Abstract
Bacteriophages are promising tools for the detection of fecal pollution in different environments, and particularly for viral pathogen risk assessment. Having similar morphological and biological characteristics, bacteriophages mimic the fate and transport of enteric viruses. Enteric bacteriophages, especially phages infecting Escherichia coli (coliphages), have been proposed as alternatives or complements to fecal indicator bacteria. Here, we provide a general overview of the potential use of enteric bacteriophages as fecal and viral indicators in different environments, as well as the available methods for their detection and enumeration, and the regulations for their application.
1. Introduction
Life on our planet cannot exist without water and it is estimated that 50% of the global human population lives close to rivers, lakes, or oceans [1]. Besides its importance in maintaining health and hygiene, water is also essential for economic and productive activities (such as agriculture, industry, tourism, transportation, etc.), recreation and leisure (swimming pools, fountains, etc.), and the preservation and restoration of natural ecosystems. Therefore, a decrease in water quality due to pollution poses a risk to human wellbeing and the natural environment.
One of the main sources of water pollution is the discharge of human and animal fecal waste. If poorly managed, effluents of wastewater treatment plants (WWTPs) and industrial and livestock wastes can spread enteric pathogens, including viruses, into aquatic environments. Though crucial for human health and development, the microbiological quality of water is difficult to control, due to the variety of existing waterborne pathogens, the lack of effective methods, and the analytical and logistics costs required to detect such a high number of pathogens (parasites, bacteria and viruses) [2]. The definition of indicator, index, and model microorganisms, more than a century ago, allowed such limitations to be overcome by ensuring a sufficiently appropriate control of water quality.
Despite their limitations, fecal indicator microorganisms represent a useful tool to monitor the microbiological quality of water, where their presence is a sign of fecal contamination and potentially the existence of pathogens. The most commonly used are fecal indicator bacteria (FIB), including total coliforms, fecal coliforms, Escherichia coli, streptococci and enterococci [3].
One of their drawbacks is that they do not provide information on the source of fecal contamination, being frequently found in the microbiota of many animals. Moreover, they correlate poorly with human viruses or parasites pathogens in natural aquatic environments and WWTPs, displaying different behavior and lower survival rates [3].
Recently, bacteriophages capable of infecting enteric bacteria have been proposed as alternative indicators of fecal and viral pollution. Bacteriophages have several advantages over bacterial indicators, as they are more abundant and generally more persistent in the environment and can provide more accurate information about viral pathogens. Bacteriophages serve as useful indicators of fecal contamination, as they are eliminated in feces, and do not replicate in a natural environment unless their host is present and metabolically active [4]. They can also indicate viral contamination, as bacteriophages infecting intestinal bacteria spread into the environment in a similar way to enteric viral pathogens and have similar fates and survival patterns [5]. Monitoring the presence of every specific viral pathogen is impracticable for routine control purposes. Besides the technical difficulties, it would be extremely time-consuming and prohibitively expensive, especially for those countries in most urgent need of efficient water quality control. Therefore, easy-to-detect bacteriophages have been proposed as indicators of fecal and viral pollution and are now included in multiple water quality regulations and guidelines worldwide [6].
Presented here is a general overview of the potential of bacteriophages as fecal pollution indicators, not only in water environments, but also in a range of solid matrices.
2. Families of Bacteriophages Used as Indicators of Fecal Pollution
Bacteriophages capable of infecting enteric bacteria are generally classified into three taxonomically diverse groups: somatic coliphages, F-specific coliphages, and bacterio-phages capable of infecting Bacteroides spp. [7,8]. Though less common, enterophages (bacteriophages capable of infecting Enterococcus spp.) also have valuable potential as indicator organisms due to their high concentrations in wastewater, similar survival rates to enteric viruses, and differential prevalence in human or animal gut microbiota [9,10]. Nevertheless, their suitability has only been tested in tropical regions and further studies are necessary [5,11].
Somatic coliphages are a heterogeneous group of bacteriophages capable of infecting E. coli and other coliform bacteria through the cell wall after becoming attached to specific receptors on the outer membrane [12]. Under optimal physiological conditions, lysis occurs approximately 30 min after attachment, and between 100 and 1000 of progeny are released per infected cell [13]. Four major families of somatic coliphages have been described in polluted wastewaters; the most abundant are the Myoviridae and Siphoviridae, followed by the Podoviridae and Microviridae [14]. The phage families differ in morphology and resistance to inactivation factors. The Microviridae phages differ genetically from the other three families in having single-stranded (ss) rather than double-stranded (ds) DNA. Microviridae phages have tailless isometric capsids of 25–30 nm; those of the Myoviridae family have capsids of up to 100 nm and a long contractile tail; the isometric capsids of Siphoviridae phages are up to 60 nm and have long non-contractile tails; and Podoviridae phages have isometric capsids of up to 65 nm with short tails. The somatic coliphages most commonly used as model organisms in research are ΦX174, T2, and T7 [8].
Due to the availability of new genetic data, phage classification by the ICTV is currently undergoing a major overhaul, with new families being described and existing ones divided. Following this reclassification, bacteriophages capable of infecting E. coli from the Caudovirales order (dsDNA viruses) now include the new families Ackermannviridae, Autographviridae, Chaseviridae, Demerecviridae, and Drexlerviridae [15,16]. As the contribution of the recently described families to phage presence in the environment has still not been clearly defined, the initial four groups of somatic coliphages (Myoviridae, Siphoviridae, Podoviridae, and Microviridae) [14] remain valid, as they are distinguished on a morphological basis. In contrast, some of the new families, defined by genetic differences, are indistinguishable morphologically (e.g., Ackermannviridae and Chaseviridae are myophages, Autographviridae are podophages, Drexlerviridae and Demerecviridae are siphophages).
Somatic coliphages are the most abundant group of indicator bacteriophages in almost all environmental samples [7]. They have less resistance to disinfectants such as UV light than other bacteriophages, but more than bacterial indicators [13]. Their potential replication in environments outside the gut was once a concern [17,18], but has proved to be negligible for several reasons: their narrow host range; the high concentrations of host and phages required; the possible interference of accompanying microbiota and other particles with the replicative process; the low metabolic activity of hosts in environmental conditions; and responses to environmental stresses possibly minimize phage infection [19,20,21]. It has also been proven that neither coliphages isolated from wastewater nor laboratory stock coliphages are capable of replicating in natural samples, even in tropical climates with more conducive temperatures. It can therefore be concluded that the proportion of somatic coliphages in a natural sample arising from replication in the environment will be practically null [4,22].
F-specific bacteriophages are the second most widespread indicator phages in the environment [7]. They are capable of infecting E. coli and other coliform bacteria through the sexual pili, encoded in the F-plasmid, which is transferable to enteric bacteria via conjugation [23]. This group includes two phage families: the Leviviridae, ssRNA phages with isometric tailless capsids of around 25 nm, and the Inoviridae, ssDNA phages with flexible filamentous capsids of 800 nm [24]. The use of RNase in culture methods permits the differentiation between F-DNA (Inoviridae) and F-RNA (Leviviridae) coliphages. Based on nucleotide analysis, the Leviviridae are divided into two genera, Levivirus and Allolevirus [25], which include four different genotypes of F-RNA phages: those of subgroups I and II belong to Levivirus and subgroups III and IV belong to Allolevirus genera [26,27]. Model coliphages representing these groups are MS2 and f2 from genotype I, GA from genotype II, Qβ from genotype III, and FI from genotype IV. The study of these different subgroups is particularly useful for identifying the origin of fecal contamination, since genotypes I and IV predominate in waters contaminated by animal residues whereas genotypes II and III are commonly associated with human contamination [28], though these associations do not always hold, and cross-reactivity has been recorded [29]. F-RNA phages are typically more abundant than F-DNA phages (for instance, 90–95% of F-specific coliphages in wastewater are F-RNA phages) and morphologically more similar to enteric viruses [30,31]. The replication of F-specific bacteriophages outside the gut is considered extremely improbable, as sexual pili cannot be synthesized under 32 °C [32].
F-specific bacteriophages can perform more accurately as indicators in samples where they predominate, such as groundwater, clay sediments, and reclaimed waters; they are also useful for monitoring water treatments such as UV disinfection [33,34,35]. In contrast, they have a low persistence in surface waters, especially in warmer climates, and are readily inactivated by heat or high pH [13]. Therefore, a combination of both types of coliphages may be preferable in some types of samples. Total coliphages can be determined by either summing the results of somatic and F-specific coliphage detection assays or using a host strain that determines both in only one assay [36].
The third group of bacteriophages proposed as indicators infect Bacteroides spp. and their concentrations in feces or fecally contaminated samples are usually lower compared to coliphages [37]. Most of these morphologically homogenous bacteriophages belong to the Siphoviridae family and infect bacteria through receptors in the cell wall [38,39]. They have a narrow host range, with a high specificity for the cell wall receptors of a particular host [40]. Infectivity seems to be limited by the amount of bacterial capsules, which hamper phage access to the receptors [41]. Bacteroides spp. strains are commonly used for microbial source tracking (MST) as they are strongly associated with a given human or animal host and differ in their capacity to recover phages from samples fecally contaminated by different species [41,42]. In general, their utility as MST markers also depends on the geographical location [37,43]. For example, in southern Europe, strain B. thetaiotaomicron GA17 can detect phages of human fecal origin, unlike other strains such as B. fragilis RYC2056 or HB13 [44] yet in the UK, a B. fragilis strain (GB-124) isolated in Brighton [45] was more efficient in this respect. Other examples of geographical variability are strains B. fragilis HSP40 [42], B. fragilis HB13 [46], and B. thetaiotaomicron ARABA 84 [47].
