Detection of Tioman Virus in Pteropus vampyrus Near Flores, Indonesia
Abstract
1. Introduction
2. Materials and Methods
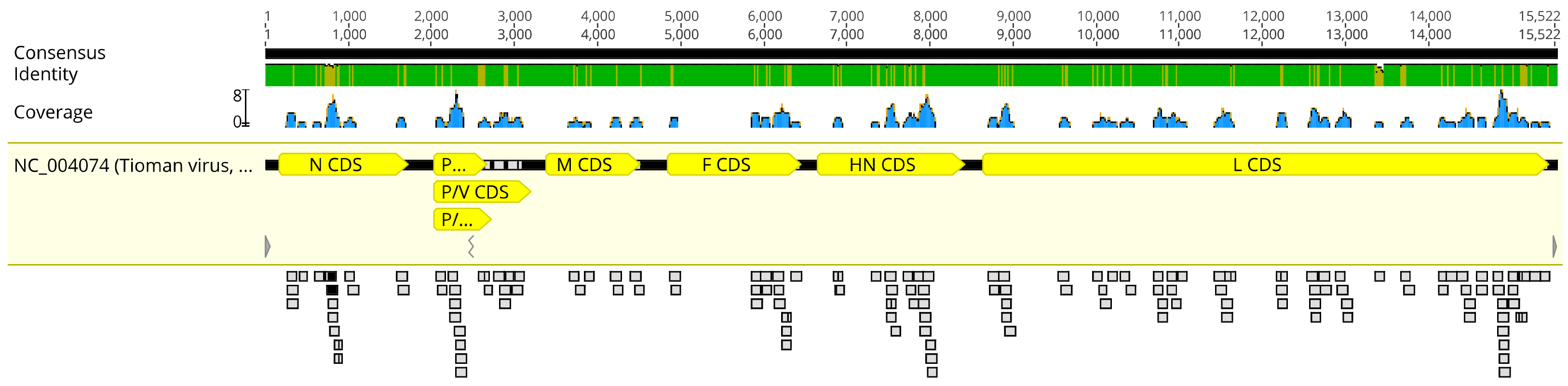
PCR, Next Generation Sequencing and Sequence Analysis
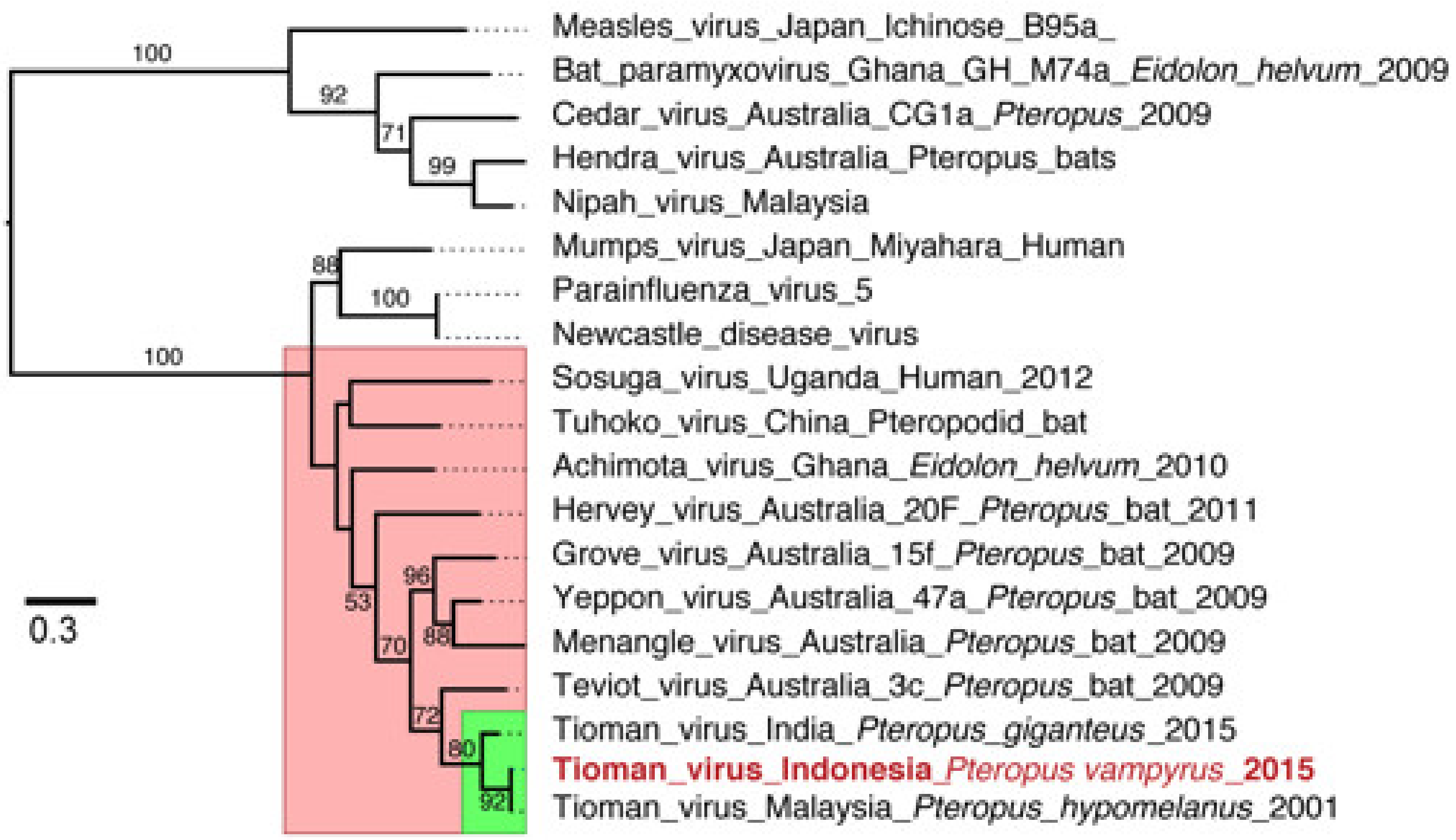
3. Results/Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Irving, A.T.; Ahn, M.; Goh, G.; Anderson, D.E.; Wang, L.F. Lessons from the host defenses of bats, a unique viral reservoir. Nature 2021, 589, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Calisher, C.H.; Childs, J.E.; Field, H.E.; Holmes, K.V.; Schountz, T. Bats: Important reservoir hosts of emerging viruses. Clin. Micro. Rev. 2006, 19, 531–545. [Google Scholar] [CrossRef]
- Hutson, A.M.; Mickleburgh, S.P. Microchiropteran Bats: Global Status Survey and Conservation Action Plan; IUCN: Cambridge, UK, 2001; Volume 56. [Google Scholar]
- Kunz, T.H.; Fenton, M.B. Bat Ecology; University of Chicago Press: Chicago, IL, USA, 2005. [Google Scholar]
- Hayman, D.T. Bats as viral reservoirs. Annu. Rev. Virol. 2016, 3, 77–99. [Google Scholar] [CrossRef] [PubMed]
