Comparative Study of SARS-CoV-2, SARS-CoV-1, MERS-CoV, HCoV-229E and Influenza Host Gene Expression in Asthma: Importance of Sex, Disease Severity, and Epithelial Heterogeneity
Abstract
1. Introduction
2. Materials and Methods
2.1. Transcriptomic Data
2.2. Clustering of Epithelial and Viral Entry Genes
2.3. Statistical Analysis and Data Visualization
2.4. Data Availability
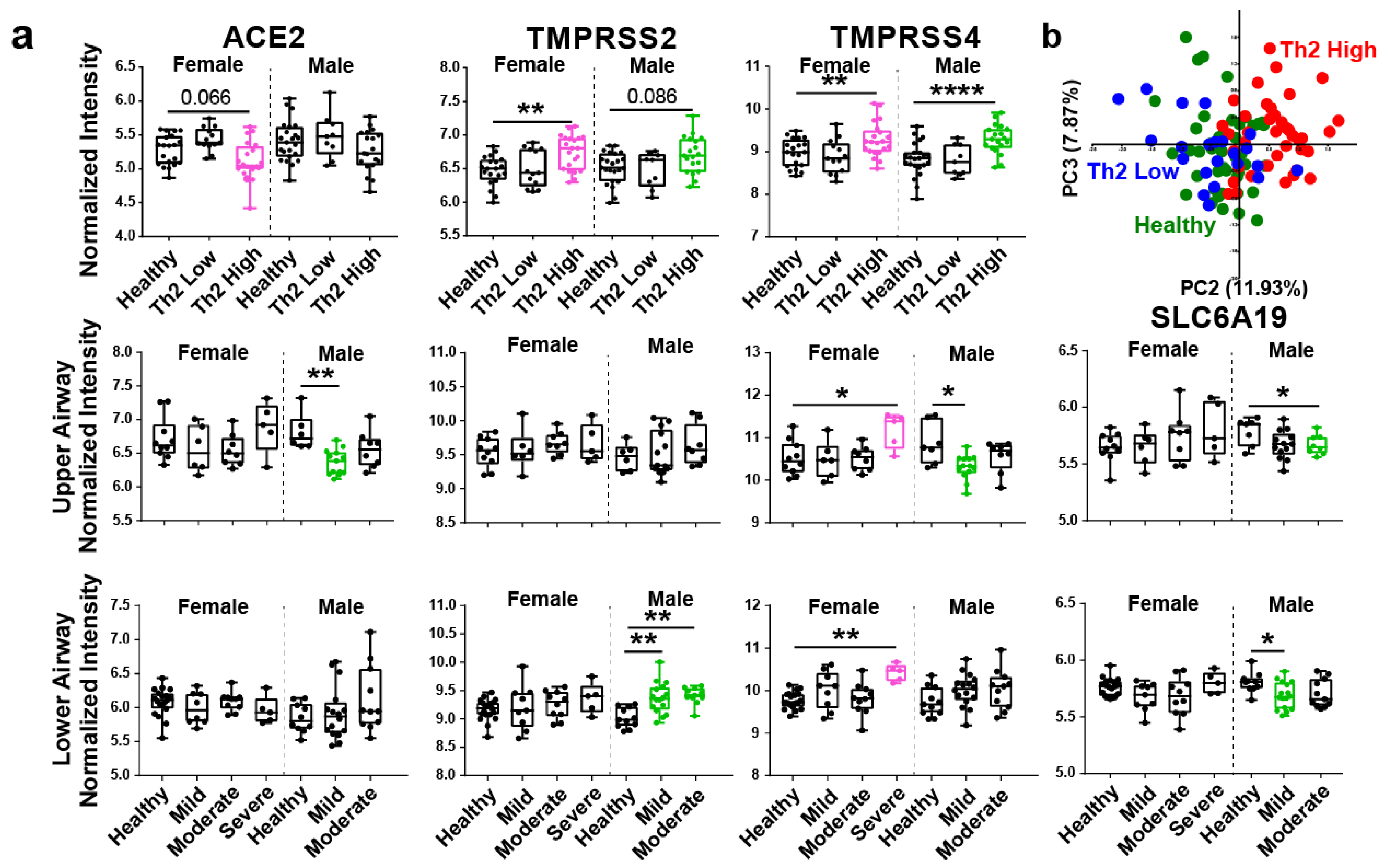
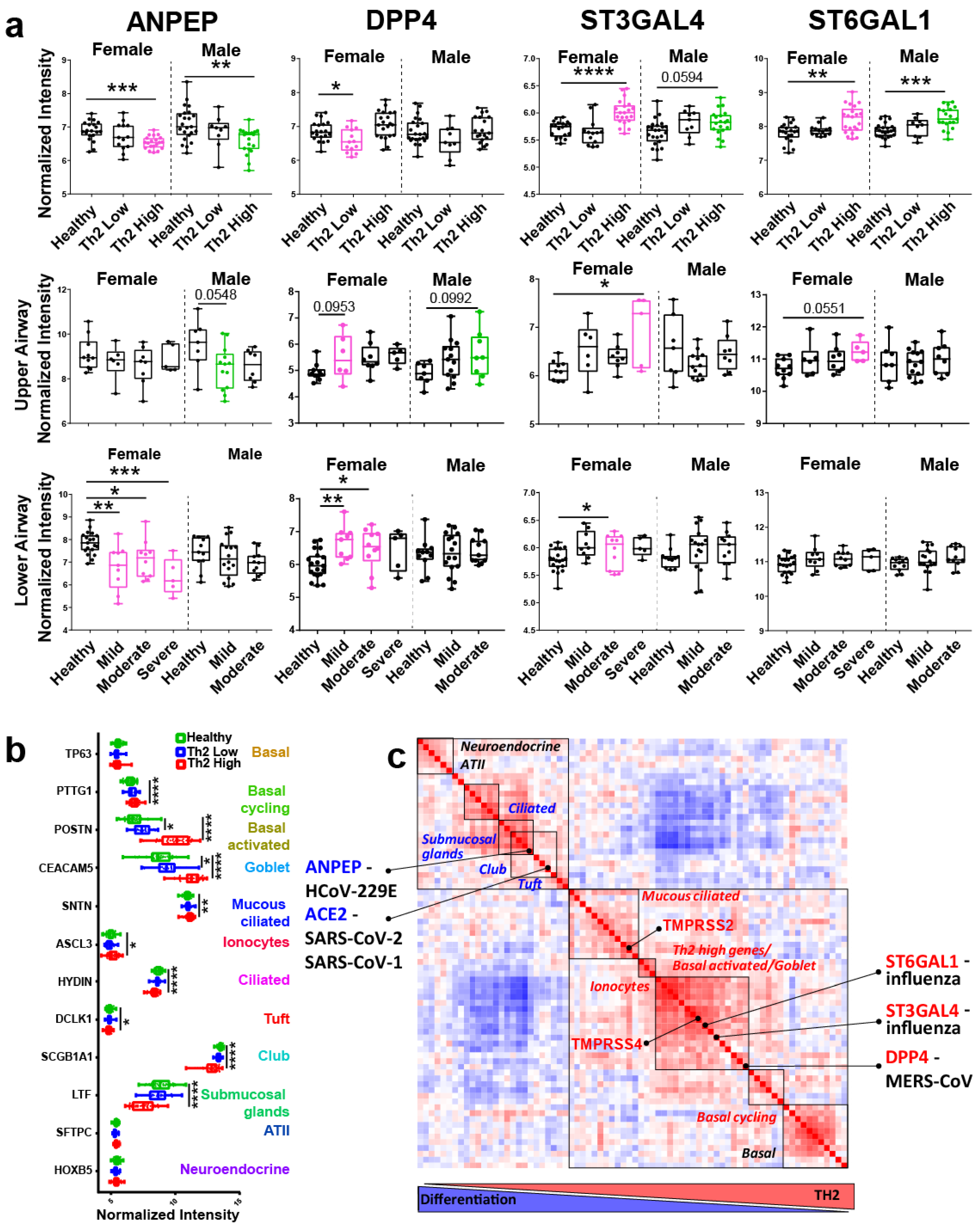
3. Results and Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, X.; Xu, S.; Yu, M.; Wang, K.; Tao, Y.; Zhou, Y.; Shi, J.; Zhou, M.; Wu, B.; Yang, Z.; et al. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan. J. Allergy Clin. Immunol. 2020, 146, 110–118. [Google Scholar] [CrossRef]
- Zhang, J.J.; Dong, X.; Cao, Y.Y.; Yuan, Y.D.; Yang, Y.B.; Yan, Y.Q.; Akdis, C.A.; Gao, Y. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy 2020, 75, 1730–1741. [Google Scholar] [CrossRef]
- Mahdavinia, M.; Foster, K.J.; Jauregui, E.; Moore, D.; Adnan, D.; Andy-Nweye, A.B.; Khan, S.; Bishehsari, F. Asthma prolongs intubation in CO VID-19. J. Allergy Clin. Immunol. Pract. 2020, 8, 2388–2391. [Google Scholar] [CrossRef] [PubMed]
- Jackson, D.J.; Busse, W.W.; Bacharier, L.B.; Kattan, M.; O’Connor, G.T.; Wood, R.A.; Visness, C.M.; Durham, S.R.; Larson, D.; Esnault, S.; et al. Association of respiratory allergy, asthma, and expression of the SARS-CoV-2 receptor ACE2. J. Allergy Clin. Immunol. 2020, 146, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Wark, P.A.; Pathinayake, P.S.; Kaiko, G.; Nichol, K.; Ali, A.; Chen, L.; Sutanto, E.N.; Garratt, L.W.; Sohal, S.S.; Lu, W.; et al. ACE2 expression is elevated in airway epithelial cells from older and male healthy individuals but reduced in asthma. Respirology 2021, 26, 442–451. [Google Scholar] [CrossRef]
- Kimura, H.; Francisco, D.; Conway, M.; Martinez, F.D.; Vercelli, D.; Polverino, F.; Billheimer, D.; Kraft, M. Type 2 inflammation modulates ACE2 and TMPRSS2 in airway epithelial cells. J. Allergy Clin. Immunol. 2020, 146, 80–88.e8. [Google Scholar] [CrossRef] [PubMed]