Other strains such as B. thetaiotaomicron CW18, B. fragilis PG76, PL122, and PZ8 have been isolated and used to detect phages as markers of bovine, porcine, and aviary fecal contamination [48,49]. Despite their low concentrations in water, Bacteroides-infecting phages are more resistant to most inactivating factors and treatments than coliphages. Their replication outside the gut is even more improbable, as the host strains are strictly anaerobic and require specific nutrients, such as hemin, that are scarcely found in the environment [42].
Metagenomic studies using sequences from fecal samples available in databases discovered the most abundant phage in the human fecal virome, named crAssphage (cross assembly phage) [50]. Sequence similarities pointed to a group of Bacteroides-infecting phages with short non-contractile tails from the Podoviridae family [50,51], later confirmed by isolation through culture methods. The morphology of the first isolated crAssphage in B. intestinalis, ΦcrAss001, was compatible with Podoviridae viruses [52]. One of the main characteristics of ΦcrAss001 is its peculiar replicative cycle; although seemingly a virulent bacteriophage, it can coexist in apparent equilibrium with its host without causing cell lysis, which might benefit both bacteria and virus in a strongly competitive environment like the gastrointestinal tract. The recently isolated crAssphage species such as ΦcrAss002 seem to follow a similar replicative pattern. Efforts to obtain lysogenic ΦcrAss001 have failed so far, though other crAssphage have integrases compatible with lysogenic cycles in their genome [53]. Therefore, expanding our knowledge about crAssphage replication could help to promote their use as fecal indicators with culture techniques [54,55]. CrAssphage have potential application as MST markers, being highly specific to humans and having an extensive geographical distribution and no seasonal variation. They are abundantly detected in human feces (constituting about 90% of the human gut virome), and in sewage throughout the year, as well as in mussels and sediments collected in areas contaminated with wastewater [56,57]. Nevertheless, crAssphage have also been found in several animal sources, so further research is required on possible animal-associated variants or specific genome regions more suitable for animal source discrimination [58,59]. They also have stronger environmental persistence than bacteria and higher concentrations than enteric viruses in sewage worldwide, allowing a more accurate description of virus removal [60,61,62]. These characteristics make crAssphage a very promising alternative as indicator microorganisms of viral fecal pollution, which could be used in MST for monitoring human fecal pollution of water [63,64,65].
3. Methods to Detect Bacteriophages
Strategies for detecting phages in samples can be culture-dependent or molecular, each with its own advantages, disadvantages, and appropriate applications.
3.1. Culture-Dependent Methods
Culture-dependent methods, available since phages were first discovered, provide qualitative or quantitative information about infectious phages in samples [66]. These methods have already been registered as standardized protocols, mainly by two regulatory bodies: the International Standardization Organization (ISO) and the United States Environment Protection Agency (Washington, DC, USA, U.S. EPA). The ISO provides standardized methodologies for detecting somatic [67], F-specific [68], and Bacteroides-infecting phages [69] (ISO 10705), each of which includes two different approaches: a spot test (a qualitative presence/absence protocol that can be adapted to quantitative results using a most probable number approximation) and a double agar layer (DAL) assay (a quantitative protocol for counting plaque-forming units (PFU) in samples). ISO methods can be easily implemented in routine microbiology laboratories without previous experience in working with phages [70], and provide optional steps for laboratories with limited equipment, and quality control assays. U.S. EPA standardized protocols for detecting somatic and F-specific coliphages [71,72] also include two different methods compatible with both types of coliphages: Method 1601 (a quantitative method based on single agar layer (SAL) assays for PFU enumeration) and Method 1602 (a qualitative method based on presence/absence assays), both of which have been successfully validated, have simplified versions [73,74,75], and have been recently revised in Methods 1642 and 1643 [76,77]. Due to the lack of a specific standardized protocol, total coliphage detection is performed using methodologies for F-specific coliphages [36].
ISO and U.S. EPA employ different host strains for the targeted phages, but their equivalent protocols usually give similar counts [36,78,79]. Host strains derived from E. coli C are reported to provide the highest counts of somatic coliphages [80]. Both regulatory bodies use nalidixic acid-resistant variants of this strain: E. coli CN13 (BCRC17137, ATCC 700609) in U.S. EPA, and E. coli WG5 (CIP 107680, ATCC 700078) in ISO methods [67,71,72]. Nalidixic acid-resistant strains were selected to minimize the growth of accompanying microbiota, which frequently interfere with the visualization of plaques. Otherwise, a previous filtration step is required, using membrane filters of 0.22 μm pore diameter made of materials that do not adsorb proteins. To detect F-specific coliphages, host strains must express the sexual pili, encoded in the F plasmid or F-derived plasmids; those in current use are Salmonella enterica WG49 (NCTC 12484, CECT 4625, ATCC 700730) [81] and E. coli HS/FAmp (ATCC 700891) [82] in ISO and U.S. EPA methods, respectively. Both strains have markers for improving strain selection and stability: Ampicilin resistance (E. coli HS/Famp) and lactose degradation capacity (S. enterica WG49) [68,71,72].
Host strains initially proposed for the F-specific protocols could also detect somatic coliphages and were suggested for the monitoring of total coliphages, although a standardized culture method has not been described. More efficient strains have been subsequently developed for this purpose: E. coli C3000 (ATCC 15597), which is mainly used in the U.S., detects lower amounts of somatic coliphages than the standardized strains, and E. coli CB390 (CECT9198), which can recover both groups of coliphages with similar efficiency to its standardized counterparts [36,83]. The standardized method for detecting Bacteroides-infecting bacteriophages uses B. fragilis RYC2056 (ATCC 700786) as a host strain, although other strains can be employed to discriminate between human and animal fecal pollution [69,84].
Standardized culture methods are simple, robust, cost-effective, and easily prepared, especially for coliphages, which do not require anaerobic growth conditions. The methods can be scaled to different sample volumes, maintaining the same proportions between medium, host strain, and sample. The material, media, reagents and labor have a similar cost to the methods currently used in routine analysis laboratories to detect fecal coliforms/E. coli. Costs may increase by 10–15% if an additional step with RNAase is required for the recovery of F-RNA and F-DNA coliphage subgroups, or due to the longer incubation times required for anaerobic Bacteroides spp. [8]. The standard methods could be optimized further to improve phage recovery and the cost/benefit relationship; modifications could include diluting the medium concentration, substituting components, and optimizing incubation protocols [79,85].
In general, standardized methods are time-consuming, requiring more than one working day to obtain reliable results (at least 18 h for coliphage plaques and more than 48 h for Bacteroides bacteriophages). To prevent potentially virally contaminated water being used for consumption, irrigation, or recreation [86,87], the results need to be obtained on the same day as the analysis. Shorter operative times and incubation periods, as well as more user-friendly handling, are also warranted by the increasing implementation of coliphages in guidelines and regulations. Several modified standardized methods have already been developed in this respect [88], the most promising being Easyphage and Quantiphage, which incorporate non-agar-based supports for plaque counting and previously prepared components for greater speed and simplicity [89,90]. Other promising modifications are based on the detection of enzymatic lysis in liquid cultures, focusing primarily on the activity of β-galactosidase [91], adenylate kinase [92], and β-glucuronidase [93]. Among these promising fast methodologies, three have commercial application: Fastphage (already validated in U.S. EPA methods), Quantiphage and Bluephage (currently in development). Fastphage and Bluephage use the activity of a liberated intracellular enzyme as an indicator of cell lysis (β-galactosidase and β-glucuronidase, respectively), the presence/absence of phages indicated by color changes, while Quantiphage incorporates cellulose supports to achieve a more rapid plaque detection [88].
3.2. Molecular Methods
Molecular methods, although fast and sensitive, have a major drawback in that they cannot provide information about infectivity, which therefore requires additional steps. Without infectivity data, viral concentration, and human health risks are often overestimated [94]. Molecular methods can be serological or involve nucleic acid-based or microelectronic sensors.
Serological techniques are rapid and can be applied in situ, but they require pre-enrichments, and antisera are less available than nucleic acid probes and primers [8]. They are mainly applied to detect F-specific coliphages, using latex agglutination or neutralization methods [95], being less suitable for the highly diverse and complex somatic coliphages, though CLAT- (Culture, Latex Agglutination, and Typing) based analyses have been developed for some specific families [96].