- Allen, T.; Murray, K.A.; Zambrana-Torrelio, C.; Morse, S.S.; Rondinini, C.; Di Marco, M.; Breit, N.; Olival, K.J.; Daszak, P. Global hotspots and correlates of emerging zoonotic diseases. Nat. Commun. 2017, 8, 1124. [Google Scholar] [CrossRef]
- Plowright, R.K.; Foley, P.; Field, H.E.; Dobson, A.P.; Foley, J.E.; Eby, P.; Daszak, P. Urban habituation, ecological connectivity and epidemic dampening: The emergence of Hendra virus from flying foxes (Pteropus spp.). Proc. R. Soc. B 2011, 278, 3703–3712. [Google Scholar] [CrossRef]
- Jones, B.A.; Grace, D.; Kock, R.; Alonso, S.; Rushton, J.; Said, M.Y.; McKeever, D.; Mutua, F.; Young, J.; McDermott, J. Zoonosis emergence linked to agricultural intensification and environmental change. Proc. Natl. Acad. Sci. USA 2013, 110, 8399–8404. [Google Scholar] [CrossRef]
- Voigt, C.C.; Kingston, T. Bats in the Anthropocene: Conservation of Bats in a Changing World; Springer Science+ Business Media: Cham, Switzerland, 2016. [Google Scholar]
- Drexler, J.F.; Corman, V.M.; Muller, M.A.; Maganga, G.D.; Vallo, P.; Binger, T.; Gloza-Rausch, F.; Cottontail, V.M.; Rasche, A.; Yordanov, S.; et al. Bats host major mammalian paramyxoviruses. Nat. Commun. 2012, 3, 796. [Google Scholar] [CrossRef] [PubMed]
