The Bee Hemolymph Metabolome: A Window into the Impact of Viruses on Bumble Bees
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect, Virus, and Injection
2.2. Hemolymph Collection
2.3. Extraction of Polar and Medium Polar Metabolites from Bee Hemolymph
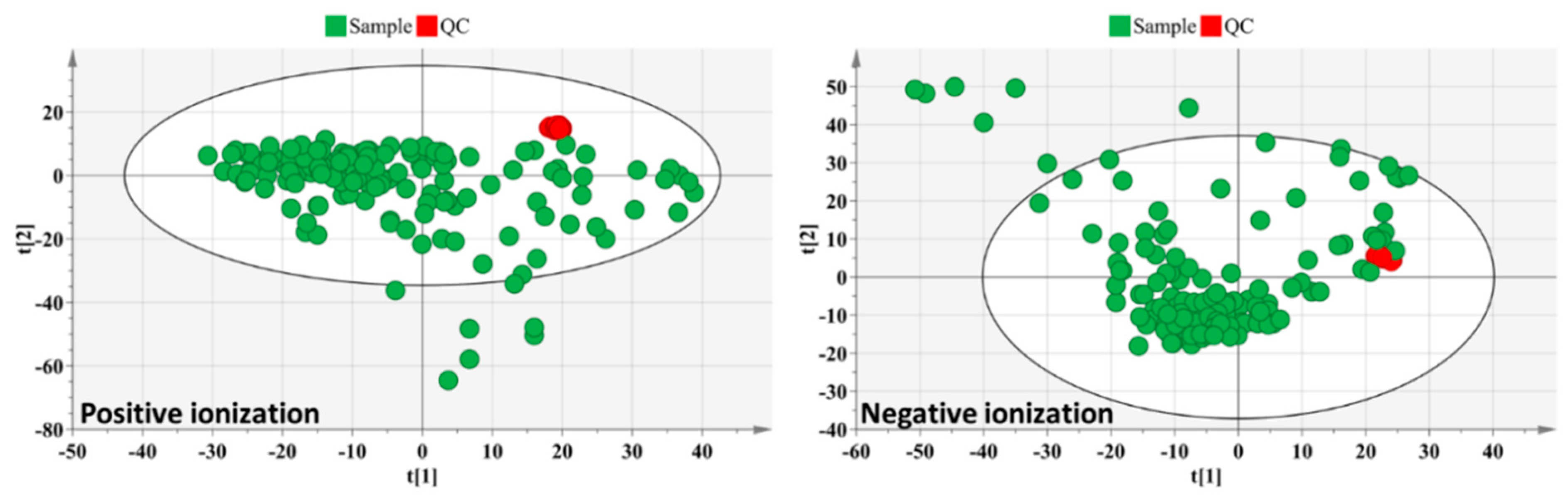
2.4. UHPLC-Q-Orbitrap-HRMS Analysis
2.5. Predictive Modelling of Viral Infection in Standardized Bees
2.5.1. Experimental Setup
2.5.2. Data Analysis and Biomarker Selection
2.6. Predictive Potential of 2 Sets of Biomarkers in Wild Bumble Bees
3. Results and Discussion
3.1. Untargeted Data Analysis
3.1.1. Predictive Modelling of Viral Infection in Non-Stressed Commercial Bees
3.1.2. Predictive Modelling of Viral Infection in Stressed Commercial Bees
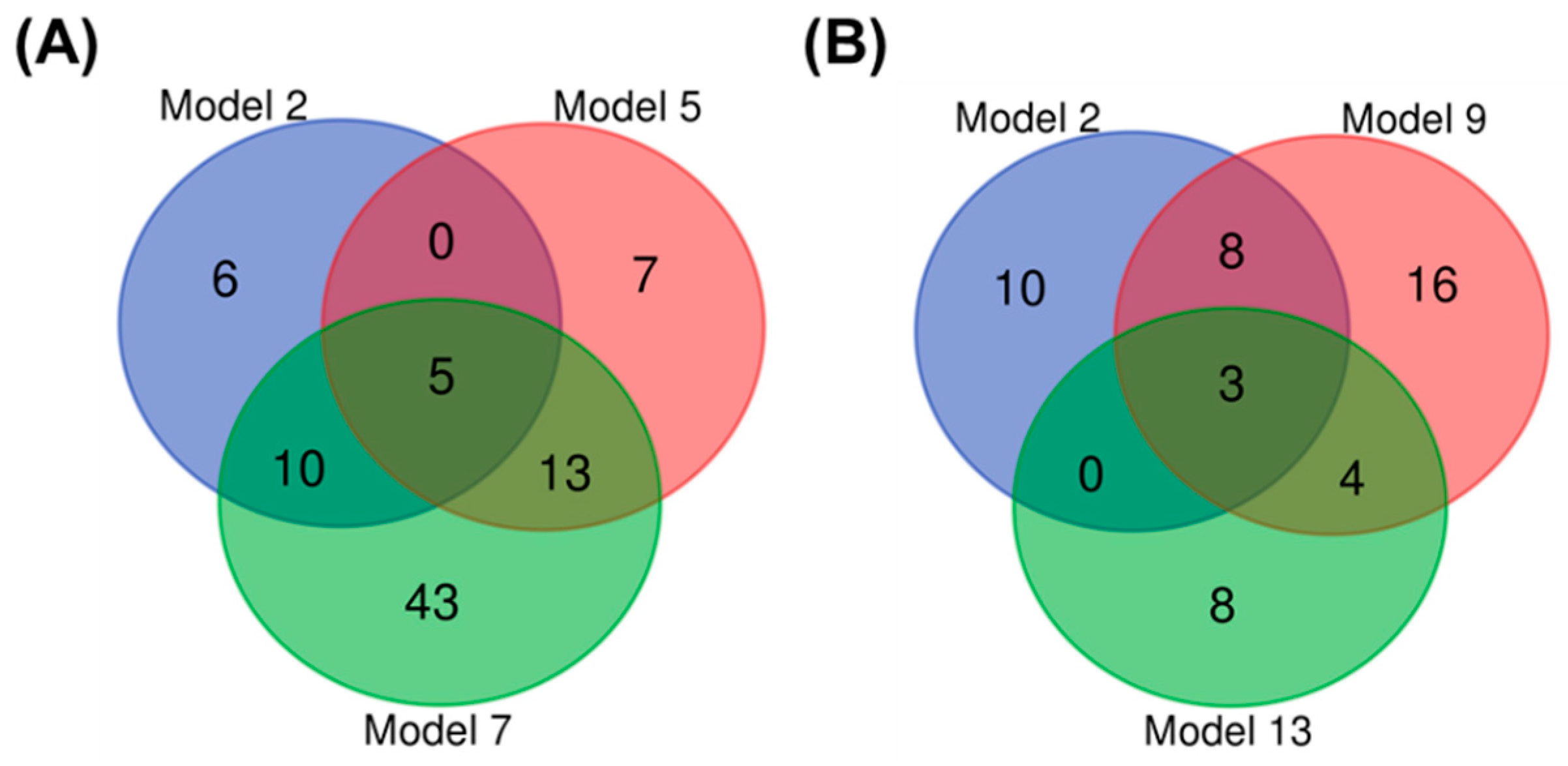
3.1.3. Biomarker Selection to Discriminate Infection in Wild Bees
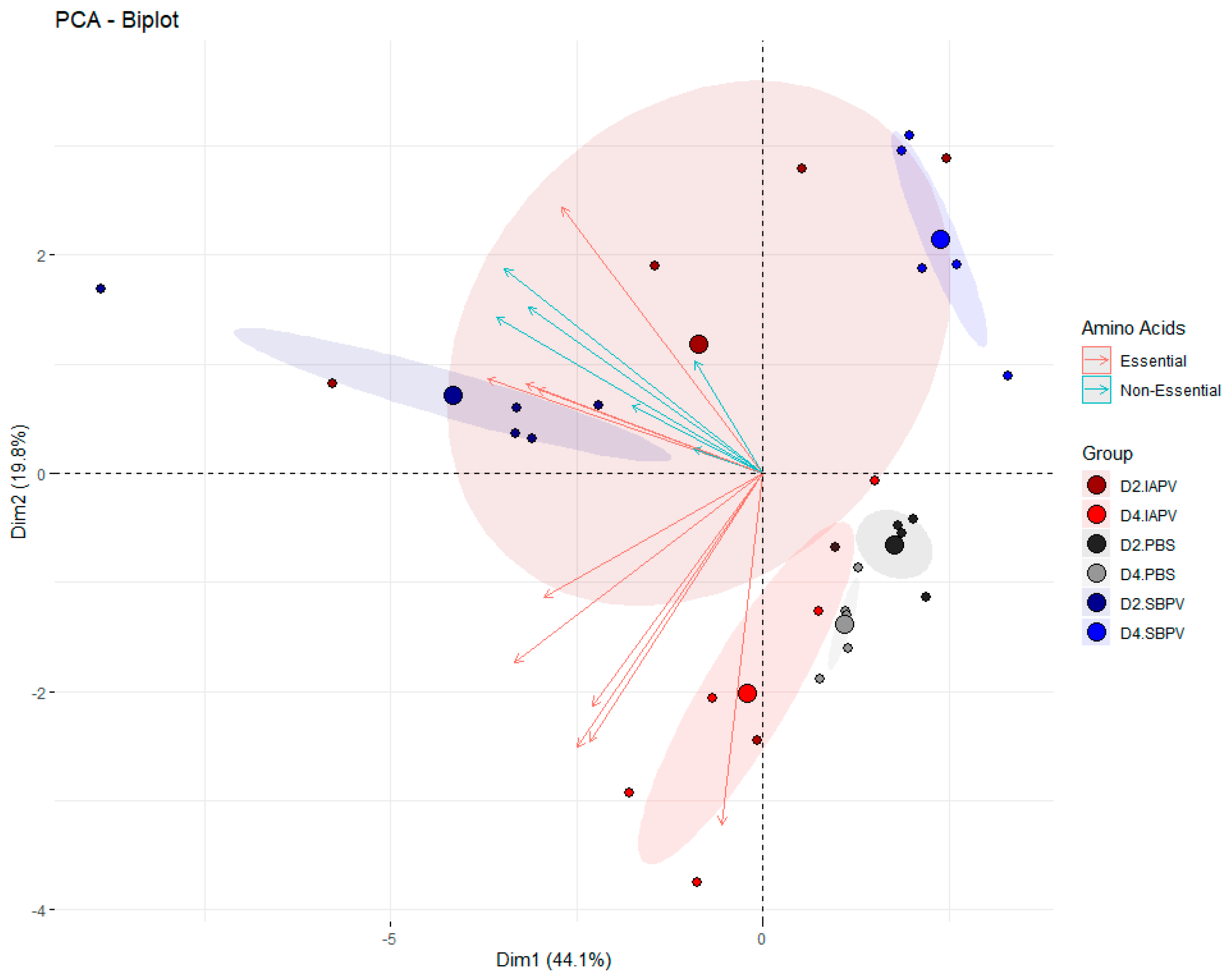
3.2. Targeted Data Analysis
3.3. Predictive Power of the Biomarker Set
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Grozinger, C.M.; Flenniken, M.L. Bee viruses: Ecology, pathogenicity, and impacts. Annu. Rev. Entomol. 2019, 64, 205–226. [Google Scholar] [CrossRef] [PubMed]
- González Varo, J.P.; Biesmeijer, J.C.; Bommarco, R.; Potts, S.G.; Schweiger, O.; Smith, H.G.; Steffan, D.I.; Szentgyörgyi, H.; Woyciechowski, M.; Vilà, M. Combined effects of global change pressures on animal-mediated pollination. Trends Ecol. Evol. 2013, 28, 524–530. [Google Scholar] [CrossRef]
- Meeus, I.; Pisman, M.; Smagghe, G.; Piot, N. Interaction effects of different drivers of wild bee decline and their influence on host-pathogen dynamics. Curr. Opin. Insect Sci. 2018, 26, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Trapp, J.; McAfee, A.; Foster, L.J. Genomics, transcriptomics and proteomics: Enabling insights into social evolution and disease challenges for managed and wild bees. Mol. Ecol. 2017, 26, 718–739. [Google Scholar] [CrossRef] [PubMed]