- Woodruff, P.G.; Modrek, B.; Choy, D.; Jia, G.; Abbas, A.R.; Ellwanger, A.; Arron, J.R.; Koth, L.L.; Fahy, J.V. T-helper Type 2–driven Inflammation Defines Major Subphenotypes of Asthma. Am. J. Respir. Crit. Care Med. 2009, 180, 388–395. [Google Scholar] [CrossRef]
- Braga, F.A.V.; Kar, G.; Berg, M.; Carpaij, O.A.; Polanski, K.; Simon, L.M.; Brouwer, S.; Gomes, T.; Hesse, L.; Jiang, J.; et al. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat. Med. 2019, 25, 1153–1163. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.-M.; Bai, P.; He, W.; Wu, F.; Liu, X.-F.; Han, D.-M.; Liu, S.; Yang, J.-K. Gender Differences in Patients With COVID-19: Focus on Severity and Mortality. Front. Public Health 2020, 8, 152. [Google Scholar] [CrossRef] [PubMed]
- Kermani, N.Z.; Song, W.J.; Badi, Y.; Versi, A.; Guo, Y.; Sun, K.; Bhavsar, P.; Howarth, P.; Dahlen, S.-E.; Sterk, P.J.; et al. Sputum ACE2, TMPRSS2 and FURIN gene expression in severe neutrophilic asthma. Respir. Res. 2021, 22, 1–10. [Google Scholar]
- Sharif-Askari, N.S.; Sharif-Askari, F.S.; Alabed, M.; Temsah, M.-H.; Al Heialy, S.; Hamid, Q.; Halwani, R. Airways Expression of SARS-CoV-2 Receptor, ACE2, and TMPRSS2 Is Lower in Children Than Adults and Increases with Smoking and COPD. Mol. Ther. Methods Clin. Dev. 2020, 18, 1–6. [Google Scholar] [CrossRef]
- Kasela, S.; NHLBI SubPopulations and InteRmediate Outcome Measures In COPD Study (SPIROMICS); Ortega, V.E.; Martorella, M.; Garudadri, S.; Nguyen, J.; Ampleford, E.; Pasanen, A.; Nerella, S.; Buschur, K.L.; et al. Genetic and non-genetic factors affecting the expression of COVID-19-relevant genes in the large airway epithelium. Genome Med. 2021, 13, 1–17. [Google Scholar] [CrossRef]
- Peters, M.C.; Sajuthi, S.; Deford, P.; Christenson, S.; Rios, C.L.; Montgomery, M.T.; Woodruff, P.G.; Mauger, D.T.; Erzurum, S.C.; Johansson, M.W.; et al. COVID-19 Related Genes in Sputum Cells in Asthma: Relationship to Demographic Features and Corti-costeroids. Am. J. Respir. Crit. Care Med. 2020, 202, 83–90. [Google Scholar] [CrossRef]
- Talbot, H.K.; Shepherd, B.E.; Crowe, J.; Griffin, M.R.; Edwards, K.M.; Podsiad, A.B.; Tollefson, S.J.; Wright, P.F.; Williams, J. The Pediatric Burden of Human Coronaviruses Evaluated for Twenty Years. Pediatr. Infect. Dis. J. 2009, 28, 682–687. [Google Scholar] [CrossRef]