- Rima, B.; Balkema-Buschmann, A.; Dundon, W.G.; Duprex, P.; Easton, A.; Fouchier, R.; Kurath, G.; Lamb, R.; Lee, B.; Rota, P. ICTV Virus Taxonomy Profile: Paramyxoviridae. J. Gen. Virol. 2019, 100, 1593–1594. [Google Scholar] [CrossRef]
- Tong, S.; Chern, S.W.; Li, Y.; Pallansch, M.A.; Anderson, L.J. Sensitive and broadly reactive reverse transcription-PCR assays to detect novel paramyxoviruses. J. Clin. Microbiol. 2008, 46, 2652–2658. [Google Scholar] [CrossRef]
- Anderson, D.E.; Wang, L.-F. New and emerging paramyxoviruses. In The Biology of Paramyxoviruses; Caister Academic Press: Norfolk, UK, 2011; pp. 435–459. [Google Scholar]
- Barr, J.; Smith, C.; Smith, I.; de Jong, C.; Todd, S.; Melville, D.; Broos, A.; Crameri, S.; Haining, J.; Marsh, G.; et al. Isolation of multiple novel paramyxoviruses from pteropid bat urine. J. Gen. Virol. 2015, 96, 24–29. [Google Scholar] [CrossRef]
- Chua, K.B.; Wang, L.F.; Lam, S.K.; Crameri, G.; Yu, M.; Wise, T.; Boyle, D.; Hyatt, A.D.; Eaton, B.T. Tioman virus, a novel paramyxovirus isolated from fruit bats in Malaysia. Virology 2001, 283, 215–229. [Google Scholar] [CrossRef]
- Simmons, N.; Cirranello, A. Bat Species of the World: A Taxonomic and Geographic Database. Available online: https://batnames.org (accessed on 15 March 2021).