- McMenamin, A.J.; Flenniken, M.L. Recently identified bee viruses and their impact on bee pollinators. Curr. Opin. Insect Sci. 2018, 26, 120–129. [Google Scholar] [CrossRef]
- Kim, B.; Kim, J.; Kim, S.J.; Kim, M.O.; Truong, A.T.; Cho, K.; Yoon, B. Detection of chronic bee paralysis virus using ultra-rapid PCR and nested ultra-rapid PCR. J. Apic. Res. 2019, 58, 133–140. [Google Scholar] [CrossRef]
- de Miranda, J.R.; Bailey, L.; Ball, B.V.; Blanchard, P.; Budge, G.E.; Chejanovskym, N.; Chen, Y.P.; Gauthier, L.; Genersch, E.; De Graaf, D.C. Standard methods for virus research in Apis mellifera. J. Apic. Res. 2013, 52, 1–56. [Google Scholar] [CrossRef]
- Galbraith, D.A.; Fuller, Z.L.; Ray, A.M.; Brockmann, A.; Frazier, M.; Gikungu, M.W.; Martinez, J.F.I.; Kapheim, K.M.; Kerby, J.T.; Kocher, S.D. Investigating the viral ecology of global bee communities with high-throughput metagenomics. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- García-Anaya, M.C.; Romo-Chacón, A.; Sáenz-Mendoza, A.I.; Pérez-Ordoñez, G.; Acosta-Muñiz, C.H. Detection of Israeli Acute Paralysis Virus (IAPV) and Apis mellifera Filamentous Virus (AmFV) in Honey Bees in Mexico. J. Apic. Res. 2018, 62, 141–144. [Google Scholar] [CrossRef]
- Goic, B.; Vodovar, N.; Mondotte, J.A.; Monot, C.; Frangeul, L.; Blanc, H.; Gausson, V.; Vera Otarola, J.; Cristofari, G.; Saleh, M.C. RNA-mediated interference and reverse transcription control the persistence of RNA viruses in the insect model Drosophila. Nat. Immunol. 2013, 14, 396–403. [Google Scholar] [CrossRef]
- Williams, T.; Virto, C.; Murillo, R.; Caballero, P. Covert infection of insects by baculoviruses. Front. Microbiol. 2017, 8, 1337. [Google Scholar] [CrossRef] [PubMed]
- McMenamin, A.J.; Genersch, E. Honey bee colony losses and associated viruses. Curr. Opin. Insect Sci. 2015, 8, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Nazzi, F.; Pennacchio, F. Honey bee antiviral immune barriers as affected by multiple stress factors: A novel paradigm to interpret colony health decline and collapse. Viruses 2018, 10, 159. [Google Scholar] [CrossRef] [PubMed]
