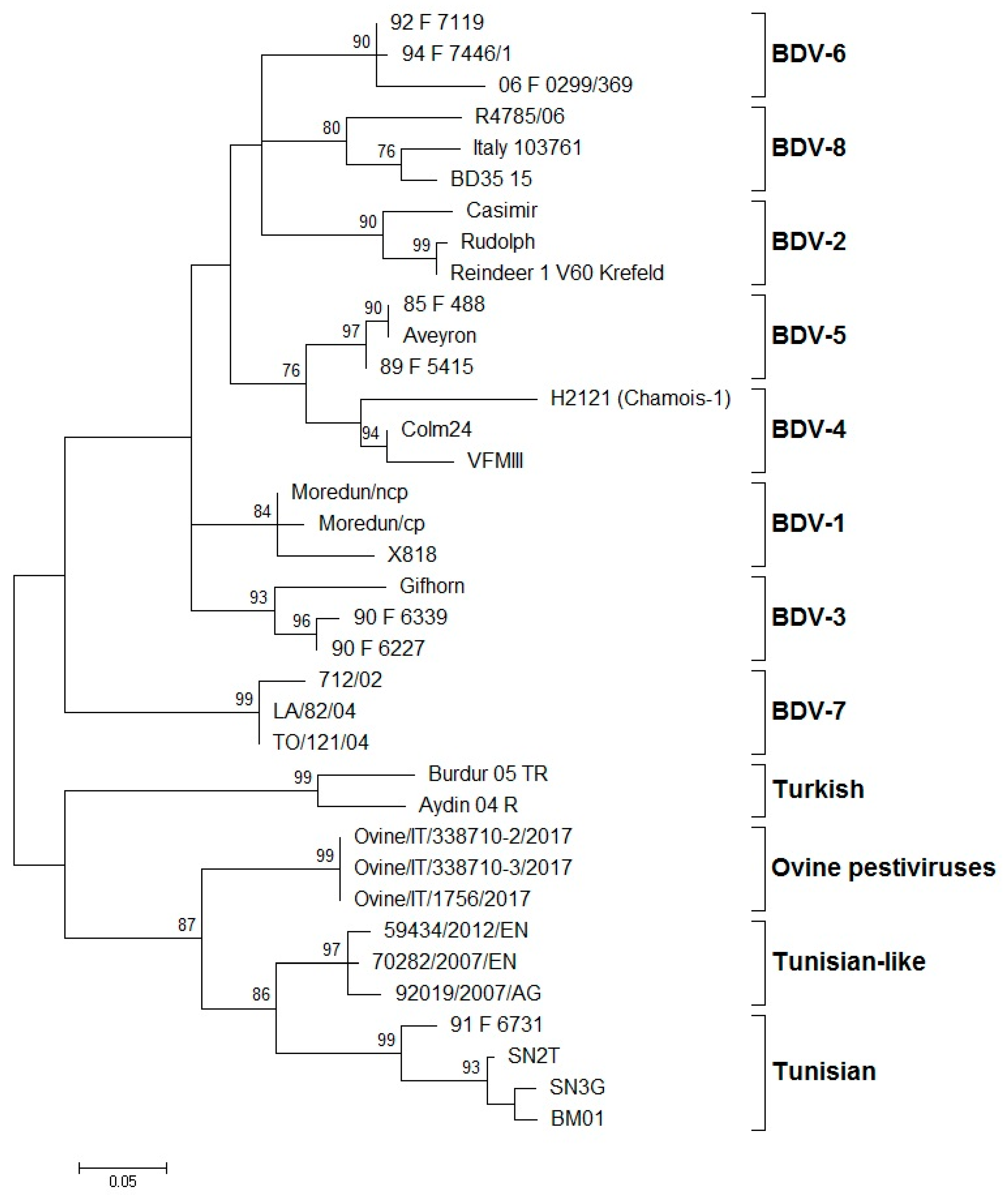

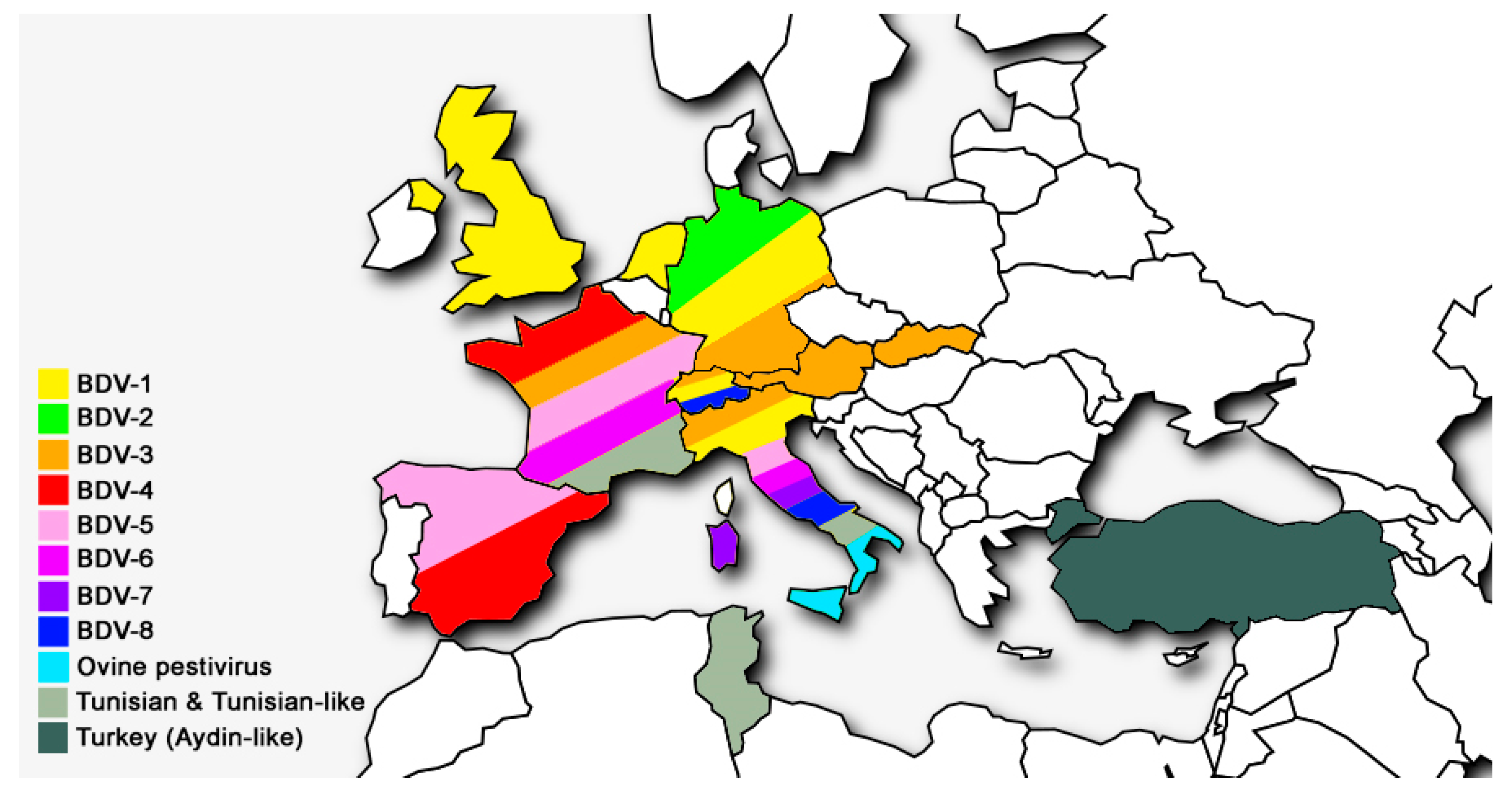
| Genotypes of BDV | Host Origin of Isolates | Country of Isolation | References |
|---|---|---|---|
| BDV-1 | Alpaca, llama, sheep, cattle, goat, pig | Australia, Germany, Italy, Japan, Mexico, Netherlands, New Zealand, Switzerland, UK, USA | [28,40,42,43] |
| BDV-2 | Reindeer, sheep, wisent | Germany | [12,44] |
| BDV-3 | Sheep, goat, cattle | Austria, China, France, Germany, India, Italy, Slovakia, Switzerland | [14,31,45,46,47] |
| BDV-4 | Chamois, sheep, pig | France, Spain | [7,13,27,48] |
| BDV-5 | Sheep, goat | France, Italy, Spain | [6,14,45] |
| BDV-6 | Sheep, chamois | France, Italy | [14,41] |
| BDV-7 | Sheep, goat | Italy | [15,49,50] |
| BDV-8 | Goat, chamois, sheep, cattle, pig | Italy, Switzerland | [16,51,52,53] |
| Turkey (Aydin-like) | Sheep, goat | Turkey | [19] |
| Tunisian and Tunisian-like | Sheep, goat | France, Italy, Tunisia | [17,18] |
| Ovine pestivirus | Sheep | Italy | [22] |
| Country of Isolation | Genomic Region | Year of Isolation | Pestivirus D | Pestivirus I | Unassigned Pestiviruses | References | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BDV-1 | BDV-2 | BDV-3 | BDV-4 | BDV-5 | BDV-6 | BDV-7 | BDV-8 | Turkey (Aydin-Like) | Tunisian and Tunisian-Like | Ovine Pestivirus | ||||
| Australia | E1, E2 | 1973 | 2 | - | - | - | - | - | - | - | - | - | - | [28,70,92] |
