Novel Virulent Bacteriophages Infecting Mediterranean Isolates of the Plant Pest Xylella fastidiosa and Xanthomonas albilineans
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Media
2.2. Environmental Samples for Phage Isolation
2.3. Phage Enrichment
2.4. Phage Isolation, Propagation and Purification
2.5. Negative Staining and Transmission Electron Microscopy (TEM)
2.6. Phage Genome Sequencing and Genetic Analysis
2.7. Phage Host Range
2.8. Phage Nomenclature and Nucleotide Sequence Accession Numbers
3. Results
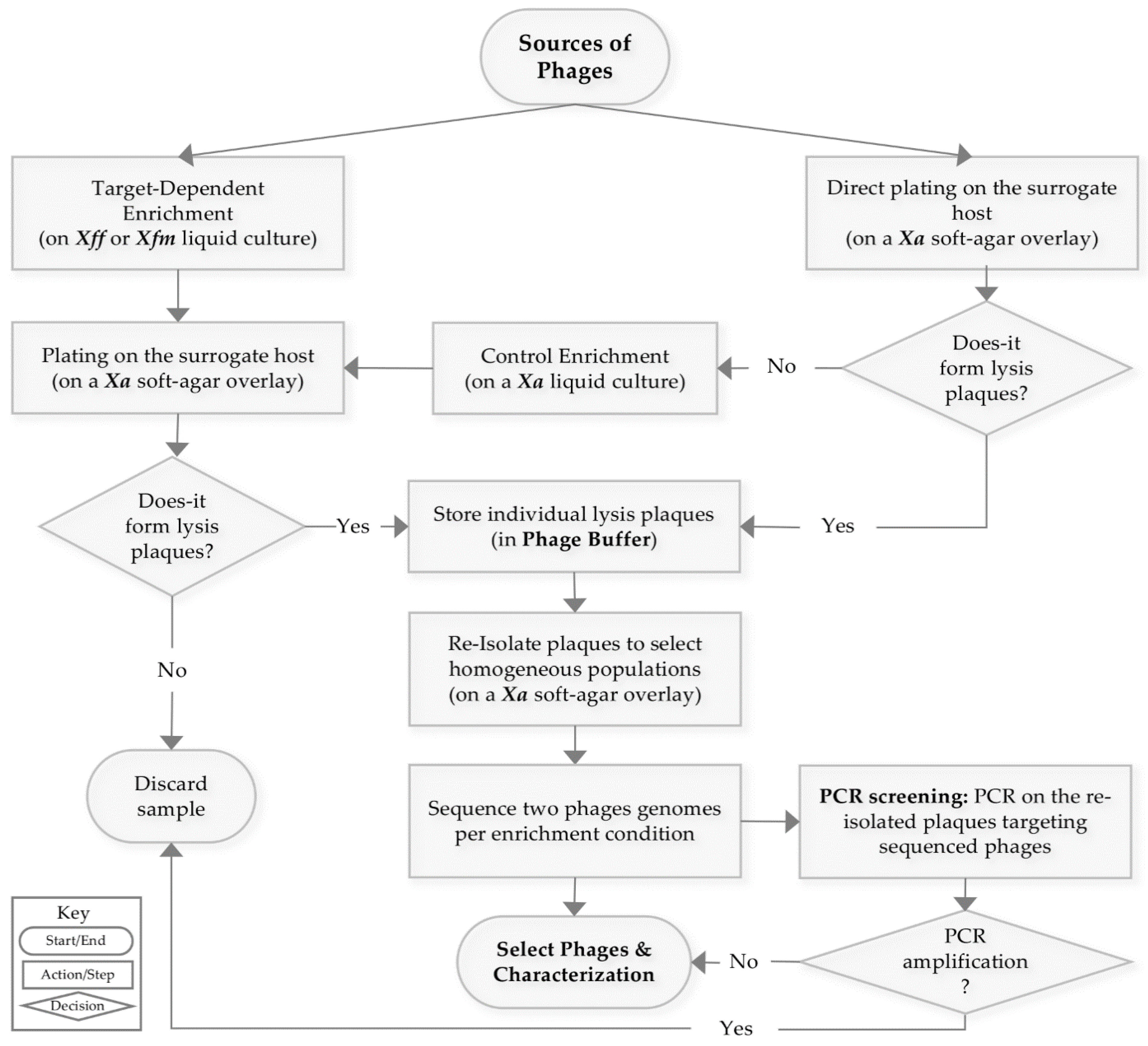
3.1. Strategy to Isolate Xf-Specific Phages
3.2. Genomic Diversity of the Phages Isolated
3.3. Genomic Characterization
3.4. Morphological Characterization
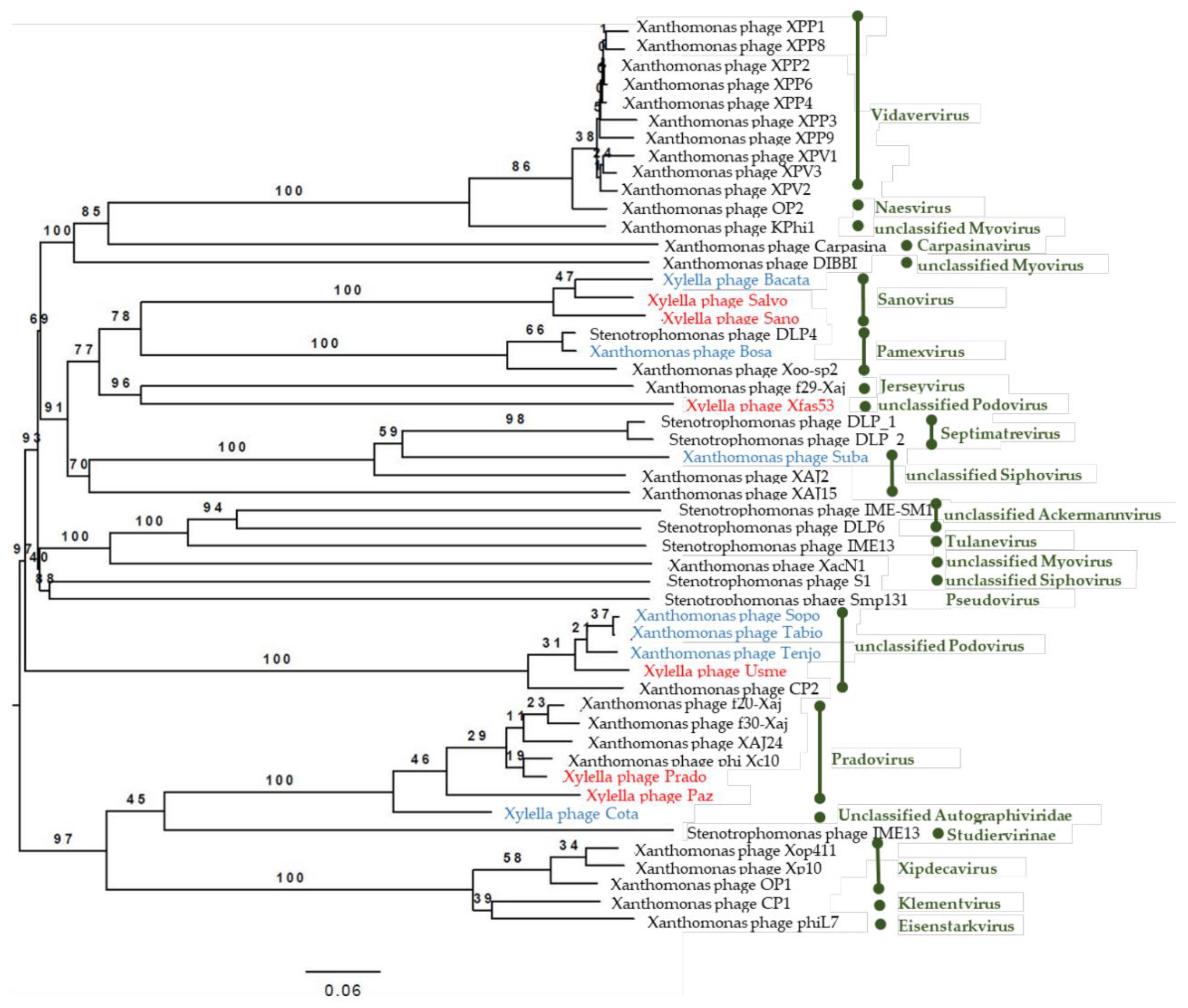
3.5. Phylogenetic Analysis of the Sequenced Phage Genomes
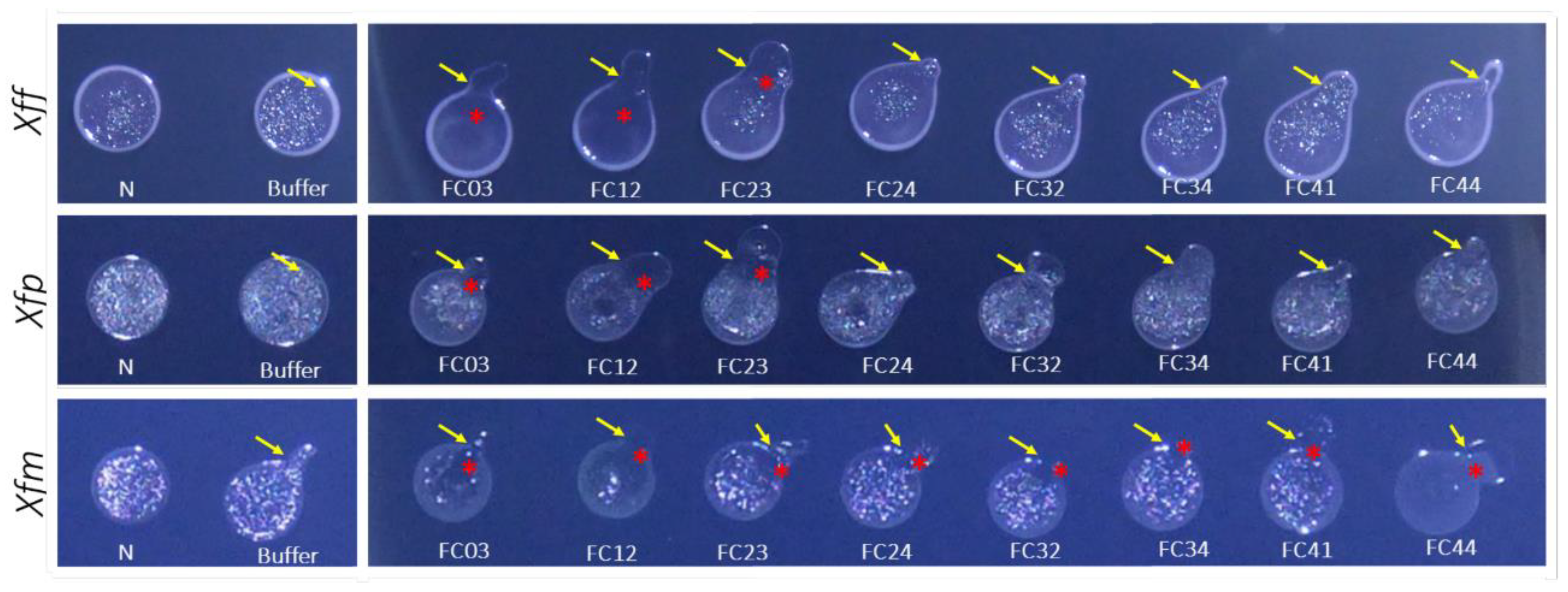
3.6. Host Range Determination
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- EFSA Panel on Plant Health (EFSA PLH Panel); Jeger, M.; Caffier, D.; Candresse, T.; Chatzivassiliou, E.; Dehnen-Schmutz, K.; Gilioli, G.; Grégoire, J.-C.; Jaques Miret, J.A.; MacLeod, A.; et al. Updated Pest Categorisation of Xylella Fastidiosa. EFSA J. 2018, 16, e05357. [Google Scholar] [CrossRef] [PubMed]