- Hails, R.S.; Ball, B.V.; Genersch, E. Infection strategies of insect viruses. In Virology and the Honey Bee; Aubert, M., Ball, B., Fries, I., Moritz, R., Milani, N., Bernardinelli, I., Eds.; European Communities: Luxembourg, 2007; pp. 255–275. [Google Scholar]
- Perakakis, N.; Yazdani, A.; Karniadakis, G.E.; Mantzoros, C. Omics, big data and machine learning as tools to propel understanding of biological mechanisms and to discover novel diagnostics and therapeutics. Metab. Clin. Exp. 2018, 87, A1–A9. [Google Scholar] [CrossRef] [PubMed]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Metabolomics: The apogee of the omics trilogy. Nat. Rev. Mol. 2012, 13, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Douglas, A.E. Omics and the metabolic function of insect–microbial symbioses. Curr. Opin. Insect Sci. 2018, 29, 1–6. [Google Scholar] [CrossRef]
- Wang, L.L.; Swevers, L.; Van Meulebroek, L.; Meeus, I.; Vanhaecke, L.; Smagghe, G. Metabolomic analysis of cricket paralysis virus infection in Drosophila S2 cells reveals divergent effects on central carbon metabolism as compared with silkworm Bm5 cells. Viruses 2020, 12, 393. [Google Scholar] [CrossRef]
- Wang, L.L.; Swevers, L.; Rombouts, C.; Meeus, I.; Van Meulebroek, L.; Vanhaecke, L.; Smagghe, G. A metabolomics approach to unravel cricket paralysis virus infection in silkworm Bm5 cells. Viruses 2019, 11, 861. [Google Scholar] [CrossRef]
- Wang, L.L.; Meeus, I.; Rombouts, C.; Van Meulebroek, L.; Vanhaecke, L.; Smagghe, G. Metabolomics-based biomarker discovery for bee health monitoring: A proof of concept study concerning nutritional stress in Bombus terrestris. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Niu, J.; Smagghe, G.; De Coninck, D.I.; Van Nieuwerburgh, F.; Deforce, D.; Meeus, I. In vivo study of Dicer-2-mediated immune response of the small interfering RNA pathway upon systemic infections of virulent and avirulent viruses in Bombus terrestris. Insect Biochem. Mol. Biol. 2015, 70, 127–137. [Google Scholar] [CrossRef]