| Austria | 5′-UTR, Npro | 2003–2014 | - | - | 29 | - | - | - | - | - | - | - | - | [30,32,36,46,79,80] |
| China | 5′-UTR, Npro | 2012 | - | - | 4 | - | - | - | - | - | - | - | - | [101,102] |
| France | 5′-UTR, Npro | 1984–2010 | - | - | 7 | 2 | 5 | 11 | - | - | - | 3 | - | [4,5,14,41,81] |
| Germany | Npro, E1, E2 | 1985–2001 | 1 | 6 | 1 | - | - | - | - | - | - | - | - | [12,39,44,82,83] |
| India | 5′-UTR, Npro, E2 | 2009–2010 | - | - | 1 | - | - | - | - | - | - | - | - | [103] |
| Italy | 5′-UTR, Npro | 2002–2018 | 3 | - | 7 | - | 1 | 1(*) | 14 | 2 | - | 3 | 4 | [15,16,18,22,23,31,45,47,49,50,52] |
| Japan | 5′-UTR, Npro, E2 | 2012 | 4 | - | - | - | - | - | - | - | - | - | - | [25,26] |
| Mexico | 5′UTR | 2016 | 3 | - | - | - | - | - | - | - | - | - | - | [37] |
| Netherlands | 5′-UTR | 1994 | 1 | - | - | - | - | - | - | - | - | - | - | [24,28] |
| New Zealand | 5′-UTR, Npro | 1990–2010 | 5 | - | - | - | - | - | - | - | - | - | - | [34,42,72] |
| Slovakia | 5′-UTR, Npro, E2 | 2007 | - | - | 1 | - | - | - | - | - | - | - | - | [85] |
| Spain | 5′-UTR, Npro, E2 | 1996–2011 | - | - | - | 83 | 5 | - | - | - | - | - | 1 | [1,6,7,13,27,48,86,87,89,90,91,92,93] |
| Switzerland | 5′-UTR, Npro | 2001–2006 | 1 | - | 1 | - | - | - | - | 5 | - | - | - | [35,51,53,92,97,99] |
| Tajikistan | 5′-UTR, Npro, E2 | 2017 | - | - | 1 | - | - | - | - | - | - | - | - | [104] |