- Alston, J.M.; Fuller, K.B.; Kaplan, J.D.; Tumber, K.P. Assessing the Returns to R&D on Perennial Crops: The Costs and Benefits of Pierce’s Disease Research in the California Winegrape Industry. Aust. J. Agric. Resour. Econ. 2015, 59, 95–115. [Google Scholar] [CrossRef]
- Tumber, K.P.; Alston, J.M.; Fuller, K. Pierce’s Disease Costs California $104 Million per Year. Calif. Agric. 2014, 68. [Google Scholar] [CrossRef]
- Schneider, K.; van der Werf, W.; Cendoya, M.; Mourits, M.; Navas-Cortés, J.A.; Vicent, A.; Lansink, A.O. Impact of Xylella Fastidiosa Subspecies Pauca in European Olives. PNAS 2020, 117, 9250–9259. [Google Scholar] [CrossRef]
- Ali, B.M.; van der Werf, W.; Oude Lansink, A. Assessment of the Environmental Impacts of Xylella Fastidiosa Subsp. Pauca in Puglia. Crop Prot. 2021, 142, 105519. [Google Scholar] [CrossRef]
- Simpson, A.J.; Reinach, F.C.; Arruda, P.; Abreu, F.A.; Acencio, M.; Alvarenga, R.; Alves, L.M.; Araya, J.E.; Baia, G.S.; Baptista, C.S.; et al. The Genome Sequence of the Plant Pathogen Xylella Fastidiosa. The Xylella Fastidiosa Consortium of the Organization for Nucleotide Sequencing and Analysis. Nature 2000, 406, 151–159. [Google Scholar] [CrossRef]