- Mayer, K.A.; Stockl, J.; Zlabinger, G.J.; Gualdoni, G.A. Hijacking the supplies: Metabolism as a novel facet of virus-host interaction. Front. Immunol. 2019, 10, 1533. [Google Scholar] [CrossRef]
- Ren, W.; Rajendran, R.; Zhao, Y.; Tan, B.; Wu, G.; Bazer, F.W.; Zhu, G.; Peng, Y.; Huang, X.; Deng, J. Amino acids as mediators of metabolic cross talk between host and pathogen. Front. Immunol. 2018, 9, 319. [Google Scholar] [CrossRef]
- Tong, L.; Nieh, J.C.; Tosi, S. Combined nutritional stress and a new systemic pesticide (flupyradifurone, Sivanto®) reduce bee survival, food consumption, flight success, and thermoregulation. Chemosphere 2019, 237, 124408. [Google Scholar] [CrossRef]
- Niu, J.; Cappelle, K.; de Miranda, J.R.; Smagghe, G.; Meeus, I. Analysis of reference gene stability after Israeli acute paralysis virus infection in bumblebees Bombus terrestris. J. Invertebr. Pathol. 2014, 115, 76–79. [Google Scholar] [CrossRef]
- Locke, B.; Forsgren, E.; Fries, I.; de Miranda, J.R. Acaricide treatment affects viral dynamics in Varroa destructor-infested honey bee colonies via both host physiology and mite control. Appl. Environ. Microbiol. 2012, 78, 227–235. [Google Scholar] [CrossRef]
- Michopoulos, F.; Lai, L.; Gika, H.; Theodoridis, G.; Wilson, I. UPLC-MS-based analysis of human plasma for metabonomics using solvent precipitation or solid phase extraction. J. Proteome Res. 2009, 8, 2114–2121. [Google Scholar] [CrossRef]
- Ulaszewska, M.M.; Weinert, C.H.; Trimigno, A.; Portmann, R.; Andres Lacueva, C.; Badertscher, R.; Brennan, L.; Brunius, C.; Bub, A.; Capozzi, F. Nutrimetabolomics: An integrative action for metabolomic analyses in human nutritional studies. Mol. Nutr. Food Res. 2019, 63, 1800384. [Google Scholar] [CrossRef] [PubMed]
- Vanden Bussche, J.; Marzorati, M.; Laukens, D.; Vanhaecke, L. Validated high resolution mass spectrometry-based approach for metabolomic fingerprinting of the human gut phenotype. Anal. Chem. 2015, 87, 10927–10934. [Google Scholar] [CrossRef] [PubMed]
