Isolation and Identification of a Novel Phlebovirus, Hedi Virus, from Sandflies Collected in China
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Virus Isolation and Identification
2.3. Virus Genome Sequencing
2.4. Phylogenetic Analysis
2.5. Electron Microscopy
2.6. Growth Curve of Isolated Virus
2.7. Species Identification of Sandfly Specimens
3. Results
3.1. Isolation of Hedi Virus
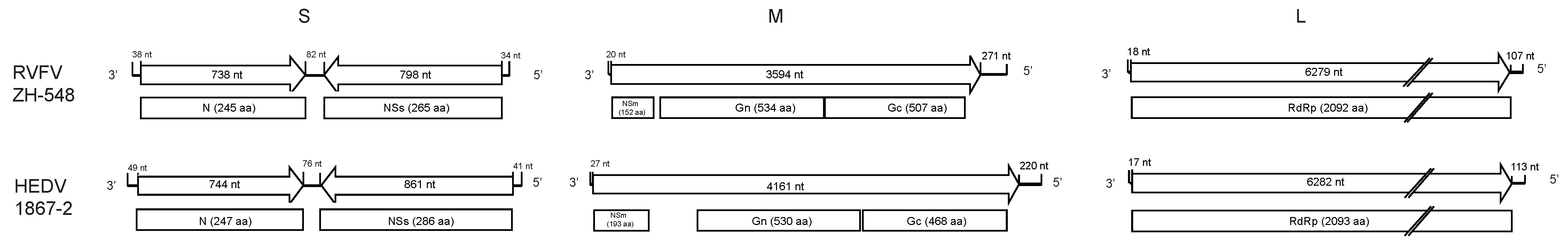
3.2. Genome Sequence Characterization of Hedi Virus
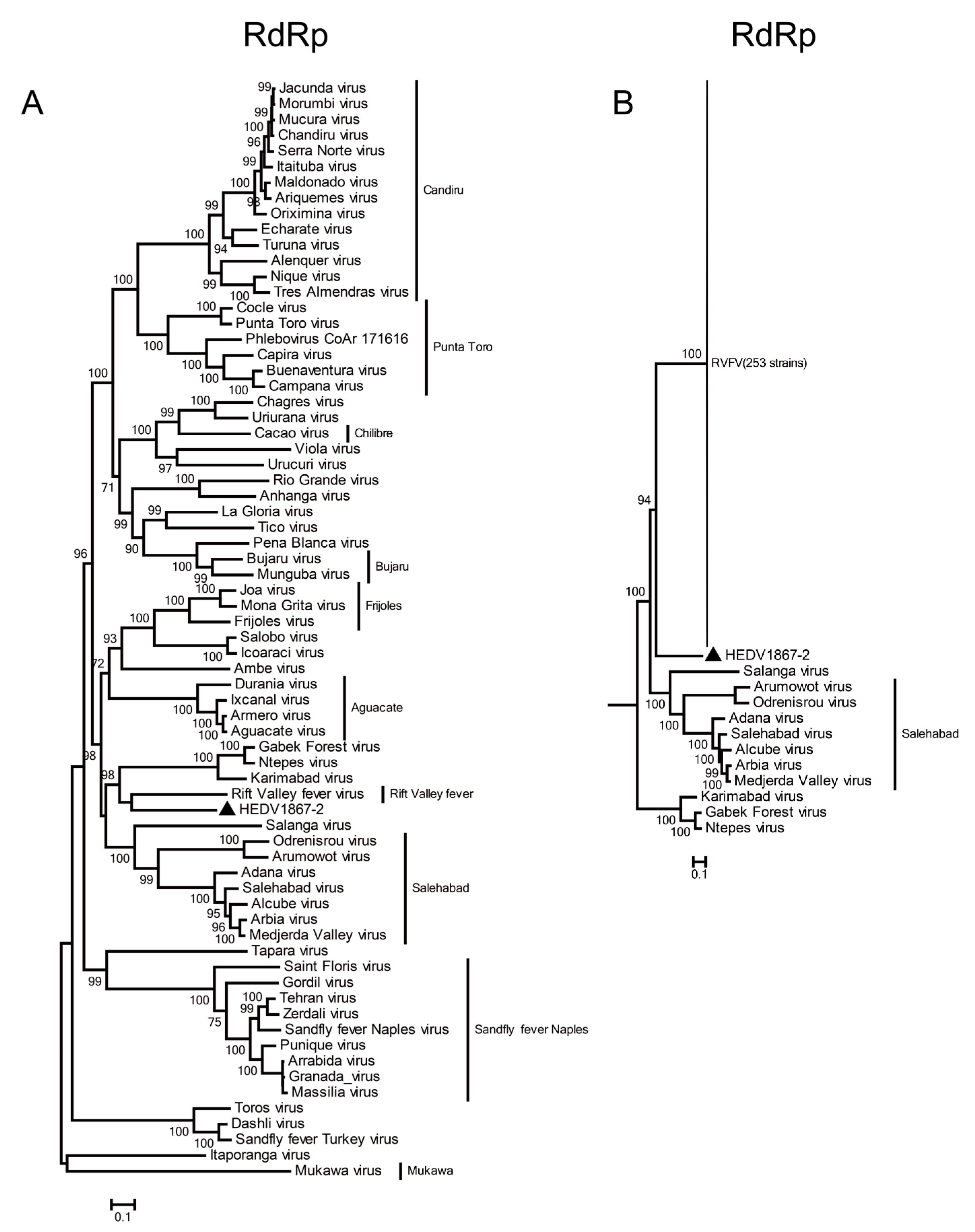
3.3. Phylogenetic Relationship of HEDV
3.4. Viral Morphology and Replication
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kuhn, J.H.; Adkins, S.; Alioto, D.; Alkhovsky, S.V.; Amarasinghe, G.K.; Anthony, S.J.; Avšič-Županc, T.; Ayllón, M.A.; Bahl, J.; Balkema-Buschmann, A.; et al. 2020 taxonomic update for phylum Negarnaviricota (Riboviria: Orthornavirae), including the large orders Bunyavirales and Mononegavirales. Arch. Virol. 2020, 165, 3023–3072. [Google Scholar] [CrossRef]
- Marklewitz, M.; Tchouassi, D.P.; Hieke, C.; Heyde, V.; Torto, B.; Sang, R.; Junglen, S. Insights into the Evolutionary Origin of Mediterranean Sandfly Fever Viruses. MSphere 2020, 5. [Google Scholar] [CrossRef]
- Yu, X.J.; Liang, M.F.; Zhang, S.Y.; Liu, Y.; Li, J.D.; Sun, Y.L.; Zhang, L.; Zhang, Q.F.; Popov, V.L.; Li, C.; et al. Fever with thrombocytopenia associated with a novel bunyavirus in China. N. Engl. J. Med. 2011, 364, 1523–1532. [Google Scholar] [CrossRef] [PubMed]
- McMullan, L.K.; Folk, S.M.; Kelly, A.J.; MacNeil, A.; Goldsmith, C.S.; Metcalfe, M.G.; Batten, B.C.; Albariño, C.G.; Zaki, S.R.; Rollin, P.E.; et al. A new phlebovirus associated with severe febrile illness in Missouri. N. Engl. J. Med. 2012, 367, 834–841. [Google Scholar] [CrossRef]
- Woods, C.W.; Karpati, A.M.; Grein, T.; McCarthy, N.; Gaturuku, P.; Muchiri, E.; Dunster, L.; Henderson, A.; Khan, A.S.; Swanepoel, R.; et al. An outbreak of Rift Valley fever in Northeastern Kenya, 1997–1998. Emerg. Infect. Dis 2002, 8, 138–144. [Google Scholar] [CrossRef]
- Pepin, M.; Bouloy, M.; Bird, B.H.; Kemp, A.; Paweska, J. Rift Valley fever virus (Bunyaviridae: Phlebovirus): An update on pathogenesis, molecular epidemiology, vectors, diagnostics and prevention. Vet. Res. 2010, 41, 61. [Google Scholar] [CrossRef]