- Van Kerkhove, M.D.; Alaswad, S.; Assiri, A.; Perera, R.A.; Peiris, M.; El Bushra, H.E.; BinSaeed, A.A. Transmissibility of MERS-CoV Infection in Closed Setting, Riyadh, Saudi Arabia, 2015. Emerg. Infect. Dis. 2019, 25, 1802–1809. [Google Scholar] [CrossRef] [PubMed]
- Van Bever, H.P.; Chng, S.Y.; Goh, D.Y. Childhood severe acute respiratory syndrome, coronavirus infections and asthma. Pediatr. Allergy Immunol. 2004, 15, 206–209. [Google Scholar] [CrossRef]
- Jia, H.P.; Look, D.C.; Shi, L.; Hickey, M.; Pewe, L.; Netland, J.; Farzan, M.; Wohlford-Lenane, C.; Perlman, S.; McCray, P.B., Jr. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differenti-ation of human airway epithelia. J. Virol. 2005, 79, 14614–14621. [Google Scholar] [CrossRef] [PubMed]
- Schleimer, R.P.; Berdnikovs, S. Etiology of epithelial barrier dysfunction in patients with type 2 inflammatory diseases. J. Allergy Clin. Immunol. 2017, 139, 1752–1761. [Google Scholar] [CrossRef]
- Wiener, R.S.; Cao, Y.X.; Hinds, A.; Ramirez, M.I.; Williams, M.C. Angiotensin converting enzyme 2 is primarily epithelial and is developmentally regulated in the mouse lung. J. Cell. Biochem. 2007, 101, 1278–1291. [Google Scholar] [CrossRef] [PubMed]
- Sungnak, W.; Huang, N.; Bécavin, C.; Berg, M.; Queen, R.; Litvinukova, M.; Talavera-López, C.; Maatz, H.; Reichart, D.; Sampaziotis, F.; et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020, 26, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Rostami, M.R.; Leopold, P.L.; Mezey, J.G.; O’Beirne, S.L.; Strulovici-Barel, Y.; Crystal, R.G. Expression of the SARS-CoV-2 ACE2 Receptor in the Human Airway Epithelium. Am. J. Respir. Crit. Care Med. 2020, 202, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, C.G.; Allon, S.J.; Nyquist, S.K.; Mbano, I.M.; Miao, V.N.; Tzouanas, C.N.; Cao, Y.; Yousif, A.S.; Bals, J.; Hauser, B.M.; et al. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell 2020, 181, 1016–1035. [Google Scholar] [CrossRef] [PubMed]
- Iwata-Yoshikawa, N.; Okamura, T.; Shimizu, Y.; Hasegawa, H.; Takeda, M.; Nagata, N. TMPRSS2 Contributes to Virus Spread and Immunopathology in the Airways of Murine Models after Coronavirus Infection. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [PubMed]