Nucleic acid methods are based on plaque hybridization, employing specific probes or, more frequently, qPCR/RTqPCR assays. They are mainly used to detect F-specific and Bacteroides-infecting phages [8], and, as with serological methods, have limited applications for somatic coliphages, though PCR and qPCR techniques have been developed for specific families or bacteriophages [96]. In plaque hybridization methods, specific probes designed for each targeted phage or phage group are applied to plaques obtained by culture [26]. RT-qPCR analyses are used to quantify the number of genome copies (GC) present in a given sample. The quantities detected by molecular methods tend to be higher compared with culture methods, as GC signals are more persistent in the environment and more resistant to treatments than infectious viruses [29]. In order to solve this discrepancy, nucleic acid amplification techniques based on the membrane or capsid integrity have been developed. However, membrane integrity does not equate with viability and therefore cannot serve as a control of the efficacy of inactivation mechanisms that do not directly target cell membranes [97,98]. PCR-based approaches can also be inhibited by organic substances such as phenolic compounds, which are occasionally present in environmental samples [99]. Molecular methods are available for F-specific coliphage detection, but solely for genogroups of F-RNA-specific coliphages or specific phages such as MS2 [27,100]. PCR-based methods have also been developed to detect certain Bacteroides phages, facilitating the recovery of phages associated with a certain animal host [101]. As demonstrated by the discovery of crAssphage, metagenomics studies of the gut bacteria open the possibility of identifying sequences of new bacteriophages that infect non-cultivable host bacteria that are specific of certain species [37]. When used as fecal indicators, crAssphage are predominantly detected and quantified in environmental samples by qPCR assays [58,102,103]. Their isolation from environmental samples by lysis plaque formation using DAL is still difficult, due to the absence of appropriate hosts and a still unexplored biological replicative cycle [61]. CrAssphage are also suitable for fecal source discrimination and have been shown in comparative MST molecular qPCR assays to have advantages over existing bacterial markers (such as HF183/BacR287) in terms of specificity, accuracy and high sensitivity [58,102,103].
Microelectronic methods involve the detection of viral particles, or the lysis of host bacteria caused by bacteriophage infection [88]. Although fast, with results being obtained in less than 1 h, their sensitivity and precision do not yet match DAL-based methods [104,105,106]. Their usage is normally restricted to the detection of a specific phage, rather than to analyze environmental samples containing different phages at varying concentrations. Consequently, no microelectronic method so far has achieved a useful or feasible application to determine infectious bacteriophages as indicators of fecal pollution in environmental samples.
Viral concentration methods to optimize detection processes have been developed in parallel with standardized methodologies. For the analysis of larger volumes, the samples need to be concentrated, especially if quantification is required, as in drinking water samples with low levels of contamination. Two methods are recommended to concentrate volumes of up to 1000 mL, depending on the turbidity of the sample. When turbidity is low, a simple, inexpensive, and practical procedure is recommended, using mixed cellulose and acetate membrane filters with a pore size of 0.45 μm after the addition of salts and pH adjustment [107,108]. When turbidity is high, flocculation with magnesium hydroxide is feasible for all three groups of fecal indicator phages [109,110]. Furthermore, phages can also be concentrated by ultrafiltration, like other viruses [8].
4. Application of Bacteriophages as Indicators of Fecal Pollution
4.1. Bacteriophages as Fecal Indicators in Water
As already mentioned, FIB are used to estimate the microbiological quality of water, but they may not be suitable or sufficiently reliable to predict the presence of enteric viruses. In general, enteric viruses have higher survival rates during wastewater and drinking water treatment than bacterial indicators and greater persistence in environmental waters [17,34,110,111,112]. Therefore, the use of bacterial indicators alone could underestimate the microbiological contamination of water and the associated human health risks. Adding at least one viral indicator to the analysis provides a more accurate assessment of water quality and promotes more confidence in its safety.
The primary origin of coliphages in water environments are human and animal feces. They can reach water through raw or treated human and animal wastewater, septic tank overflow, sewer leakage, and the spread of solid waste (sewage sludge, slurry, manure, and the feces of pets, farmed animals, and wild animals) [7]. As early as 1948, Guelin [113] already saw the potential of coliphages as indicators of enteric microorganisms in water, observing their good correlation with the numbers of coliform bacteria in fresh and marine water. Since then, many other studies have assessed the potential of bacteriophages as indicators of fecal contamination in different water environments (Figure 1):
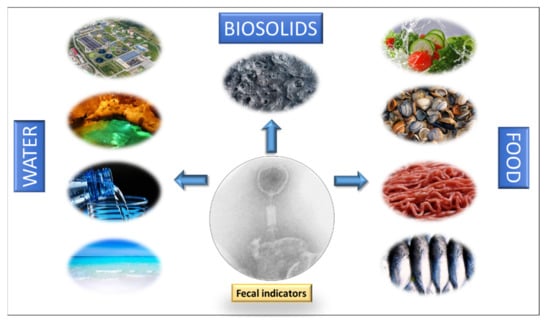
Figure 1.
Fields of application of bacteriophages as fecal indicators.
Wastewater treatment plants: Bacteriophages are considered to be useful tools to evaluate the efficacy of wastewater treatment plants [112,114,115,116] because their reduction by certain pathogen removal methods is similar to that of human enteric viruses [114,117] whereas the reduction of traditional FIB is significantly higher [5]. Coliphage concentration in wastewater shows no seasonality and remains consistently high throughout the year worldwide, as occurs with bacterial indicators [114,118,119]. Coliphage densities in wastewater are quite variable but the lower density of F-specific coliphages compared to somatic coliphages in both treated and untreated wastewater sources [120], potentially limits their use as indicators in this environment.
Drinking water: The presence of coliphages or Bacteroides-infecting phages in drinking water sources is a likely indicator of fecal contamination or an inadequate treatment [121]. Generally, the levels of bacterial indicators, viruses, and bacteriophages in these sources are low and seldom detected after treatment. The few reports describing the potential of phages to assess the quality of drinking water suggest once again that they outperform conventional FIB, undergoing less reduction after different drinking water treatments [122,123,124,125,126].
Recreational water: The sanitary quality of recreational waters is monitored using FIB according to the EU Directive 2006/7/EC [127], but alternative indicators, such as Clostridium perfringens and bacteriophages, have also been proposed [111]. Somatic coliphages are found in recreational water [128], and at beaches with unknown sources of fecal contamination, the presence of coliphages correlates with the occurrence of diseases more often than the presence of FIB [129,130]. It has been reported that in waters with detectable coliphages there was an increased incidence of gastrointestinal illness among bathers when fecal pollution was likely present, but not otherwise [131]. Compared with enterococci, the correlation was similar for somatic coliphages and even higher for F-specific coliphages [131]. These findings indicate that coliphages may be suitable for application as indicators of bathing water quality.
Groundwater: Groundwater constitutes an important fraction of the water used for household and municipal supplies, agriculture and landscape irrigation, and industry. Contaminants reaching surface waters also affect groundwater, the routes including failure in septic systems, leaking sewer lines and passage through soils and fissures. A study shows that one bacterial indicator and one phage indicator provide more information than two bacterial indicators when assessing the microbiological quality of groundwater [34]. However, as shown in Table 1, to date, only one regulation (2006) includes bacteriophages as indicators of enteric viral pollution in groundwater, reflecting that further research is needed in this area.

Table 1.
Current guidelines and regulations around the globe that include bacteriophages as indicators of fecal pollution [132,133,134,135,136,137,138,139,140,141,142,143,144].
4.2. Bacteriophages as Fecal Indicators in Solid Matrices
Solid or semisolid matrices play an important role in the persistence and dispersion of pathogens in the water cycle, as they can contain large amounts of pathogens, especially viruses, if contaminated with fecal waste [6,145].
To optimize the use of bacteriophages as indicators in solid matrices (Figure 1), standardized methodologies for their extraction, detection, and enumeration need to be developed. The current methods differ according to the matrix, which hinders the comparison of results. Nevertheless, in general, studies indicate that somatic coliphages are found in solid matrices at higher levels than traditional FIB and F-specific RNA coliphages. In addition, they persist longer in soils and sediments and are more resistant to sludge and manure treatments. For an extensive review of this question, and more data on solid matrices, see Martín-Díaz et al. [6].
4.3. Bacteriophages as Fecal Indicators in Food
Coliphages can be found in food when fecally contaminated water is used to grow vegetables and fruits, in meat processing, or to farm shellfish. Bivalve shellfish are regularly implicated in foodborne viral disease outbreaks because there is no effective way to rid them of viral contamination without changing their sensory characteristics. Instead, efforts are focused on preventing contamination. Shellfish accumulate and concentrate bacteriophages and viruses through their feeding process, and their depuration systems are more efficient for eliminating bacteria than viruses. Accordingly, Blanco-Picazo et al. found somatic coliphages in 70% of the tested shellfish samples but no E. coli [146]. However, the utility of phages as routine indicators of viral pollution in shellfish, in contaminated sites, or under normal growing conditions, is controversial; studies with conflicting results have been reviewed [147]. Regardless, the use of bacterial indicators alone is clearly insufficient to prevent viral disease outbreaks stemming from shellfish consumption [148] and more accurate data about enteric viruses could be provided by the addition of a viral indicator.
Regarding fish, a comparative study found somatic coliphages in Atlantic, farmed and frozen fish, with 30%, 10%, and 20% of the samples testing positive, respectively; in contrast E. coli was only found in 10% of the Atlantic fish samples [146].
Enteric bacteriophages have been proposed as potential fecal indicators in different types of meat. Hsu et al. found somatic coliphages in 88% of ground meat and poultry meat samples and F-specific coliphages in 63%. They also evaluated the risk of fecal contamination at three control points (evisceration, washing and chilling) and observed that the reduction of F-specific coliphages during these processing steps matched that of FIB [149]. Somatic coliphages have also been reported in minced pork, minced chicken, and ham, with 60%, 100%, and 40% of the samples testing positive, respectively. No E. coli was found in ham samples, and only 30% of minced pork and 90% of minced chicken samples tested positive for this bacterial indicator [150].