- Arevalo, R.L.M.; Amador, L.I.; Almeida, F.C.; Giannini, N.P. Evolution of body mass in bats: Insights from a large supermatrix phylogeny. J. Mamm. Evol. 2020, 27, 123–138. [Google Scholar] [CrossRef]
- Tsang, S.M.; Wiantoro, S. Indonesian flying foxes: Research and conservation status update. Trebuia 2019, 46, 103–113. [Google Scholar] [CrossRef]
- Tsang, S.M.; Wiantoro, S.; Veluz, M.J.; Simmons, N.B.; Lohman, D.J. Low levels of population structure among geographically distant populations of Pteropus vampyrus (Chiroptera: Pteropodidae). Acta Chiropt. 2018, 20, 59–71. [Google Scholar] [CrossRef]
- Tsang, S.M.; Wiantoro, S.; Simmons, N.B. New records of flying foxes (Chiroptera: Pteropus sp.) from Seram, Indonesia, with notes on ecology and conservation status. Am. Mus. Novit. 2015, 3842, 1–23. [Google Scholar] [CrossRef]
- Eaton, B.T.; Broder, C.C.; Middleton, D.; Wang, L.-F. Hendra and Nipah viruses: Different and dangerous. Nat. Rev. Microbiol. 2006, 4, 23–35. [Google Scholar] [CrossRef]
- Tsang, S.M.; Wiantoro, S.; Veluz, M.J.; Sugita, N.; Nguyen, Y.L.; Simmons, N.B.; Lohman, D.J. Dispersal out of Wallacea spurs diversification of Pteropus flying foxes, the world’s largest bats (Mammalia: Chiroptera). J. Biogeogr. 2020, 47, 527–537. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Bioinformatics, Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Beier, S.; Flade, I.; Gorska, A.; El-Hadidi, M.; Mitra, S.; Ruscheweyh, H.J.; Tappu, R. MEGAN Community Edition -Interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput. Biol. 2016, 12, e1004957. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Yaiw, K.C.; Crameri, G.; Wang, L.; Chong, H.T.; Chua, K.B.; Tan, C.T.; Goh, K.J.; Shamala, D.; Wong, K.T. Serological evidence of possible human infection with Tioman virus, a newly described paramyxovirus of bat origin. J. Infect. Dis. 2007, 196, 884–886. [Google Scholar] [CrossRef] [PubMed]
- Yaiw, K.C.; Bingham, J.; Crameri, G.; Mungall, B.; Hyatt, A.; Yu, M.; Eaton, B.; Shamala, D.; Wang, L.F.; Thong Wong, K. Tioman virus, a paramyxovirus of bat origin, causes mild disease in pigs and has a predilection for lymphoid tissues. J. Virol. 2008, 82, 565–568. [Google Scholar] [CrossRef][Green Version]
- Anderson, D.E.; Marsh, G.A. Bat paramyxoviruses. In Bats and Viruses; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; pp. 99–126. [Google Scholar]