- Hatesuer, B.; Bertram, S.; Mehnert, N.; Bahgat, M.M.; Nelson, P.S.; Pöhlman, S.; Schughart, K. Tmprss2 Is Essential for Influenza H1N1 Virus Pathogenesis in Mice. PLOS Pathog. 2013, 9, e1003774. [Google Scholar] [CrossRef] [PubMed]
- Coronavirus (COVID-19) Related Deaths by Ethnic Group, England and Wales: 2 March 2020 to 10 April 2020. United Kingdom Office for National Statistics. 2020. Available online: https://www.ons.gov.uk/peoplepopulationandcommunity/birthsdeathsandmarriages/deaths/articles/coronavirusrelateddeathsbyethnicgroupenglandandwales/2march2020to10april2020 (accessed on 1 September 2020).
- Forno, E.; Celedón, J.C. Asthma and Ethnic Minorities: Socieoeconomic Status and Beyond. Curr. Opin. Allergy Clin. Immunol. 2009, 9, 154–160. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coden, M.E.; Loffredo, L.F.; Abdala-Valencia, H.; Berdnikovs, S. Comparative Study of SARS-CoV-2, SARS-CoV-1, MERS-CoV, HCoV-229E and Influenza Host Gene Expression in Asthma: Importance of Sex, Disease Severity, and Epithelial Heterogeneity. Viruses 2021, 13, 1081. https://doi.org/10.3390/v13061081
Coden ME, Loffredo LF, Abdala-Valencia H, Berdnikovs S. Comparative Study of SARS-CoV-2, SARS-CoV-1, MERS-CoV, HCoV-229E and Influenza Host Gene Expression in Asthma: Importance of Sex, Disease Severity, and Epithelial Heterogeneity. Viruses. 2021; 13(6):1081. https://doi.org/10.3390/v13061081
Chicago/Turabian StyleCoden, Mackenzie E., Lucas F. Loffredo, Hiam Abdala-Valencia, and Sergejs Berdnikovs. 2021. "Comparative Study of SARS-CoV-2, SARS-CoV-1, MERS-CoV, HCoV-229E and Influenza Host Gene Expression in Asthma: Importance of Sex, Disease Severity, and Epithelial Heterogeneity" Viruses 13, no. 6: 1081. https://doi.org/10.3390/v13061081
APA StyleCoden, M. E., Loffredo, L. F., Abdala-Valencia, H., & Berdnikovs, S. (2021). Comparative Study of SARS-CoV-2, SARS-CoV-1, MERS-CoV, HCoV-229E and Influenza Host Gene Expression in Asthma: Importance of Sex, Disease Severity, and Epithelial Heterogeneity. Viruses, 13(6), 1081. https://doi.org/10.3390/v13061081