In a study of animal feeds, Maciorowski et al. analyzed animal feeds, feed ingredients, and poultry diets for the presence of coliphages, finding somatic and F-specific coliphages in all the tested samples, even after 14 months of storage at −20 °C [151].
Bacteriophages can also be used as fecal indicators in vegetables, as they have been found in lettuce and cucumber [152]. In lettuce, the number of samples positive for E. coli was slightly higher compared to somatic coliphages (50% and 40%, respectively); however, 20% of cucumber samples were positive for coliphages and none contained E. coli [152].
All these data suggest bacteriophages perform well as fecal indicators in food (Figure 1) as they seem to remain longer in the different food matrices than bacteria.
5. Regulations and Future Perspectives
Bacteriophages, specifically coliphages, have been included as viral indicators of fecal pollution in several water quality policies and guidelines over the last two decades, as bacterial indicators have proven to be ineffective for predicting viral outbreaks in water and food samples [153]. It is of particular importance that EU regulations for drinking and reclaimed water have recently incorporated coliphages as parameters of microbial quality. [132,133]. The number of regulations including bacteriophages can therefore be expected to increase dramatically in the next decade, after member states of the EU adopt this legislation. The current regulations and guidelines around the whole that include bacteriophages as indicator organisms can be found in Table 1. Moreover, a consequence of the COVID-19 pandemic is that interest in the control of viral contamination is likely to increase, due to health concerns and the growing public awareness of viral infection, though SARS-CoV2 is not a waterborne pathogen [154,155].
The inclusion of coliphages in regulations means that standardized techniques for their enumeration need to be improved and optimized, to enable faster and simpler testing. Considering the ongoing research in this field, it seems likely that streamlined user-friendly kits providing results in a few hours at very reasonable costs will become available in the near future [83].
Regarding crAssphage, research is expected to grow in the coming years as new phages from this family are isolated and host strains are described [48,49]. Studies of particular interest will be focusing on the replication cycle of crAss-like bacteriophages, their high persistence in the human gut microbiota, their prevalence in wastewater and other aquatic environments, their significance for human intestinal physiology and disease, and the development of culture techniques. Evidence from this research will help to elucidate the true value and suitability of crAssphage as a fecal indicator and MST marker.
6. Conclusions
Bacteriophages are attractive as promising alternative fecal indicators to assess the risk of viral contamination in natural and built environments. Further research is needed to facilitate their application, including the development of improved standardized methods, but there is no doubt that they are already a valuable complement to existing methodologies.
Author Contributions
Conceptualization, D.T.-A., L.R.-R.; writing—original draft preparation, D.T.-A., L.R.-R.; writing—review and editing, D.T.-A., A.R.B., M.M., L.R.-R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Spanish Ministerio de Innovación y Ciencia (AGL2016-75536-P) the Agencia Estatal de Investigación (AEI), the European regional fund (ERF) and the Generalitat de Catalunya (2017SGR170). D.T-A. has a grant from the Ministerio de Ciencia e Inovación of the Spanish Government (FPU-998758–2016) and L.R.-R. is a Serra Húnter Lecturer.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kummu, M.; de Moel, H.; Ward, P.J.; Varis, O. How close do we live to water? A global analysis of population distance to freshwater bodies. PLoS ONE 2011, 6, e20578. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Long, S.C.; Das, D.; Dorner, S.M. Are microbial indicators and pathogens correlated? A statistical analysis of 40 years of research. J. Water Health 2011, 9, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Ashbolt, N.J.; Grabow, W.O.K.; Snozzi, M. Indicators of microbial water quality. In Water Quality: Guidelines, Standards and Health; IWA Publishing: London, UK, 2001; pp. 289–315. [Google Scholar] [CrossRef]
- Jofre, J. Is the replication of somatic coliphages in water environments significant? J. Appl. Microbiol. 2009, 106, 1059–1069. [Google Scholar] [CrossRef]
- McMinn, B.R.; Ashbolt, N.J.; Korajkic, A. Bacteriophages as indicators of faecal pollution and enteric virus removal. Lett. Appl. Microbiol. 2017, 65, 11–26. [Google Scholar] [CrossRef]
- Martín-Díaz, J.; Lucena, F.; Blanch, A.R.; Jofre, J. Review: Indicator bacteriophages in sludge, biosolids, sediments and soils. Environ. Res. 2020, 182, 109133. [Google Scholar] [CrossRef] [PubMed]
- Jofre, J.; Lucena, F.; Blanch, A.; Muniesa, M. Coliphages as Model Organisms in the Characterization and Management of Water Resources. Water 2016, 8, 199. [Google Scholar] [CrossRef]
- Jebri, S.; Muniesa, M.; Jofre, J. General and host-associated bacteriophage indicators of faecal pollution. In Water and Sanitation for the 21st Century: Health and Microbiological Aspects of Excreta and Wastewater Management (Global Water Pathogen Project); Farnleitner, A., Blanch, A., Eds.; Michigan State University Press: E. Lansing, MI, USA; UNESCO: Paris, France, 2017. [Google Scholar]
- Bonilla, N.; Santiago, T.; Marcos, P.; Urdaneta, M.; Santo Domingo, J.; Toranzos, G.A. Enterophages, a group of phages infecting Enterococcus faecalis, and their potential as alternate indicators of human faecal contamination. Water Sci. Technol. 2010, 61, 293–300. [Google Scholar] [CrossRef]
- Santiago-Rodriguez, T.M.; Marcos, P.; Monteiro, S.; Urdaneta, M.; Santos, R.; Toranzos, G.A. Evaluation of Enterococcus-infecting phages as indices of fecal pollution. J. Water Health 2013, 11, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Chyerochana, N.; Kongprajug, A.; Somnark, P.; Leelapanang Kamphaengthong, P.; Mongkolsuk, S.; Sirikanchana, K. Distributions of enterococci and human-specific bacteriophages of enterococci in a tropical watershed. Int. J. Hyg. Environ. Health 2020, 226, 113482. [Google Scholar] [CrossRef]
- Muniesa, M.; Mocé-Llivina, L.; Katayama, H.; Jofre, J. Bacterial host strains that support replication of somatic coliphages. Antonie Leeuwenhoek 2003, 83, 305–315. [Google Scholar] [CrossRef]
- Jofre, J. Chapter 11 Indicators of Waterborne Enteric Viruses. In Human Viruses in Water Perspectives in Medical Virology; Bosch, A., Ed.; Elsevier: Amsterdam, The Netherlands, 2007; Volume 17, pp. 227–249. ISBN 9780444521576. [Google Scholar]
- Muniesa, M.; Lucena, F.; Jofre, J. Study of the potential relationship between the morphology of infectious somatic coliphages and their persistence in the environment. J. Appl. Microbiol. 1999, 87, 402–409. [Google Scholar] [CrossRef]
- Dion, M.B.; Oechslin, F.; Moineau, S. Phage diversity, genomics and phylogeny. Nat. Rev. Microbiol. 2020, 18, 125–138. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Dolja, V.V.; Krupovic, M.; Varsani, A.; Wolf, Y.I.; Yutin, N.; Zerbini, F.M.; Kuhn, J.H. Global Organization and Proposed Megataxonomy of the Virus World. Microbiol. Mol. Biol. Rev. 2020, 84, 1–33. [Google Scholar] [CrossRef] [PubMed]
- Borrego, J.J.; Córnax, R.; Moriñigo, M.A.; Martínez-Manzanares, E.; Romero, P. Coliphages as an indicator of faecal pollution in water. their survival and productive infectivity in natural aquatic environments. Water Res. 1990, 24, 111–116. [Google Scholar] [CrossRef]
- Seeley, N.D.; Primrose, S.B. The Effect of Temperature on the Ecology of Aquatic Bacteriophages. J. Gen. Virol. 1980, 46, 87–95. [Google Scholar] [CrossRef]
- Cornax, R.; Moriñigo, M.A.; Balebona, M.C.; Castro, D.; Borrego, J.J. Significance of several bacteriophage groups as indicators of sewage pollution in marine waters. Water Res. 1991, 25, 673–678. [Google Scholar] [CrossRef]