- De Paepe, E.; Van Meulebroek, L.; Rombouts, C.; Huysman, S.; Verplanken, K.; Lapauw, B.; Wauters, J.; Hemeryck, L.Y.; Vanhaecke, L. A validated multi-matrix platform for metabolomic fingerprinting of human urine, feces and plasma using ultra-high performance liquid-chromatography coupled to hybrid orbitrap high-resolution mass spectrometry. Anal. Chim. Acta 2018, 1033, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Frank, E.; Hall, M.; Trigg, L.; Holmes, G.; Witten, I.H. Data mining in bioinformatics using Weka. Bioinformatics 2004, 20, 2479–2481. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.Y.; Lee, H.S.; Kang, D.G.; Kim, N.S.; Cha, M.H.; Bang, O.S.; Ryu, D.H.; Hwang, G.S. 1H-NMR-based metabolomics study of cerebral infarction. Stroke 2011, 42, 1282–1288. [Google Scholar] [CrossRef] [PubMed]
- Dolezal, A.G.; Toth, A.L. Feedbacks between nutrition and disease in honey bee health. Curr. Opin. Insect Sci. 2018, 26, 114–119. [Google Scholar] [CrossRef]
- Wright, G.A.; Nicolson, S.W.; Shafir, S. Nutritional physiology and ecology of honey bees. Annu. Rev. Entomol. 2018, 63, 327–344. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, G.R. The biochemistry of insect hemolymph. Annu. Rev. Entomol. 1961, 6, 75–102. [Google Scholar] [CrossRef]
- Wyatt, G.R.; Kalf, G.F. The chemistry of insect hemolymph: II. Trehalose and other carbohydrates. J. Gen. Physiol. 1957, 40, 833–847. [Google Scholar] [CrossRef]
- Paoli, P.P.; Donley, D.; Stabler, D.; Saseendranath, A.; Nicolson, S.W.; Simpson, S.J.; Wright, G.A. Nutritional balance of essential amino acids and carbohydrates of the adult worker honeybee depends on age. Amino Acids 2014, 46, 1449–1458. [Google Scholar] [CrossRef] [PubMed]
- Mounce, B.C.; Olsen, M.E.; Vignuzzi, M.; Connor, J.H. Polyamines and their role in virus infection. Microbiol. Mol. Biol. Rev. 2017, 81, e00029-17. [Google Scholar] [CrossRef] [PubMed]