- Falahi Charkhabi, N.; Booher, N.J.; Peng, Z.; Wang, L.; Rahimian, H.; Shams-Bakhsh, M.; Liu, Z.; Liu, S.; White, F.F.; Bogdanove, A.J. Complete Genome Sequencing and Targeted Mutagenesis Reveal Virulence Contributions of Tal2 and Tal4b of Xanthomonas Translucens Pv. Undulosa ICMP11055 in Bacterial Leaf Streak of Wheat. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Pieretti, I.; Royer, M.; Barbe, V.; Carrere, S.; Koebnik, R.; Cociancich, S.; Couloux, A.; Darrasse, A.; Gouzy, J.; Jacques, M.-A.; et al. The Complete Genome Sequence of Xanthomonas Albilineans Provides New Insights into the Reductive Genome Evolution of the Xylem-Limited Xanthomonadaceae. BMC Genom. 2009, 10, 616. [Google Scholar] [CrossRef]
- Gerlin, L.; Cottret, L.; Cesbron, S.; Taghouti, G.; Jacques, M.-A.; Genin, S.; Baroukh, C. Genome-Scale Investigation of the Metabolic Determinants Generating Bacterial Fastidious Growth. mSystems 2020, 5. [Google Scholar] [CrossRef]
- Pierce, N.B.; Newton, B. The California Vine Disease: A Preliminary Report of Investigations; G.P.O.: Washington, DC, USA, 1892.