- Muniesa, M.; Jofre, J. Factors influencing the replication of somatic coliphages in the water environment. Antonie Van Leeuwenhoek 2004, 86, 65–76. [Google Scholar] [CrossRef]
- Raivio, T. Identifying your enemies—Could envelope stress trigger microbial immunity? Mol. Microbiol. 2011, 79, 557–561. [Google Scholar] [CrossRef]
- Hernández-Delgado, E.A.; Toranzos, G.A. In situ replication studies of somatic and male-specific coliphages in a tropical pristine river. Water Sci. Technol. 1995, 31, 247–250. [Google Scholar] [CrossRef]
- Davis, J.E.; Sinsheimer, R.L.; Strauss, J.H. Bacteriophage MS2-another RNA phage. Science 1961, 134, 1427. [Google Scholar]
- Fauquet, C.M.; Fargette, D. International Committee on Taxonomy of Viruses and the 3,142 unassigned species. Virol. J. 2005, 2, 1–10. [Google Scholar] [CrossRef]
- King, A.M.Q.; Adams, M.J.; Carstens, E.B.; Lefkowitz, E.J. Virus taxonomy. In Ninth Report of the International Committee on Taxonomy of Viruse; Elsevier: Amsterdam, The Netherlands, 2012; pp. 486–487. [Google Scholar]
- Hsu, F.C.; Shieh, Y.S.C.; Van Duin, J.; Beekwilder, M.J.; Sobsey, M.D. Genotyping male-specific RNA coliphages by hybridization with oligonucleotide probes. Appl. Environ. Microbiol. 1995, 61, 3960–3966. [Google Scholar] [CrossRef]
- Ogorzaly, L.; Gantzer, C. Development of real-time RT-PCR methods for specific detection of F-specific RNA bacteriophage genogroups: Application to urban raw wastewater. J. Virol. Methods 2006, 138, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Jofre, J.; Stewart, J.R.; Grabow, W. Phage Methods. In Microbial Source Tracking: Methods, Applications, and Case Studies; Hagedorn, C., Blanch, A.R., Harwood, V.J., Eds.; Springer: New York, NY, USA, 2011; pp. 137–156. ISBN 978-1-4419-9386-1. [Google Scholar]
- Hartard, C.; Rivet, R.; Banas, S.; Gantzer, C. Occurrence of and sequence variation among F-specific RNA bacteriophage subgroups in feces and wastewater of urban and animal origins. Appl. Environ. Microbiol. 2015, 81, 6505–6515. [Google Scholar] [CrossRef] [PubMed]
- Doré, W.J.; Henshilwood, K.; Lees, D.N. Evaluation of F-specific RNA bacteriophage as a candidate human enteric virus indicator for bivalve molluscan shellfish. Appl. Environ. Microbiol. 2000, 66, 1280–1285. [Google Scholar] [CrossRef] [PubMed]
- Hartard, C.; Leclerc, M.; Rivet, R.; Maul, A.; Loutreul, J.; Banas, S.; Boudaud, N.; Gantzera, C. F-specific RNA bacteriophages, especially members of subgroup II, should be reconsidered as good indicators of viral pollution of oysters. Appl. Environ. Microbiol. 2018, 84, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Woody, M.A.; Cliver, D.O. Replication of coliphage Qβ as affected by host cell number, nutrition, competition from insusceptible cells and non-RNA coliphages. J. Appl. Microbiol. 1997, 82, 431–440. [Google Scholar] [CrossRef]
- Montemayor, M.; Costan, A.; Lucena, F.; Jofre, J.; Muñoz, J.; Dalmau, E.; Mujeriego, R.; Sala, L. The combined performance of UV light and chlorine during reclaimed water disinfection. Water Sci. Technol. 2008, 57, 935–940. [Google Scholar] [CrossRef] [PubMed]
- Lucena, F.; Ribas, F.; Duran, A.E.; Skraber, S.; Gantzer, C.; Campos, C.; Morón, A.; Calderón, E.; Jofre, J. Occurrence of bacterial indicators and bacteriophages infecting enteric bacteria in groundwater in different geographical areas. J. Appl. Microbiol. 2006, 101, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Leclerc, H.; Edberg, S.; Pierzo, V.; Delattre, J.M. Bacteriophages as indicators of enteric viruses and public health risk in groundwaters. J. Appl. Microbiol. 2000, 88, 5–21. [Google Scholar] [CrossRef]
- Guzmán, C.; Mocé-Llivina, L.; Lucena, F.; Jofre, J. Evaluation of Escherichia coli host strain CB390 for simultaneous detection of somatic and F-specific coliphages. Appl. Environ. Microbiol. 2008, 74, 531–534. [Google Scholar] [CrossRef] [PubMed]
- Jofre, J.; Blanch, A.R.; Lucena, F.; Muniesa, M. Bacteriophages infecting Bacteroides as a marker for microbial source tracking. Water Res. 2014, 55, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Queralt, N.; Jofre, J.; Araujo, R.; Muniesa, M. Homogeneity of the morphological groups of bacteriophages infecting Bacteroides fragilis strain HSP40 and strain RYC2056. Curr. Microbiol. 2003, 46, 163–168. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, M.R.; Rose, J.B. Application of Bacteroides fragilis phage as an alternative indicator of sewage pollution in Tampa Bay, Florida. Estuaries Coasts 2006, 29, 246–256. [Google Scholar] [CrossRef]
- Puig, A.; Araujo, R.; Jofre, J.; Frias-Lopez, J. Identification of cell wall proteins of Bacteroides fragilis to which bacteriophage B40-8 binds specifically. Microbiology 2001, 147, 281–288. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Klieve, A.V.; Gregg, K.; Bauchop, T. Isolation and characterization of lytic phages from Bacterioides ruminicola ss brevis. Curr. Microbiol. 1991, 23, 183–187. [Google Scholar] [CrossRef]
- Tartera, C.; Jofre, J. Bacteriophages active against Bacteroides fragilis in sewage-polluted waters. Appl. Environ. Microbiol. 1987, 53, 1632–1637. [Google Scholar] [CrossRef] [PubMed]
- Payan, A.; Ebdon, J.; Taylor, H.; Gantzer, C.; Ottoson, J.; Papageorgiou, G.T.; Blanch, A.R.; Lucena, F.; Jofre, J.; Muniesa, M. Method for isolation of Bacteroides bacteriophage host strains suitable for tracking sources of fecal pollution in water. Appl. Environ. Microbiol. 2005, 71, 5659–5662. [Google Scholar] [CrossRef]
- Hagedorn, C.; Blanch, A.R.; Harwood, V.J. Microbial Source Tracking: Methods, Applications, and Case Studies; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2011; ISBN 144199386X. [Google Scholar]
- Ebdon, J.; Muniesa, M.; Taylor, H. The application of a recently isolated strain of Bacteroides (GB-124) to identify human sources of faecal pollution in a temperate river catchment. Water Res. 2007, 41, 3683–3690. [Google Scholar] [CrossRef]
- Venegas, C.; Diez, H.; Blanch, A.R.; Jofre, J.; Campos, C. Microbial source markers assessment in the Bogotá River basin (Colombia). J. Water Health 2015, 13, 801–810. [Google Scholar] [CrossRef]
- Wicki, M.; Auckenthaler, A.; Felleisen, R.; Tanner, M.; Baumgartner, A. Novel Bacteroides host strains for detection of human- and animal-specific bacteriophages in water. J. Water Health 2011, 9, 159–168. [Google Scholar] [CrossRef]
- Gómez-Doñate, M.; Payán, A.; Cortés, I.; Blanch, A.R.; Lucena, F.; Jofre, J.; Muniesa, M. Isolation of bacteriophage host strains of Bacteroides species suitable for tracking sources of animal faecal pollution in water. Environ. Microbiol. 2011, 13, 1622–1631. [Google Scholar] [CrossRef]
- Sánchez-Alfonso, A.C.; Venegas, C.; Díez, H.; Méndez, J.; Blanch, A.R.; Jofre, J.; Campos, C. Microbial indicators and molecular markers used to differentiate the source of faecal pollution in the Bogotá River (Colombia). Int. J. Hyg. Environ. Health 2020, 225, 113450. [Google Scholar] [CrossRef] [PubMed]
- Dutilh, B.E.; Cassman, N.; McNair, K.; Sanchez, S.E.; Silva, G.G.Z.; Boling, L.; Barr, J.J.; Speth, D.R.; Seguritan, V.; Aziz, R.K.; et al. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat. Commun. 2014, 5, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Yutin, N.; Makarova, K.S.; Gussow, A.B.; Krupovic, M.; Segall, A.; Edwards, R.A.; Koonin, E.V. Discovery of an expansive bacteriophage family that includes the most abundant viruses from the human gut. Nat. Microbiol. 2018, 3, 38–46. [Google Scholar] [CrossRef]
- Shkoporov, A.N.; Khokhlova, E.V.; Fitzgerald, C.B.; Stockdale, S.R.; Draper, L.A.; Ross, R.P.; Hill, C. ΦCrAss001 represents the most abundant bacteriophage family in the human gut and infects Bacteroides intestinalis. Nat. Commun. 2018, 9, 1–8. [Google Scholar] [CrossRef]
- Koonin, E.V.; Yutin, N. The crAss-like Phage Group: How Metagenomics Reshaped the Human Virome. Trends Microbiol. 2020, 28, 349–359. [Google Scholar] [CrossRef]