- Firpo, M.R.; Mounce, B.C. Diverse functions of polyamines in virus infection. Biomolecules 2020, 10, 628. [Google Scholar] [CrossRef]
- Balint, R.; Cohen, S.S. The incorporation of radiolabeled polyamines and methionine into turnip yellow mosaic virus in protoplasts from infected plants. Virology 1985, 144, 181–193. [Google Scholar] [CrossRef]
- Dolezal, A.G.; Carrillo Tripp, J.; Judd, T.M.; Allen Miller, W.; Bonning, B.C.; Toth, A.L. Interacting stressors matter: Diet quality and virus infection in honeybee health. R. Soc. Open Sci. 2019, 6, 181803. [Google Scholar] [CrossRef]
| Model | Comparison | Model Specification | Numbers of Model Components (to + tp) a | Model Characteristics b | p-value Cross-Validated ANOVA c | Permutation d |
|---|---|---|---|---|---|---|
| Virus | ||||||
| 1 | 25% diet: virus (n = 25) vs. non-virus (n = 30) | Virus | - e | - | - | No |
| 2 | 50% diet: virus (n = 25) vs. non-virus (n = 30) | Virus | 1 + 4 + 0 | R2Y = 0.97; Q2 = 0.741 | 6.10 × 10−10 | Good |
| 3 | All: virus (n = 50) vs. non-virus (n = 60) | Virus | - | - | - | No |
| IAPV | ||||||
| 4 | 25% diet: IAPV (n = 10) vs. PBS (n = 10) | IAPV | - | - | - | No |
| 5 | 50% diet: IAPV (n = 10) vs. PBS (n = 10) | IAPV | 1 + 3 + 0 | R2Y = 0.995; Q2 = 0.906 | 2.80 × 10−5 | Good |
| 6 | All: 4DPI (n = 10) vs. 2DPI (n = 10) | Time | 1 + 5 + 0 | R2Y = 0.999; Q2 = 0.875 | 1.75 × 10−3 | Good |
| 7 | All: IAPV (n = 20) vs. PBS (n = 20) | IAPV | 1 + 5 + 0 | R2Y = 0.993; Q2 = 0.905 | 1.01 × 10−10 | Good |
| SBPV | ||||||
| 8 | 25% diet: SBPV (n = 15) vs. PBS (n = 15) | SBPV | - | - | - | No |
| 9 | 50% diet: SBPV (n = 15) vs. PBS (n = 15) | SBPV | 1 + 4 + 0 | R2Y = 0.995; Q2 = 0.931 | 6.49 × 10−9 | Good |
| 10 | All: 4DPI (n = 10) vs. 2DPI (n = 10) | Time | 1 + 4 + 0 | R2Y = 0.997; Q2 = 0.883 | 3.81 × 10−4 | Good |
| 11 | All: 14DPI (n = 10) vs. 2DPI (n = 10) | Time | 1 + 2 + 0 | R2Y = 0.970; Q2 = 0.871 | 4.11 × 10−5 | Good |
| 12 | All: 14DPI (n = 10) vs. 4DPI (n = 10) | Time | 1 + 7 + 0 | R2Y = 1.000; Q2 = 0.863 | 2.00 × 10−2 | Good |