- Ikegami, T.; Makino, S. The pathogenesis of Rift Valley fever. Viruses 2011, 3, 493–519. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, K.L.; Banyard, A.C.; McElhinney, L.; Johnson, N.; Horton, D.L.; Hernández-Triana, L.M.; Fooks, A.R. Rift Valley fever virus: A review of diagnosis and vaccination, and implications for emergence in Europe. Vaccine 2015, 33, 5520–5531. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Li, X.; Gao, X.; Fu, S.; Wang, H.; Li, M.; Lu, Z.; Zhu, W.; Lu, X.; Wang, L.; et al. Arboviruses and their related infections in China: A comprehensive field and laboratory investigation over the last 3 decades. Rev. Med. Virol. 2018, 28. [Google Scholar] [CrossRef] [PubMed]
- Xu, J. Reverse microbial etiology: A research field for predicting and preventing emerging infectious diseases caused by an unknown microorganism. J. Biosaf. Biosecur. 2019, 1, 19–21. [Google Scholar] [CrossRef]
- Wang, J.; Fu, S.; Xu, Z.; Cheng, J.; Shi, M.; Fan, N.; Song, J.; Tian, X.; Cheng, J.; Ni, S.; et al. Emerging Sand Fly-Borne Phlebovirus in China. Emerg. Infect. Dis. 2020, 26, 2435–2438. [Google Scholar] [CrossRef]
- Xu, Z.; Yang, W.; Feng, Y.; Li, Y.; Fu, S.; Li, X.; Song, J.; Zhang, H.; Zhang, Y.; Liu, W.J.; et al. Isolation and Identification of a Highly Divergent Kaeng Khoi Virus from Bat Flies (Eucampsipoda sundaica) in China. Vector Borne Zoonotic Dis. 2019, 19, 73–80. [Google Scholar] [CrossRef]
- Sánchez-Seco, M.P.; Echevarría, J.M.; Hernández, L.; Estévez, D.; Navarro-Marí, J.M.; Tenorio, A. Detection and identification of Toscana and other phleboviruses by RT-nested-PCR assays with degenerated primers. J. Med. Virol. 2003, 71, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Marklewitz, M.; Dutari, L.C.; Paraskevopoulou, S.; Page, R.A.; Loaiza, J.R.; Junglen, S. Diverse novel phleboviruses in sandflies from the Panama Canal area, Central Panama. J. Gen. Virol. 2019, 100, 938–949. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Lei, W.; Guo, X.; Fu, S.; Feng, Y.; Tao, X.; Gao, X.; Song, J.; Yang, Z.; Zhou, H.; Liang, G. The genetic characteristics and evolution of Tembusu virus. Vet. Microbiol. 2017, 201, 32–41. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Wang, J.; Fan, N.; Fu, S.; Cheng, J.; Wu, B.; Xu, Z.; Song, J.; Tian, X.; Li, Y.; He, Y.; et al. Isolation and characterization of Wuxiang virus from sandflies collected in Yangquan County, Shanxi Province, China. Vector Borne Zoonotic Dis. 2021. [Google Scholar] [CrossRef] [PubMed]
- Reguera, J.; Malet, H.; Weber, F.; Cusack, S. Structural basis for encapsidation of genomic RNA by La Crosse Orthobunyavirus nucleoprotein. Proc. Natl. Acad. Sci. USA 2013, 110, 7246–7251. [Google Scholar] [CrossRef] [PubMed]