- Guerin, E.; Shkoporov, A.N.; Stockdale, S.R.; Comas, J.C.; Khokhlova, E.V.; Clooney, A.G.; Daly, K.M.; Draper, L.A.; Stephens, N.; Scholz, D.; et al. Isolation and characterisation of ΦcrAss002, a crAss-like phage from the human gut that infects Bacteroides xylanisolvens. Microbiome 2021, 9, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Hryckowian, A.J.; Merrill, B.D.; Porter, N.T.; Van Treuren, W.; Nelson, E.J.; Garlena, R.A.; Russell, D.A.; Martens, E.C.; Sonnenburg, J.L. Bacteroides thetaiotaomicron-Infecting Bacteriophage Isolates Inform Sequence-Based Host Range Predictions. Cell Host Microbe 2020, 28, 371–379. [Google Scholar] [CrossRef]
- Crank, K.; Li, X.; North, D.; Ferraro, G.B.; Iaconelli, M.; Mancini, P.; La Rosa, G.; Bibby, K. CrAssphage abundance and correlation with molecular viral markers in Italian wastewater. Water Res. 2020, 184, 116161. [Google Scholar] [CrossRef]
- Farkas, K.; Adriaenssens, E.M.; Walker, D.I.; McDonald, J.E.; Malham, S.K.; Jones, D.L. Critical Evaluation of CrAssphage as a Molecular Marker for Human-Derived Wastewater Contamination in the Aquatic Environment. Food Environ. Virol. 2019, 11, 113–119. [Google Scholar] [CrossRef]
- García-Aljaro, C.; Ballesté, E.; Muniesa, M.; Jofre, J. Determination of crAssphage in water samples and applicability for tracking human faecal pollution. Microb. Biotechnol. 2017, 10, 1775–1780. [Google Scholar] [CrossRef]
- Li, Y.; Gordon, E.; Shean, R.C.; Idle, A.; Deng, X.; Greninger, A.L.; Delwart, E. CrAssphage and its bacterial host in cat feces. Sci. Rep. 2021, 11, 815. [Google Scholar] [CrossRef]
- Wu, Z.; Greaves, J.; Arp, L.; Stone, D.; Bibby, K. Comparative fate of CrAssphage with culturable and molecular fecal pollution indicators during activated sludge wastewater treatment. Environ. Int. 2020, 136, 105452. [Google Scholar] [CrossRef] [PubMed]
- Bivins, A.; Crank, K.; Greaves, J.; North, D.; Wu, Z.; Bibby, K. Cross-assembly phage and pepper mild mottle virus as viral water quality monitoring tools—potential, research gaps, and way forward. Curr. Opin. Environ. Sci. Health 2020, 16, 54–61. [Google Scholar] [CrossRef]
- Ballesté, E.; Pascual-Benito, M.; Martín-Díaz, J.; Blanch, A.R.; Lucena, F.; Muniesa, M.; Jofre, J.; García-Aljaro, C. Dynamics of crAssphage as a human source tracking marker in potentially faecally polluted environments. Water Res. 2019, 155, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Stachler, E.; Akyon, B.; De Carvalho, N.A.; Ference, C.; Bibby, K. Correlation of crAssphage qPCR Markers with Culturable and Molecular Indicators of Human Fecal Pollution in an Impacted Urban Watershed. Environ. Sci. Technol. 2018, 52, 7505–7512. [Google Scholar] [CrossRef] [PubMed]
- Edwards, R.A.; Vega, A.A.; Norman, H.M.; Ohaeri, M.; Levi, K.; Dinsdale, E.A.; Cinek, O.; Aziz, R.K.; McNair, K.; Barr, J.J.; et al. Global phylogeography and ancient evolution of the widespread human gut virus crAssphage. Nat. Microbiol. 2019, 4, 1727–1736. [Google Scholar] [CrossRef]
- Ballesté, E.; Blanch, A.R.; Mendez, J.; Sala-comorera, L.; Maunula, L.; Monteiro, S.; Farnleitner, A.H.; Tiehm, A.; Jofre, J.; García-aljaro, C.; et al. Bacteriophages Are Good Estimators of Human Viruses Present in Water. Front. Microbiol. 2021, 12, 973. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.H. Bacteriophages. In Bacteriophages; Inter-Science Publishers: New York, NY, USA; London, UK, 1959; p. 592. [Google Scholar]
- ISO 10705-2: Water Quality. Detection and Enumeration of Bacteriophages—Part 2: Enumeration of Somatic Coliphages; International Standardisation Organisation: Geneva, Switzerland, 2000; Available online: https://www.iso.org/standard/20127.html (accessed on 13 May 2021).
- ISO 10705-1: Water Quality—Detection and Enumeration of Bacteriophages—Part 1: Enumeration of F-Specific RNA Bacteriophages. International Standardisation Organisation: Geneva, Switzerland, 1995; Available online: https://www.iso.org/standard/18794.html (accessed on 13 May 2021).
- ISO 10705-4: Water Quality. Detection and Enumeration of Bacteriophages—Part 4: Enumeration of Bacteriophages Infecting Bacteroides Fragilis; International Standardization Organization: Geneva, Switzerland, 2001; Available online: https://www.iso.org/standard/27892.html (accessed on 13 May 2021).
- Mooijman, K.A.; Ghameshlou, Z.; Bahar, M.; Jofre, J.; Havelaar, A.H. Enumeration of bacteriophages in water by different laboratories of the European Union in two interlaboratory comparison studies. J. Virol. Methods 2005, 127, 60–68. [Google Scholar] [CrossRef]
- US EPA. Method 1601: Male-Specific (F+) and Somatic Coliphage in Water by Two-Step Enrichment Procedure April 2001. EPA Document 821-R-01-030; U.S. Environmental Protection Agency: Washington, DC, USA, 2001; p. 40. [Google Scholar]
- US EPA. Method 1602: Male-Specific (F+) and Somatic Coliphage in Water by Single Agar Layer (SAL) Procedure. EPA 821-R-01-029; Office of Water, Engineering and Analysis Division: Washington, DC, USA, 2001. [Google Scholar]
- US EPA. USEPA Manual of Methods for Virology; U.S. Environmental Protection Agency: Washington, DC, USA, 2001; ISBN 9781578085712. [Google Scholar]
- US EPA. Results of the Interlaboratory Validation of EPA Method 1601 for Presence/Absence of Male Specific (F+) and Somatic Coliphages in Water by Two-Step Enrichment. EPA 821-R-03-015; U.S. Environmental Protection Agency: Washington, DC, USA, 2003. [Google Scholar]
- US EPA. Results of the Interlaboratory Validation of EPA Method 1602 for Enumeration of Male Specific (F+) and Somatic Coliphage in Water by Single Agar Layer (SAL); EPA 821-R-03-016; U.S. Environmental Protection Agency: Washington, DC, USA, 2003. [Google Scholar]
- US EPA. Method 1642: Male-specific (F+) and Somatic Coliphage in Recreational Waters and Wastewater by Ultrafiltration (UF) and Single Agar Layer (SAL) Procedure; Office of Water, Engineering and Analysis Division: Washington, DC, USA, 2018; pp. 1–40. [Google Scholar]
- US EPA. Method 1643: Male-Specific (F+) and Somatic Coliphage in Secondary (No Disinfection) Wastewater by the Single Agar Layer (SAL) Procedure; Office of Water, Engineering and Analysis Division: Washington, DC, USA, 2018; pp. 1–40. [Google Scholar]
- Grabow, W.; Holtzhausen, C.; de Villiers, J. Research on Bacteriophages as Indicators of Water Quality; Department of Medical Virology, Faculty of Medicine University of Pretoria: Pretoria, South Africa, 1993; Volume 321. [Google Scholar]
- Toribio-Avedillo, D.; Martín-Díaz, J.; Blanco-Picazo, P.; Blanch, A.R.; Muniesa, M. F-specific coliphage detection by the Bluephage method. Water Res. 2020, 184, 116215. [Google Scholar] [CrossRef] [PubMed]
- Havelaar, A.H.; Hogeboom, W.M. Factors affecting the enumeration of coliphages in sewage and sewage-polluted waters. Antonie Leeuwenhoek 1983, 49, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Havelaar, A.H.; Hogeboom, W.M. A method for the enumeration of male-specific bacteriophages in sewage. J. Appl. Bacteriol. 1984, 56, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Debartorlomeis, J.; Cabelli, V.J. Evaluation of an Escherichia coli Host Strain for Enumeration of of F Male-Specific Bacteriophages. Appl. Environ. Microbiol. 1991, 57, 1301–1305. [Google Scholar] [CrossRef] [PubMed]
- Bailey, E.S.; Price, M.; Casanova, L.M.; Sobsey, M.D. E. coli CB390: An alternative E. coli host for simultaneous detection of somatic and F+ coliphage viruses in reclaimed and other waters. J. Virol. Methods 2017, 250, 25–28. [Google Scholar] [CrossRef] [PubMed]
- Blanch, A.R.; Belanche-Muñoz, L.; Bonjoch, X.; Ebdon, J.; Gantzer, C.; Lucena, F.; Ottoson, J.; Kourtis, C.; Iversen, A.; Kühn, I.; et al. Integrated analysis of established and novel microbial and chemical methods for microbial source tracking. Appl. Environ. Microbiol. 2006, 72, 5915–5926. [Google Scholar] [CrossRef]
- Toribio-Avedillo, D.; Méndez, J.; Muniesa, M.; Blanch, A.R. Evaluation of New Components in Modified Scholten’s Medium for the Detection of Somatic Coliphages. Food Environ. Virol. 2020, 12, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Boehm, A.B.; Yamahara, K.M.; Love, D.C.; Peterson, B.M.; Mcneill, K.; Nelson, K.L. Covariation and photoinactivation of traditional and novel indicator organisms and human viruses at a sewage-impacted marine beach. Environ. Sci. Technol. 2009, 43, 8046–8052. [Google Scholar] [CrossRef]