- Iehle, C.; Razafitrimo, G.; Razainirina, J.; Andriaholinirina, N.; Goodman, S.M.; Faure, C.; Georges-Courbot, M.C.; Rousset, D.; Reynes, J.M. Henipavirus and Tioman virus antibodies in pteropodid bats, Madagascar. Emerg. Infect. Dis. 2007, 13, 159–161. [Google Scholar] [CrossRef] [PubMed]
- Yadav, P.; Sarkale, P.; Patil, D.; Shete, A.; Kokate, P.; Kumar, V.; Jain, R.; Jadhav, S.; Basu, A.; Pawar, S. Isolation of Tioman virus from Pteropus giganteus bat in North-East region of India. Infect. Genet. Evol. 2016, 45, 224–229. [Google Scholar] [CrossRef]
- Breed, A.C.; Yu, M.; Barr, J.A.; Crameri, G.; Thalmann, C.M.; Wang, L.-F. Prevalence of henipavirus and rubulavirus antibodies in pteropid bats, Papua New Guinea. Emerg. Infect. Dis. 2010, 16, 1997. [Google Scholar] [CrossRef] [PubMed]
- Boardman, W.S.; Baker, M.L.; Boyd, V.; Crameri, G.; Peck, G.R.; Reardon, T.; Smith, I.G.; Caraguel, C.G.; Prowse, T.A. Seroprevalence of three paramyxoviruses; Hendra virus, Tioman virus, Cedar virus and a rhabdovirus, Australian bat lyssavirus, in a range expanding fruit bat, the Grey-headed flying fox (Pteropus poliocephalus). PLoS ONE 2020, 15, e0232339. [Google Scholar] [CrossRef]
- Marsh, G.A.; Wang, L.-F. Henipaviruses: Deadly zoonotic paramyxoviruses of bat origin. In The Role of Animals in Emerging Viral Diseases; Elsevier: Amsterdam, The Netherlands, 2014; pp. 125–142. [Google Scholar]
- Lomolino, M.V. The unifying, fundamental principles of biogeography: Understanding island life. Front. Biogeogr. 2016, 8, e29920. [Google Scholar] [CrossRef]
- Peel, A.J.; Baker, K.S.; Hayman, D.T.; Broder, C.C.; Cunningham, A.A.; Fooks, A.R.; Garnier, R.; Wood, J.L.; Restif, O. Support for viral persistence in bats from age-specific serology and models of maternal immunity. Sci. Rep. 2018, 8, 1–11. [Google Scholar]
- Subudhi, S.; Rapin, N.; Misra, V. Immune system modulation and viral persistence in bats: Understanding viral spillover. Viruses 2019, 11, 192. [Google Scholar] [CrossRef] [PubMed]
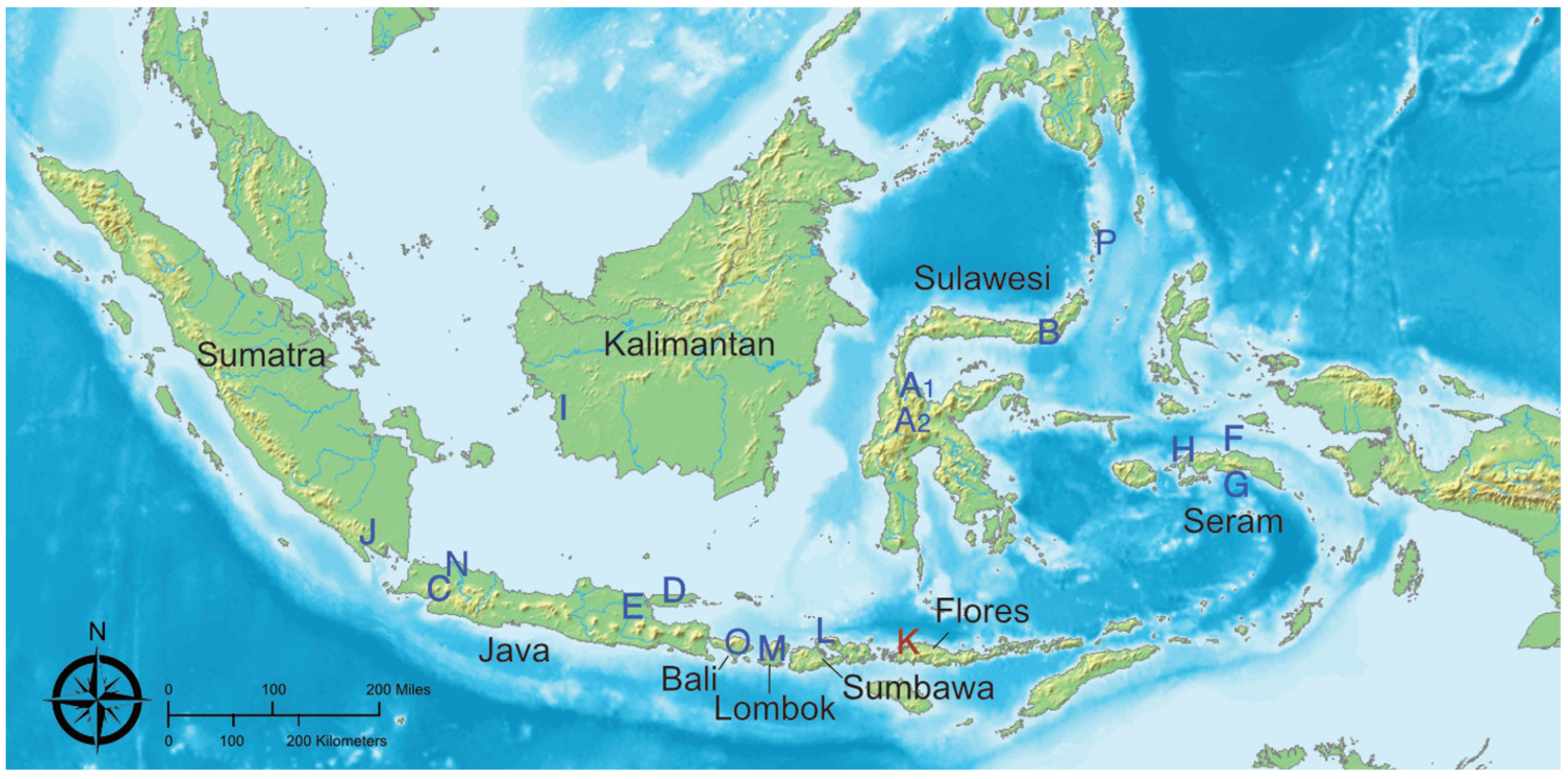
| Species | Sampling Location | Site Code | No. bats | Kidney | Liver | Lung | Oral Swab | Rectal Swab | Small Intestine |
|---|---|---|---|---|---|---|---|---|---|
| Acerodon celebensis | Central Sulawesi | A1 | 6 | - | 6 | - | - | 3 | - |