- Davis, M.J.; Purcell, A.H.; Thomson, S.V. Pierce’s Disease of Grapevines: Isolation of the Causal Bacterium. Science 1978, 199, 75–77. [Google Scholar] [CrossRef]
- Wells, J.M.; Raju, B.C.; Hung, H.-Y.; Weisburg, W.G.; Mandelco-Paul, L.; Brenner, D.J. Xylella Fastidiosa Gen. Nov., Sp. Nov: Gram-Negative, Xylem-Limited, Fastidious Plant Bacteria Related to Xanthomonas Spp. Int. J. Syst. Evol. Microbiol. 1987, 37, 136–143. [Google Scholar] [CrossRef]
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 Plant Pathogenic Bacteria in Molecular Plant Pathology: Top 10 Plant Pathogenic Bacteria. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef]
- Jacques, M.-A.; Arlat, M.; Boulanger, A.; Boureau, T.; Carrère, S.; Cesbron, S.; Chen, N.W.G.; Cociancich, S.; Darrasse, A.; Denancé, N.; et al. Using Ecology, Physiology, and Genomics to Understand Host Specificity in Xanthomonas. Annu. Rev. Phytopathol. 2016, 54, 163–187. [Google Scholar] [CrossRef] [PubMed]
- Bragard, C.; Dehnen-Schmutz, K.; Serio, F.D.; Gonthier, P.; Jacques, M.-A.; Miret, J.A.J.; Justesen, A.F.; MacLeod, A.; Magnusson, C.S.; Milonas, P.; et al. Effectiveness of in Planta Control Measures for Xylella Fastidiosa. EFSA J. 2019, 17, e05666. [Google Scholar] [CrossRef]
- Dupas, E.; Briand, M.; Jacques, M.-A.; Cesbron, S. Novel Tetraplex QPCR Assays for Simultaneous Detection and Identification of Xylella Fastidiosa Subspecies in Plant Tissues. bioRxiv 2019, 699371. [Google Scholar] [CrossRef]
- Directive européene 2006/7/EC Directive 2006/7/EC of the European Parliament and of the Council of 15 February 2006 Concerning the Management of Bathing Water Quality and Repealing Directive 76/160/EEC. Off. J. Eur. Union 2006, L64, 37–51.
- Andrea Luvisi; Francesca Nicolì; Luigi De Bellis Sustainable Management of Plant Quarantine Pests: The Case of Olive Quick Decline Syndrome. Sustainability 2017, 9, 659. [CrossRef]
- EFSA Panel on Plant Health (PLH) Treatment Solutions to Cure Xylella Fastidiosa Diseased Plants. EFSA J. 2016, 14. [CrossRef]
- Scortichini, M.; Chen, J.; Caroli, M.D.; Dalessandro, G.; Pucci, N.; Modesti, V.; L’aurora, A.; Petriccione, M.; Zampella, L.; Mastrobuoni, F.; et al. A Zinc, Copper and Citric Acid Biocomplex Shows Promise for Control of Xylella Fastidiosa Subsp. Pauca in Olive Trees in Apulia Region (Southern Italy). Phytopathol. Mediterr. 2018, 57, 48–72. [Google Scholar] [CrossRef]
- Hopkins, D.L. Biological Control of Pierce’s Disease in the Vineyard with Strains of Xylella Fastidiosa Benign to Grapevine. Plant Dis. 2005, 89, 1348–1352. [Google Scholar] [CrossRef]
- Baccari, C.; Antonova, E.; Lindow, S. Biological Control of Pierce’s Disease of Grape by an Endophytic Bacterium. Phytopathology 2019, 109, 248–256. [Google Scholar] [CrossRef]
- Vandamme, E.J.; Mortelmans, K. A Century of Bacteriophage Research and Applications: Impacts on Biotechnology, Health, Ecology and the Economy!: A Century of Bacteriophage Research and Applications. J. Chem. Technol. Biotechnol. 2019, 94, 323–342. [Google Scholar] [CrossRef]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 2018, 23, 795. [Google Scholar] [CrossRef]
- Żaczek, M.; Weber-Dąbrowska, B.; Górski, A. Phages in the Global Fruit and Vegetable Industry. J. Appl. Microbiol. 2015, 118, 537–556. [Google Scholar] [CrossRef]