| 13 | All: SBPV (n = 30) vs. PBS (n = 30) | SBPV | 1 + 9 + 0 | R2Y = 0.996; Q2 = 0.694 | 3.66×10−5 | Good |
| IAPV vs. SBPV | ||||||
| 14 | 25% diet: SBPV (n = 10) vs. IAPV (n = 10) | SBPV/IAPV | 1 + 4 + 0 | R2Y = 0.996; Q2 = 0.827 | 2.77 × 10−3 | Good |
| 15 | 50% diet: SBPV (n = 10) vs. IAPV (n = 10) | SBPV/IAPV | 1 + 1 + 0 | R2Y = 0.958; Q2 = 0.849 | 5.14 × 10−6 | Good |
| 16 | All: SBPV (n = 30) vs. IAPV (n = 20) | SBPV/IAPV | 1 + 5 + 0 | R2Y = 0.984; Q2 = 0.811 | 5.67 × 10−10 | Good |
| Features | Discrimination | Sensitivity | Precision | AUC |
|---|---|---|---|---|
| Untargeted (n = 8) | Virus vs. Non-virus | 77.5% | 77.6% | 0.892 |
| IAPV vs. Non-virus | 86.7% | 88.9% | 0.955 | |
| SBPV vs. Non-virus | 80.0% | 79.5 | 0.815 | |
| Amino acids + polyamines (n = 21) | Virus vs. Non-virus | 90.0% | 90.0% | 0.945 |
| IAPV vs. Non-virus | 93.3% | 93.3% | 0.985 | |
| SBPV vs. Non-virus | 90.0% | 89.9% | 0.875 | |
| Combined (n = 29) | Virus vs. Non-virus | 90.0% | 90.0% | 0.948 |
| IAPV vs. Non-virus | 90.0% | 89.9% | 0.985 | |
| SBPV vs. Non-virus | 83.3% | 83.1% | 0.930 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Van Meulebroek, L.; Vanhaecke, L.; Smagghe, G.; Meeus, I. The Bee Hemolymph Metabolome: A Window into the Impact of Viruses on Bumble Bees. Viruses 2021, 13, 600. https://doi.org/10.3390/v13040600
Wang L, Van Meulebroek L, Vanhaecke L, Smagghe G, Meeus I. The Bee Hemolymph Metabolome: A Window into the Impact of Viruses on Bumble Bees. Viruses. 2021; 13(4):600. https://doi.org/10.3390/v13040600
Chicago/Turabian StyleWang, Luoluo, Lieven Van Meulebroek, Lynn Vanhaecke, Guy Smagghe, and Ivan Meeus. 2021. "The Bee Hemolymph Metabolome: A Window into the Impact of Viruses on Bumble Bees" Viruses 13, no. 4: 600. https://doi.org/10.3390/v13040600
APA StyleWang, L., Van Meulebroek, L., Vanhaecke, L., Smagghe, G., & Meeus, I. (2021). The Bee Hemolymph Metabolome: A Window into the Impact of Viruses on Bumble Bees. Viruses, 13(4), 600. https://doi.org/10.3390/v13040600