| Acerodon celebensis | Gorontalo | B | 6 | - | 6 | 2 | 4 | 2 | - |
| Acerodon mackloti | Lombok | M | 2 | 2 | 2 | 2 | 3 | 2 | 2 |
| Chironax melanocephalus | Central Sulawesi | A2 | 1 | - | 1 | - | - | - | - |
| Cynopterus sp. | Central Sulawesi | A2 | 5 | - | 4 | - | - | 2 | - |
| Dobsonia cf. peronii | Lombok | M | 1 | - | - | - | 1 | 1 | - |
| Dobsonia moluccensis | Maluku | H | 1 | - | 1 | - | - | - | - |
| Dobsonia sp. | Seram, Maluku | G | 2 | - | 1 | - | 2 | 2 | - |
| Dobsonia viridis | Maluku | H | 3 | - | 3 | - | - | 2 | - |
| Eonycteris spelaea | West Kalimantan | I | 2 | - | 2 | - | - | - | - |
| Macroglossus minimus | Central Sulawesi | A2 | 2 | - | 2 | - | - | 1 | - |
| Macroglossus minimus | Seram, Maluku | H | 1 | - | 1 | - | - | - | - |
| Nyctimene sp. | Seram, Maluku | G,H | 6 | - | 2 | - | 4 | 4 | - |
| Pteropus alecto | Central Sulawesi | A1 | 12 | - | 11 | 4 | 2 | 5 | - |
| Pteropus alecto | Gorontalo | B | 4 | - | 4 | - | 2 | 1 | - |
| Pteropus chrysoproctus | Seram, Maluku | H | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| Pteropus hypomelanus | East Java | E | 3 | 2 | 2 | 2 | - | 3 | 2 |
| Pteropus hypomelanus | Sangihe Islands | P | 3 | - | 3 | - | 3 | 2 | - |
| Pteropus lombocensis | Lombok | M | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Pteropus ocularis | Seram, Maluku | F | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Pteropus temminckii | Seram Maluku | F,G | 3 | 3 | 3 | 2 | 3 | 3 | 3 |
| Pteropus vampyrus | Bali | O | 1 | - | - | - | 1 | 1 | - |
| Pteropus vampyrus | East Java | E | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Pteropus vampyrus | Flores | K | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Pteropus vampyrus | South Sumatra | J | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Pteropus vampyrus | West Java | C | 4 | 1 | 4 | 2 | 4 | 4 | 1 |
| Pteropus vampyrus | West Kalimantan | I | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Pteropus vampyrus | Sumbawa | L | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Rousettus amplexicaudatus | Seram, Maluku | H | 2 | - | 1 | - | 1 | 1 | - |
| Rousettus celebensis | Central Sulawesi | A2 | 1 | - | 1 | - | - | - | - |
| Rousettus linduensis | Central Sulawesi | A2 | 1 | - | 1 | - | - | - | - |
| Rousettus sp. | Central Sulawesi | A2 | 4 | - | 3 | - | - | 2 | - |