| Tunisia | 5′-UTR, Npro | 1995–2000 | - | - | - | - | - | - | - | - | - | 10 | - | [17,92] |
| Turkey | 5′-UTR, Npro | 2004 | - | - | - | - | - | - | - | - | 2 | - | - | [19,20,21] |
| United Kingdom | 5′-UTR, Npro, E2, E1 | 1976–2008 | 28 | - | - | - | - | - | - | - | - | - | - | [12,33,72] |
| USA | 5′-UTR, Erns, E1 | 1978–2012 | 2 | - | - | - | - | - | - | - | - | - | - | [43,65,105] |
| Total Number | 50 | 6 | 52 | 85 | 11 | 12 | 14 | 7 | 2 | 16 | 5 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Righi, C.; Petrini, S.; Pierini, I.; Giammarioli, M.; De Mia, G.M. Global Distribution and Genetic Heterogeneity of Border Disease Virus. Viruses 2021, 13, 950. https://doi.org/10.3390/v13060950
Righi C, Petrini S, Pierini I, Giammarioli M, De Mia GM. Global Distribution and Genetic Heterogeneity of Border Disease Virus. Viruses. 2021; 13(6):950. https://doi.org/10.3390/v13060950
Chicago/Turabian StyleRighi, Cecilia, Stefano Petrini, Ilaria Pierini, Monica Giammarioli, and Gian Mario De Mia. 2021. "Global Distribution and Genetic Heterogeneity of Border Disease Virus" Viruses 13, no. 6: 950. https://doi.org/10.3390/v13060950
APA StyleRighi, C., Petrini, S., Pierini, I., Giammarioli, M., & De Mia, G. M. (2021). Global Distribution and Genetic Heterogeneity of Border Disease Virus. Viruses, 13(6), 950. https://doi.org/10.3390/v13060950