- Sieiro, C.; Areal-Hermida, L.; Pichardo-Gallardo, Á.; Almuiña-González, R.; de Miguel, T.; Sánchez, S.; Sánchez-Pérez, Á.; Villa, T.G. A Hundred Years of Bacteriophages: Can Phages Replace Antibiotics in Agriculture and Aquaculture? Antibiotics 2020, 9, 493. [Google Scholar] [CrossRef]
- Buttimer, C.; McAuliffe, O.; Ross, R.P.; Hill, C.; O’Mahony, J.; Coffey, A. Bacteriophages and Bacterial Plant Diseases. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Ahern, S.J.; Das, M.; Bhowmick, T.S.; Young, R.; Gonzalez, C.F. Characterization of Novel Virulent Broad-Host-Range Phages of Xylella Fastidiosa and Xanthomonas. J. Bacteriol. 2014, 196, 459–471. [Google Scholar] [CrossRef]
- Das, M.; Bhowmick, T.S.; Ahern, S.J.; Young, R.; Gonzalez, C.F. Control of Pierce’s Disease by Phage. PLoS ONE 2015, 10, e0128902. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, C.F.; Ahern, S.J.; Das, M.; Young, I.R.F.; Bhowmick, T.S. Methods and Compositions for Treatment and Control of Plant Disease. U.S. Patent No. 10,499,650, 10 December 2015. [Google Scholar]
- PM 7/24 (2) Xylella Fastidiosa. EPPO Bull. 2016, 46, 463–500. [CrossRef]
- Lopes, S.A.; Torres, S.C.Z. An Effective and Low-Cost Culture Medium for Isolation and Growth of Xylella Fastidiosa from Citrus and Coffee Plants. Curr. Microbiol. 2006, 53, 467–469. [Google Scholar] [CrossRef] [PubMed]
- Saux, M.F.-L.; Bonneau, S.; Essakhi, S.; Manceau, C.; Jacques, M.-A. Aggressive Emerging Pathovars of Xanthomonas Arboricola Represent Widespread Epidemic Clones Distinct from Poorly Pathogenic Strains, as Revealed by Multilocus Sequence Typing. Appl. Environ. Microbiol. 2015, 81, 4651–4668. [Google Scholar] [CrossRef]
- La Situation de Xylella en France et en Europe | Alim’agri. Available online: http://agriculture.gouv.fr/la-situation-de-xylella-en-france-et-en-europe (accessed on 27 February 2018).
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Li, Y.; Hu, Y.; Bolund, L.; Wang, J. State of the Art de Novo Assembly of Human Genomes from Massively Parallel Sequencing Data. Hum. Genom. 2010, 4, 271–277. [Google Scholar] [CrossRef][Green Version]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de Novo Short Read Assembly Using de Bruijn Graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations Using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Briand, M.; Bouzid, M.; Hunault, G.; Legeay, M.; Saux, M.F.-L.; Barret, M. A Rapid and Simple Method for Assessing and Representing Genome Sequences Relatedness. bioRxiv 2019, 569640. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. VICTOR: Genome-Based Phylogeny and Classification of Prokaryotic Viruses. Bioinformatics 2017, 33, 3396–3404. [Google Scholar] [CrossRef] [PubMed]
- Göker, M.; García-Blázquez, G.; Voglmayr, H.; Tellería, M.T.; Martín, M.P. Molecular Taxonomy of Phytopathogenic Fungi: A Case Study in Peronospora. PLoS ONE 2009, 4, e6319. [Google Scholar] [CrossRef]
- Adriaenssens, E.; Brister, J.R. How to Name and Classify Your Phage: An Informal Guide. Viruses 2017, 9, 70. [Google Scholar] [CrossRef]
- El Haddad, L.; Ben Abdallah, N.; Plante, P.-L.; Dumaresq, J.; Katsarava, R.; Labrie, S.; Corbeil, J.; St-Gelais, D.; Moineau, S. Improving the Safety of Staphylococcus Aureus Polyvalent Phages by Their Production on a Staphylococcus Xylosus Strain. PLoS ONE 2014, 9, e102600. [Google Scholar] [CrossRef]