- Girones, R.; Ferrús, M.A.; Alonso, J.L.; Rodriguez-Manzano, J.; Calgua, B.; de Abreu Corrêa, A.; Hundesa, A.; Carratala, A.; Bofill-Mas, S. Molecular detection of pathogens in water—The pros and cons of molecular techniques. Water Res. 2010, 44, 4325–4339. [Google Scholar] [CrossRef]
- Blanch, A.R.; Lucena, F.; Muniesa, M.; Jofre, J. Fast and easy methods for the detection of coliphages. J. Microbiol. Methods 2020, 173, 105940. [Google Scholar] [CrossRef]
- Fung, D.Y.C.; Fujioka, R.; Vijayavel, K.; Sato, D.; Bishop, D. Evaluation of Fung double tube test for Clostridium perfringens and EasyPhage test for F-specific RNA coliphages as rapid screening tests for fecal contamination in recreational waters of Hawaii. J. Rapid Methods Autom. Microbiol. 2007, 15, 217–229. [Google Scholar] [CrossRef]
- Rames, E.; Macdonald, J. The QuantiPhage assay: A novel method for the rapid colorimetric detection of coliphages using cellulose pad materials. Water Res. 2019, 149, 98–110. [Google Scholar] [CrossRef]
- Salter, R.S.; Durbin, G.W.; Conklin, E.; Rosen, J.; Clancy, J. Proposed modifications of environmental protection agency method 1601 for detection of coliphages in drinking water, with same-day fluorescence-based detection and evaluation by the performance-based measurement system and alternative test protocol valida. Appl. Environ. Microbiol. 2010, 76, 7803–7810. [Google Scholar] [CrossRef]
- Guzmán Luna, C.; Costán-Longares, A.; Lucena, F.; Jofre, J. Detection of somatic coliphages through a bioluminescence assay measuring phage mediated release of adenylate kinase and adenosine 5′-triphosphate. J. Virol. Methods 2009, 161, 107–113. [Google Scholar] [CrossRef]
- Muniesa, M.; Ballesté, E.; Imamovic, L.; Pascual-Benito, M.; Toribio-Avedillo, D.; Lucena, F.; Blanch, A.R.; Jofre, J. Bluephage: A rapid method for the detection of somatic coliphages used as indicators of fecal pollution in water. Water Res. 2018, 128, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Farkas, K.; Mannion, F.; Hillary, L.S.; Malham, S.K.; Walker, D.I. Emerging technologies for the rapid detection of enteric viruses in the aquatic environment. Curr. Opin. Environ. Sci. Health 2020, 16, 1–6. [Google Scholar] [CrossRef]
- Love, D.C.; Sobsey, M.D. Simple and rapid F+ coliphage culture, latex agglutination, and typing assay to detect and source track fecal contamination. Appl. Environ. Microbiol. 2007, 73, 4110–4118. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.S. Somatic Coliphage Families As Potential Indicators of Enteric Viruses In Water and Methods for Their Detection; University of North Carolina at Chapel Hill: Chapel Hill, NC, USA, 2009. [Google Scholar]
- García-Aljaro, C.; Blanch, A.R.; Campos, C.; Jofre, J.; Lucena, F. Pathogens, faecal indicators and human-specific microbial source-tracking markers in sewage. J. Appl. Microbiol. 2019, 126, 701–717. [Google Scholar] [CrossRef]
- Leifels, M.; Cheng, D.; Sozzi, E.; Shoults, D.C.; Wuertz, S.; Mongkolsuk, S.; Sirikanchana, K. Capsid integrity quantitative PCR to determine virus infectivity in environmental and food applications—A systematic review. Water Res. 2021, 11, 100080. [Google Scholar] [CrossRef]
- Matheson, C.D.; Gurney, C.; Esau, N.; Lehto, R. Assessing PCR inhibition from humic substances. Open Enzym. Inhib. J. 2010, 3, 38–45. [Google Scholar] [CrossRef]
- O’Connell, K.P.; Bucher, J.R.; Anderson, P.E.; Cao, C.J.; Khan, A.S.; Gostomski, M.V.; Valdes, J.J. Real-time fluorogenic reverse transcription-PCR assays for detection of bacteriophage MS2. Appl. Environ. Microbiol. 2006, 72, 478–483. [Google Scholar] [CrossRef] [PubMed]
- Ogilvie, L.A.; Caplin, J.; Dedi, C.; Diston, D.; Cheek, E.; Bowler, L.; Taylor, H.; Ebdon, J.; Jones, B.V. Comparative (meta)genomic analysis and ecological profiling of human gut-specific bacteriophage φB124-14. PLoS ONE 2012, 7, e35053. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Lobos, A.; Senkbeil, J.; Peraud, J.; Gallard, J.; Harwood, V.J. Evaluation of the novel crAssphage marker for sewage pollution tracking in storm drain outfalls in Tampa, Florida. Water Res. 2018, 131, 142–150. [Google Scholar] [CrossRef]
- Stachler, E.; Kelty, C.; Sivaganesan, M.; Li, X.; Bibby, K.; Shanks, O.C. Quantitative CrAssphage PCR Assays for Human Fecal Pollution Measurement. Environ. Sci. Technol. 2017, 51, 9146–9154. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, P.; Rodriguez, S.D.; Vlassiouk, I.; Hansen, I.A.; Smirnov, S.N. Simple and Versatile Detection of Viruses Using Anodized Alumina Membranes. ACS Sensors 2016, 1, 488–492. [Google Scholar] [CrossRef]
- García-Aljaro, C.; Muñoz-Berbel, X.; Jenkins, A.T.A.; Blanch, A.R.; Muñoz, F.X. Surface Plasmon Resonance Assay for Real-Time Monitoring of Somatic Coliphages in Wastewaters. Appl. Environ. Microbiol. 2008, 74, 4054–4058. [Google Scholar] [CrossRef]
- Zago, M.; Scaltriti, E.; Fornasari, M.E.; Rivetti, C.; Grolli, S.; Giraffa, G.; Ramoni, R.; Carminati, D. Epifluorescence and atomic force microscopy: Two innovative applications for studying phage-host interactions in Lactobacillus helveticus. J. Microbiol. Methods 2012, 88, 41–46. [Google Scholar] [CrossRef]
- Sobsey, M.D.; Schwab, K.J.; Handzel, T.R. Simple membrane filter method to concentrate and enumerate male-specific RNA coliphages. J. Am. Water Work. Assoc. 1990, 82, 52–59. [Google Scholar] [CrossRef]
- Méndez, J.; Audicana, A.; Isern, A.; Llaneza, J.; Moreno, B.; Tarancón, M.L.; Jofre, J.; Lucena, F. Standardised evaluation of the performance of a simple membrane filtration-elution method to concentrate bacteriophages from drinking water. J. Virol. Methods 2004, 117, 19–25. [Google Scholar] [CrossRef]
- Schulze, E.; Lenk, J. Concentration of coliphages from drinking water by Mg(OH)2 flocculation. Naturwissenschaften 1983, 70, 612–613. [Google Scholar] [CrossRef]
- Contreras-Coll, N.; Lucena, F.; Mooijman, K.; Havelaar, A.; Pierzo, V.; Boque, M.; Gawler, A.; Höller, C.; Lambiri, M.; Mirolo, G.; et al. Occurrence and levels of indicator bacteriophages in bathing waters throughout Europe. Water Res. 2002, 36, 4963–4974. [Google Scholar] [CrossRef]
- Korajkic, A.; McMinn, B.R.; Harwood, V.J. Relationships between microbial indicators and pathogens in recreational water settings. Int. J. Environ. Res. Public Health 2018, 15, 2842. [Google Scholar] [CrossRef] [PubMed]
- Lucena, F.; Duran, A.E.; Morón, A.; Calderón, E.; Campos, C.; Gantzer, C.; Skraber, S.; Jofre, J. Reduction of bacterial indicators and bacteriophages infecting faecal bacteria in primary and secondary wastewater treatments. J. Appl. Microbiol. 2004, 97, 1069–1076. [Google Scholar] [CrossRef] [PubMed]
- Guelin, A. Etude quantitative de bacteriophages de la mer. Ann. Inst. Pasteur 1948, 74, 104–112. [Google Scholar]
- Dias, E.; Ebdon, J.; Taylor, H. The application of bacteriophages as novel indicators of viral pathogens in wastewater treatment systems. Water Res. 2018, 129, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Yahya, M.; Hmaied, F.; Jebri, S.; Jofre, J.; Hamdi, M. Bacteriophages as indicators of human and animal faecal contamination in raw and treated wastewaters from Tunisia. J. Appl. Microbiol. 2015, 118, 1217–1225. [Google Scholar] [CrossRef]
- Lucena, F.; Jofre, J. Potential Use of Bacteriophages as Indicators of Water Quality and Wastewater Treatment Processes. In Bacteriophages in the Control of Food- and Waterborne Pathogens; ASM Press: Washington, DC, USA, 2014; pp. 103–118. [Google Scholar]
- Rose, J. Reduction of Pathogens, Indicator Bacteria, and Alternative Indicators by Wastewater Treatment and Reclamation Processes. Water Intell. Online 2015, 4, 9781780404370. [Google Scholar] [CrossRef]
- Muniesa, M.; Lucena, F.; Blanch, A.R.; Payán, A.; Jofre, J. Use of abundance ratios of somatic coliphages and bacteriophages of Bacteroides thetaiotaomicron GA17 for microbial source identification. Water Res. 2012, 46, 6410–6418. [Google Scholar] [CrossRef]