- Reguera, J.; Gerlach, P.; Rosenthal, M.; Gaudon, S.; Coscia, F.; Günther, S.; Cusack, S. Comparative Structural and Functional Analysis of Bunyavirus and Arenavirus Cap-Snatching Endonucleases. PLoS Pathog. 2016, 12, e1005636. [Google Scholar] [CrossRef] [PubMed]
- Gerlach, P.; Malet, H.; Cusack, S.; Reguera, J. Structural Insights into Bunyavirus Replication and Its Regulation by the vRNA Promoter. Cell 2015, 161, 1267–1279. [Google Scholar] [CrossRef]
- Gogrefe, N.; Reindl, S.; Günther, S.; Rosenthal, M. Structure of a functional cap-binding domain in Rift Valley fever virus L protein. PloS Pathog. 2019, 15, e1007829. [Google Scholar] [CrossRef]
- Marklewitz, M.; Palacios, G.; Ebihara, H.; Kuhn, J.; Junglen, S. Create four new genera, create seventy nine new species, rename/move seven species, rename/move three genera and abolish one genus in the family Phenuiviridae. In Order Bunyavirales; ICTV: Berlin, Germany, 2019. [Google Scholar]
- Smith, M.R.; Schirtzinger, E.E.; Wilson, W.C.; Davis, A.S. Rift Valley Fever Virus: Propagation, Quantification, and Storage. Curr. Protoc. Microbiol. 2019, 55, e92. [Google Scholar] [CrossRef] [PubMed]
- Weingartl, H.M.; Zhang, S.; Marszal, P.; McGreevy, A.; Burton, L.; Wilson, W.C. Rift Valley fever virus incorporates the 78 kDa glycoprotein into virions matured in mosquito C6/36 cells. PLoS ONE 2014, 9, e87385. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Z.; Fan, N.; Hou, X.; Wang, J.; Fu, S.; Song, J.; Shi, M.; Liang, G. Isolation and Identification of a Novel Phlebovirus, Hedi Virus, from Sandflies Collected in China. Viruses 2021, 13, 772. https://doi.org/10.3390/v13050772
Xu Z, Fan N, Hou X, Wang J, Fu S, Song J, Shi M, Liang G. Isolation and Identification of a Novel Phlebovirus, Hedi Virus, from Sandflies Collected in China. Viruses. 2021; 13(5):772. https://doi.org/10.3390/v13050772
Chicago/Turabian StyleXu, Ziqian, Na Fan, Xin Hou, Jing Wang, Shihong Fu, Jingdong Song, Mang Shi, and Guodong Liang. 2021. "Isolation and Identification of a Novel Phlebovirus, Hedi Virus, from Sandflies Collected in China" Viruses 13, no. 5: 772. https://doi.org/10.3390/v13050772
APA StyleXu, Z., Fan, N., Hou, X., Wang, J., Fu, S., Song, J., Shi, M., & Liang, G. (2021). Isolation and Identification of a Novel Phlebovirus, Hedi Virus, from Sandflies Collected in China. Viruses, 13(5), 772. https://doi.org/10.3390/v13050772