- González-Menéndez, E.; Arroyo-López, F.N.; Martínez, B.; García, P.; Garrido-Fernández, A.; Rodríguez, A. Optimizing Propagation of Staphylococcus Aureus Infecting Bacteriophage VB_SauM-PhiIPLA-RODI on Staphylococcus Xylosus Using Response Surface Methodology. Viruses 2018, 10, 153. [Google Scholar] [CrossRef] [PubMed]
- David, H.L.; Clavel, S.; Clement, F. Adsorption and Growth of the Bacteriophage D29 in Selected Mycobacteria. Ann. L’institut Pasteur Virol. 1980, 131, 167–184. [Google Scholar] [CrossRef]
- Santos, S.B.; Fernandes, E.; Carvalho, C.M.; Sillankorva, S.; Krylov, V.N.; Pleteneva, E.A.; Shaburova, O.V.; Nicolau, A.; Ferreira, E.C.; Azeredo, J. Selection and Characterization of a Multivalent Salmonella Phage and Its Production in a Nonpathogenic Escherichia Coli Strain. Appl. Environ. Microbiol. 2010, 76, 7338–7342. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.A.; Ogawa, M.; Kawasaki, T.; Fujie, M.; Yamada, T. Characterization of Bacteriophages Cp1 and Cp2, the Strain-Typing Agents for Xanthomonas Axonopodis Pv. Citri. Appl. Environ. Microbiol. 2014, 80, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Peters, D.; McCutcheon, J.; Stothard, P.; Dennis, J. Novel Stenotrophomonas Maltophilia Temperate Phage DLP4 Is Capable of Lysogenic Conversion. BMC Genom. 2019, 20, 300. [Google Scholar] [CrossRef]
- Balogh, B. Characterization and Use of Bacteriophages Associated with Citrus Bacterial Pathogens for Disease Control. Ph.D. Thesis, University of Florida, Gainesville, FL, USA, 2006. [Google Scholar]
- Ackermann, H.W. Frequency of Morphological Phage Descriptions in the Year 2000. Brief Review. Arch. Virol. 2001, 146, 843–857. [Google Scholar] [CrossRef]
- Farris, J.S. Estimating Phylogenetic Trees from Distance Matrices. Am. Nat. 1972, 106, 645–668. [Google Scholar] [CrossRef]
- Turner, D.; Kropinski, A.M.; Adriaenssens, E.M. A Roadmap for Genome-Based Phage Taxonomy. Viruses 2021, 13, 506. [Google Scholar] [CrossRef]
- Abedon, S.T. Lysis from Without. Bacteriophage 2011, 1, 46–49. [Google Scholar] [CrossRef]
- Merda, D.; Bonneau, S.; Guimbaud, J.-F.; Durand, K.; Brin, C.; Boureau, T.; Lemaire, C.; Jacques, M.-A.; Fischer-Le Saux, M. Recombination-Prone Bacterial Strains Form a Reservoir from Which Epidemic Clones Emerge in Agroecosystems. Environ. Microbiol. Rep. 2016, 8, 572–581. [Google Scholar] [CrossRef]
- Chen, J.; Civerolo, E.L. Morphological Evidence for Phages in Xylella Fastidiosa. Virol. J. 2008, 5, 75. [Google Scholar] [CrossRef]
- de Mello Varani, A.; Souza, R.C.; Nakaya, H.I.; de Lima, W.C.; Paula de Almeida, L.G.; Kitajima, E.W.; Chen, J.; Civerolo, E.; Vasconcelos, A.T.R.; Van Sluys, M.-A. Origins of the Xylella Fastidiosa Prophage-like Regions and Their Impact in Genome Differentiation. PLoS ONE 2008, 3, e4059. [Google Scholar] [CrossRef]
- Almeida, R.P.P.; Purcell, A.H. Transmission of Xylella Fastidiosa to Grapevines by Homalodisca Coagulata (Hemiptera: Cicadellidae). J. Econ. Entomol. 2003, 96, 264–271. [Google Scholar] [CrossRef] [PubMed]
- Bhowmick, T.S.; Das, M.; Heinz, K.M.; Krauter, P.C.; Gonzalez, C.F. Transmission of Phage by Glassy-Winged Sharpshooters, a Vector of Xylella Fastidiosa. Bacteriophage 2016, 6, e1218411. [Google Scholar] [CrossRef] [PubMed]
- Chanishvili, N. Phage Therapy--History from Twort and d’Herelle through Soviet Experience to Current Approaches. Adv. Virus Res. 2012, 83, 3–40. [Google Scholar] [CrossRef] [PubMed]