- Jofre, J.; Lucena, F.; Blanch, A.R. Coliphages as a Complementary Tool to Improve the Management of Urban Wastewater Treatments and Minimize Health Risks in Receiving Waters. Water 2021, 13, 1110. [Google Scholar] [CrossRef]
- Grabow, W.O.K. Bacteriophages: Update on application as models for viruses in water. Water SA 2001, 27, 251–268. [Google Scholar] [CrossRef]
- Armon, R.; Kott, Y. Distribution comparison between coliphages and phages of anaerobic bacteria (Bacteroides fragilis) in water sources, and their reliability as fecal pollution indicators in drinking water. Water Sci. Technol. 1995, 31, 215–222. [Google Scholar] [CrossRef]
- Méndez, J.; Audicana, A.; Cancer, M.; Isern, A.; Llaneza, J.; Moreno, B.; Navarro, M.; Tarancón, M.L.; Valero, F.; Ribas, F.; et al. Assessment of drinking water quality using indicator bacteria and bacteriophages. J. Water Health 2004, 2, 201–214. [Google Scholar] [CrossRef]
- Armon, R. Bacteriophage Monitoring in Drinking Water: Do They Fulfil the Index or Indicator Function? Water Sci. Technol. 1993, 27, 463–470. [Google Scholar] [CrossRef]
- Heffron, J.; McDermid, B.; Maher, E.; McNamara, P.J.; Mayer, B.K. Mechanisms of virus mitigation and suitability of bacteriophages as surrogates in drinking water treatment by iron electrocoagulation. Water Res. 2019, 163, 114877. [Google Scholar] [CrossRef] [PubMed]
- Abbaszadegan, M.; Monteiro, P.; Nwachuku, N.; Alum, A.; Ryu, H. Removal of adenovirus, calicivirus, and bacteriophages by conventional drinking water treatment. J. Environ. Sci. Health Part A Toxic Hazard. Subst. Environ. Eng. 2008, 43, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Jofre, J.; Olle, E.; Ribas, F.; Vidal, A.; Lucena, F. Potential usefulness of bacteriophages that infect Bacteroides fragilis as model organisms for monitoring virus removal in drinking water treatment plants. Appl. Environ. Microbiol. 1995, 61, 3227–3231. [Google Scholar] [CrossRef]
- Decision, C. Directive 2006/7/EC of the European Parliament and of the Council of 15 February 2006 concerning the management of bathing water quality and repealing Directive 76/160/EEC. Off. J. Eur. Union 2006, L 064, 37–51. [Google Scholar]
- Bonadonna, L.; Briancesco, R.; Suffredini, E.; Coccia, A.; Della Libera, S.; Carducci, A.; Verani, M.; Federigi, I.; Iaconelli, M.; Bonanno Ferraro, G.; et al. Enteric viruses, somatic coliphages and Vibrio species in marine bathing and non-bathing waters in Italy. Mar. Pollut. Bull. 2019, 149, 110570. [Google Scholar] [CrossRef]
- Colford, J.M.; Wade, T.J.; Schiff, K.C.; Wright, C.C.; Griffith, J.F.; Sandhu, S.K.; Burns, S.; Sobsey, M.; Lovelace, G.; Weisberg, S.B. Water Quality Indicators and the Risk of Illness at Beaches With Nonpoint Sources of Fecal Contamination. Epidemiology 2007, 18, 27–35. [Google Scholar] [CrossRef]
- Abdelzaher, A.M.; Wright, M.E.; Ortega, C.; Hasan, A.R.; Shibata, T.; Solo-Gabriele, H.M.; Kish, J.; Withum, K.; He, G.; Elmir, S.M.; et al. Daily measures of microbes and human health at a non-point source marine beach. J. Water Health 2011, 9, 443–457. [Google Scholar] [CrossRef]
- Benjamin-Chung, J.; Arnold, B.F.; Wade, T.J.; Schiff, K.; Griffith, J.F.; Dufour, A.P.; Weisberg, S.B.; Colford, J.M. Coliphages and Gastrointestinal Illness in Recreational Waters: Pooled Analysis of Six Coastal Beach Cohorts. Epidemiology 2017, 28, 644–652. [Google Scholar] [CrossRef] [PubMed]
- The European Parliament and the Council of the European Union Regulation (EU). 2020/741, Minimum requirements for water reuse. Off. J. Eur. Union 2020, 2019, L 177/32–L 177/55. [Google Scholar]
- The European Parliament and the Council of the European Union Directive (EU). 2020/2184, EU (revised) Drinking Water Directive. Off. J. Eur. Commun. 2020, 2019, 1–62. [Google Scholar]
- WHO. Guidelines for the Safe Use of Wastewater, Excreta and Greywater, 3rd ed.; World Health Organization: Geneva, Switzerland, 2006; Volume 1, ISBN 9241546824. [Google Scholar]
- US EPA. National Primary Drinking Water Regulations: Groundwater Rule. Final Rule; 40 CFR Parts 9, 141 and 142. Federal Register; U.S. Environmental Protection Agency: Washington, DC, USA, 2006; Volume 71. [Google Scholar]
- US EPA. Review of Coliphages as Possible Indicators of Fecal Contamination for Ambient Water Quality; EPA 820-R-15-098; U.S. Environmental Protection Agency: Washington, DC, USA, 2015. [Google Scholar]
- Republique Française. Arrêté du 25 Juin 2014 Modifiant l’Arrêté du 2 Août 2010 Relatif à l’Utilisation d’Eaux issues du Traitement d’Épuration des eaux Résiduaires Urbaines Pour l’Irrigation de Cultures ou d’Espaces Verts; Republique Française: Paris, France, 2014. [Google Scholar]
- Republica de Colombia. Decreto no 1287. Criterios Para el Uso de Biosólidos Generados en Plantas de Tratamiento de Aguas Residuales Municipales; Republica de Colombia: Bogota, Colombia, 2014. [Google Scholar]
- Queensland Government. Queensland Water Recycling Guidelines; Queensland Government: Brisbane, Australia, 2005. [Google Scholar]
- North Carolina Administration. North. Carolina Administrative Code 15A NCAC 2U; North Carolina Administration: Raleigh, NC, USA, 2011. [Google Scholar]
- NHMRC. NRMMC Australian Drinking Water Guidelines Paper 6 National Water Quality Management Strategy; NHMRC: Canberra, Australia, 2011. [Google Scholar]
- Bureau of Indian Standards (BIS). Indian Standard Drinking Water Specification (Second Revision); Bureau of Indian Standards (BIS): New Delhi, India, 2012. [Google Scholar]
- Anonymous. Loi sur la Qualité de l’Environnement: Règlement sur la Qualité de l’eau Potable c. Q-2, r.18.1.1; Éditeur Officiel du Québec: Québec, QC, Canada, 2001. [Google Scholar]
- World Health Organisation. Guidelines for Drinking-Water Quality, 4th ed.; World Health Organisation: Geneva, Switzerland, 2017; ISBN 978-92-4-154995-0. [Google Scholar]
- Guzmán, C.; Jofre, J.; Montemayor, M.; Lucena, F. Occurrence and levels of indicators and selected pathogens in different sludges and biosolids. J. Appl. Microbiol. 2007, 103, 2420–2429. [Google Scholar] [CrossRef]
- Blanco-Picazo, P.; Roscales, G.; Toribio-Avedillo, D.; Gómez-Gómez, C.; Avila, C.; Ballesté, E.; Muniesa, M.; Rodríguez-Rubio, L. Antibiotic resistance genes in phage particles from antarctic and mediterranean seawater ecosystems. Microorganisms 2020, 8, 1293. [Google Scholar] [CrossRef]
- Hodgson, K.R.; Torok, V.A.; Turnbull, A.R. Bacteriophages as enteric viral indicators in bivalve mollusc management. Food Microbiol. 2017, 65, 284–293. [Google Scholar] [CrossRef]
- European Food Safety Authority The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 2018, 16, e05500. [CrossRef]
- Hsu, F.C.; Shieh, Y.C.C.; Sobsey, M.D. Enteric bacteriophages as potential fecal indicators in ground beef and poultry meat. J. Food Prot. 2002, 65, 93–99. [Google Scholar] [CrossRef]
- Gómez-Gómez, C.; Blanco-Picazo, P.; Brown-Jaque, M.; Quirós, P.; Rodríguez-Rubio, L.; Cerdà-Cuellar, M.; Muniesa, M. Infectious phage particles packaging antibiotic resistance genes found in meat products and chicken feces. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Maciorowski, K.G.; Pillai, S.D.; Ricke, S.C. Presence of bacteriophages in animal feed as indicators of fecal contamination. J. Environ. Sci. Health Part B Pestic. Food Contam. Agric. Wastes 2001, 36, 699–708. [Google Scholar] [CrossRef] [PubMed]
- Larrañaga, O.; Brown-Jaque, M.; Quirós, P.; Gómez-Gómez, C.; Blanch, A.R.; Rodríguez-Rubio, L.; Muniesa, M. Phage particles harboring antibiotic resistance genes in fresh-cut vegetables and agricultural soil. Environ. Int. 2018, 115, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Bányai, K.; Estes, M.K.; Martella, V.; Parashar, U.D. Viral gastroenteritis. Lancet 2018, 392, 175–186. [Google Scholar] [CrossRef]
- Amoah, I.D.; Kumari, S.; Bux, F. Coronaviruses in wastewater processes: Source, fate and potential risks. Environ. Int. 2020, 143, 105962. [Google Scholar] [CrossRef] [PubMed]
- Yeo, C.; Kaushal, S.; Yeo, D. Enteric involvement of coronaviruses: Is faecal–oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol. Hepatol. 2020, 5, 335–337. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).