- Holtappels, D.; Fortuna, K.; Lavigne, R.; Wagemans, J. The Future of Phage Biocontrol in Integrated Plant Protection for Sustainable Crop Production. Curr. Opin. Biotechnol. 2021, 68, 60–71. [Google Scholar] [CrossRef]


| Strain/Genotype | Abbreviation | Collection Code | NCBI |
|---|---|---|---|
| Xylella fastidiosa subsp. fastidiosa | Xff | CFBP 7970 | PRJNA417585 |
| Xylella fastidiosa subsp. pauca | Xfp | CFBP 8402 | PRJNA383475 |
| Xylella fastidiosa subsp. multiplex | Xfm | CFBP 8418 | PRJNA314986 |
| Xanthomonas albilineans | Xa | CFBP 2523 | PRJNA338244 |
| Xanthomonas arboricola pv. juglandis | Xaj | CFBP 2528 | PRJNA275685 |
| Xanthomonas citri pv. citri | Xcci | CFBP 3369 | PRJNA254373 |
| Xanthomonas vesicatoria | Xv | CFBP 2537 | PRJNA60019 |
| Xanthomonas campestris pv. campestris | Xcc | CFBP 5241 | PRJNA296 |
| Xanthomonas translucens pv. cerealis | Xtc | CFBP 2541 | PRJNA268946 |
| Phage Name | Full Nomenclature | Accession Number | Raw Sequence ID |
|---|---|---|---|
| FC03-Usme FC08-Olaya | Xylella phage Usme Xanthomonas phage Olaya | LR743523 MW802488 | ERR5548206 SRX10492030 |
| FC12-Bacata FC15-Bolivar FC17-Usaquen | Xylella phage Bacata Xanthomonas phage Bolivar Xanthomonas phage Usaquen | LR743524 MW822535 MW822536 | ERR5548205 SRX10492031 SRX10492032 |
| FC23-Cota FC25-Alcala | Xylella phage Cota Xanthomonas phage Alcala | LR778216 MW822537 | ERR5548199 SRX10492033 |
| FC28-Sopo | Xanthomonas phage Sopo | LR743529 | ERR5548204 |
| FC30-Tabio | Xanthomonas phage Tabio | LR743528 | ERR5548203 |
| FC39-Tenjo | Xanthomonas phage Tenjo | LR743531 | ERR5548202 |
| FC41-Suba | Xanthomonas phage Suba | LR743530 | ERR5548201 |
| FC44-Bosa FC47-Fontebon FC57-Sumapaz | Xanthomonas phage Bosa Xanthomonas phage Fontebon Xanthomonas phage Sumapaz | LR743532 MW822538 MW822539 | ERR5548200 SRX10492034 SRX10492035 |
| PCR Groups a | ||||||||
|---|---|---|---|---|---|---|---|---|
| Source | Conditions | Isolated Phages | Sequenced Phage Genomes | FC03 | FC44 | FC12 | FC23 | Potential New Phages |
| Plant extracts | Enrichment on Xff | NA b | - | - | - | - | - | - |
| Enrichment on Xfm | NA | - | - | - | - | - | - | |
| Enrichment on Xa | NA | - | - | - | - | - | - | |
| Insect extracts | Enrichment on Xff | 6 | LR743523 MW802488 | 6 | - | - | - | 0 |
| Enrichment on Xfm | 6 | MW822538 MW822539 | 6 | - | - | - | 0 | |
| Enrichment on Xa | 12 | MW822535 MW822536 | 11 | 1 | - | - | 0 | |
| Sewage watersample | Enrichment on Xff | 6 | LR743524 (x2) c | - | 2 | 4 | - | 0 |
| Enrichment on Xfm | 6 | LR778216 MW822537 | 1 | 1 | - | 3 | 1 | |
| Direct plaquing on Xa | 12 | LR743531 LR743532 | 4 | 4 | 2 | - | 1 + LR743530 d | |
| Runoff waters sample | Enrichment on Xff | NA | - | - | - | - | - | - |
| Enrichment on Xfm | NA | - | - | - | - | - | - | |
| Direct plaquing on Xa | 8 | LR743529 LR743528 | 4 | - | - | - | 4 | |
| Phage | Size (bp) | %GC | CDS | BLAST Nearest Organism | Coverage (%) | Identity (%) | E-Value | Order | Family | Genus | Specie |
|---|---|---|---|---|---|---|---|---|---|---|---|
| FC03-Usme a | 43721 | 66.6 | 58 | X. axonopodis phage CP2 | 83 | 92.11% | 0.0 | Caudovirales | Podovirus | Unclassified Podovirus | Usmevirus |
| FC12-Bacata | 56232 | 62.9 | 73 | X. fastidiosa phage Salvo | 89 | 96.29% | 0.0 | Caudovirales | Siphovirus | Sanovirus | Bacatavirus |
| FC23-Cota | 42307 | 58.8 | 52 | none | NA | NA | NA | NA | NA | NA | Tejovirus |
| FC28-Sopo | 43861 | 66.9 | 58 | X. axonopodis phage CP2 | 82 | 93.96% | 0.0 | Caudovirales | Podovirus | Unclassified Podovirus | Bacatavirus |
| FC30-Tabio | 43458 | 66.9 | 58 | X. axonopodis phage CP2 | 83 | 95.39% | 0.0 | Caudovirales | Podovirus | Unclassified Podovirus | Bacatavirus |
| FC39-Tenjo | 43850 | 67.2 | 58 | X. axonopodis phage CP2 | 83 | 94.70% | 0.0 | Caudovirales | Podovirus | Unclassified Podovirus | Bacatavirus |
| FC41-Suba | 45510 | 52.3 | 71 | none | NA | NA | NA | NA | NA | NA | Subavirus |
| FC44-Bosa | 63828 | 64.8 | 81 | Stenotrophomonas phage DLP4 | 99% | 97.40 % | 0.0 | Caudovirales | Siphovirus | Unclassified Siphovirus | Bosavirus |
| Phage | Head Shape & Diameter (nm) | Tail Shape & Length (nm) | Morphotype a | |
|---|---|---|---|---|
| FC03-Usme (n = 15) | Isometric, 61.7 ± 2 | *NA | Podovirus | C1 |
| FC12-Bacata (n = 14) | Isometric, 66.5 ± 2 | Long, Flexible, 220.5 ± 5 | Siphovirus | B1 |
| FC23-Cota (n = 14) | Isometric, 64.1 ± 1 | *NA | Podovirus | C1 |
| FC24-Teja (n = 15) | Isometric, 65.3 ± 2 | *NA | Podovirus | C1 |
| FC28-Sopo (n = 6) | Isometric, 65.4 ± 2 | *NA | Podovirus | C1 |
| FC30-Tabio (n = 6) | Isometric, 63.6 ± 2 | *NA | Podovirus | C1 |
| FC34-Cajica (n = 15) | Isometric, 64.4 ± 2 | *NA | Podovirus | C1 |
| FC39-Tenjo (n = 6) | Isometric, 60.6 ± 2 | *NA | Podovirus | C1 |
| FC41-Suba (n = 15) | Isometric, 61.2 ± 2 | Long, Flexible, 199.5 ± 4 | Siphovirus | B1 |
| FC44-Bosa (n = 12) | Heterometric, 60.1 ± 1 × 88.8 ± 2 | Long, Flexible, 140.5 ± 7 | Siphovirus | B3 |
| CFBP | Strain | FC03-Usme | FC12-Bacata | FC23-Cota | FC24-Teja | FC32-Tabio | FC34-Tenjo | FC41-Suba | FC44-Bosa |
|---|---|---|---|---|---|---|---|---|---|
| 7970 | X. fastidiosa subsp. fastidiosa | ||||||||
| 8402 | X. fastidiosa subsp. pauca | ||||||||
| 8418 | X. fastidiosa subsp. multiplex | ||||||||
| 2523 | X. albilineans | ||||||||
| 2528 | X. arboricola pv. juglandis | ||||||||
| 3369 | X. citri pv. citri | ||||||||
| 2537 | X. vesicatoria | ||||||||
| 5241 | X. campestris pv. campestris | ||||||||
| 2541 | X. translucens pv. cerealis |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clavijo-Coppens, F.; Ginet, N.; Cesbron, S.; Briand, M.; Jacques, M.-A.; Ansaldi, M. Novel Virulent Bacteriophages Infecting Mediterranean Isolates of the Plant Pest Xylella fastidiosa and Xanthomonas albilineans. Viruses 2021, 13, 725. https://doi.org/10.3390/v13050725
Clavijo-Coppens F, Ginet N, Cesbron S, Briand M, Jacques M-A, Ansaldi M. Novel Virulent Bacteriophages Infecting Mediterranean Isolates of the Plant Pest Xylella fastidiosa and Xanthomonas albilineans. Viruses. 2021; 13(5):725. https://doi.org/10.3390/v13050725
Chicago/Turabian StyleClavijo-Coppens, Fernando, Nicolas Ginet, Sophie Cesbron, Martial Briand, Marie-Agnès Jacques, and Mireille Ansaldi. 2021. "Novel Virulent Bacteriophages Infecting Mediterranean Isolates of the Plant Pest Xylella fastidiosa and Xanthomonas albilineans" Viruses 13, no. 5: 725. https://doi.org/10.3390/v13050725
APA StyleClavijo-Coppens, F., Ginet, N., Cesbron, S., Briand, M., Jacques, M.-A., & Ansaldi, M. (2021). Novel Virulent Bacteriophages Infecting Mediterranean Isolates of the Plant Pest Xylella fastidiosa and Xanthomonas albilineans. Viruses, 13(5), 725. https://doi.org/10.3390/v13050725