| Syconycteris australis | Seram, Maluku | G,H | 4 | - | 2 | - | 2 | 3 | - |
| Total | 95 | 23 | 81 | 29 | 47 | 59 | 23 |
| Taxon | Forward Unpaired Reads | Reverse Unpaired Reads | Forward Paired Reads | Reverse Paired Reads | Total |
|---|---|---|---|---|---|
| Eukaryota | 4,569,303 (78.7%) | 803,357 (82.0%) | 27,331,682 (85.3%) | 27,643,874 (86.3%) | 60,348,216 (85.0%) |
| Vertebrata | 4,436,518 (75.3%) | 788,376 (80.5%) | 26,699,724 (83.3%) | 27,103,904 (84.6%) | 59,028,522 (83.2%) |
| Platyhelminthes | 5949 (0.1%) | 111 (<0.1%) | 28,122 (<0.1%) | 5914 (<0.1%) | 40,096 (<0.1%) |
| Bacteria | 93,262 (1.6%) | 11,778 (1.2%) | 516,241 (1.6%) | 507,629 (1.6%) | 1128,910 (1.6%) |
| Fungi | 76 (<0.1%) | 5 (<0.1%) | 392 (<0.1%) | 355 (<0.1%) | 828 (<0.1%) |
| Viruses | 446 (<0.1%) | 79 (<0.1%) | 2293 (<0.1%) | 2230 (<0.1%) | 5048 (<0.1%) |
| Herpesvirales | 3 (<0.1%) | 0 (0%) | 72 (<0.1%) | 52 (<0.1%) | 127 (<0.1%) |
| Retroviridae | 97 (<0.1%) | 28 (<0.1%) | 889 (<0.1%) | 893 (<0.1%) | 1907 (<0.1%) |
| Poxviridae | 2 (<0.1%) | 1 (<0.1%) | 16 (<0.1%) | 16 (<0.1%) | 35 (<0.1%) |
| Paramyxoviridae | 5 (<0.1%) | 4 (<0.1%) | 70 (<0.1%) | 68 (<0.1%) | 147 (<0.1%) |
| Not assigned | 1,011,911 (17.2%) | 127,330 (13.0%) | 2,916,552 (9.1%) | 2,586,915 (8.1%) | 6,645,708 (9.4%) |
| Total Reads | 5,890,265 | 979,360 | 32,056,464 | 32,030,156 | 70,956,245 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsang, S.M.; Low, D.H.W.; Wiantoro, S.; Smith, I.; Jayakumar, J.; Simmons, N.B.; Vijaykrishna, D.; Lohman, D.J.; Mendenhall, I.H. Detection of Tioman Virus in Pteropus vampyrus Near Flores, Indonesia. Viruses 2021, 13, 563. https://doi.org/10.3390/v13040563
Tsang SM, Low DHW, Wiantoro S, Smith I, Jayakumar J, Simmons NB, Vijaykrishna D, Lohman DJ, Mendenhall IH. Detection of Tioman Virus in Pteropus vampyrus Near Flores, Indonesia. Viruses. 2021; 13(4):563. https://doi.org/10.3390/v13040563
Chicago/Turabian StyleTsang, Susan M., Dolyce H. W. Low, Sigit Wiantoro, Ina Smith, Jayanthi Jayakumar, Nancy B. Simmons, Dhanasekaran Vijaykrishna, David J. Lohman, and Ian H. Mendenhall. 2021. "Detection of Tioman Virus in Pteropus vampyrus Near Flores, Indonesia" Viruses 13, no. 4: 563. https://doi.org/10.3390/v13040563
APA StyleTsang, S. M., Low, D. H. W., Wiantoro, S., Smith, I., Jayakumar, J., Simmons, N. B., Vijaykrishna, D., Lohman, D. J., & Mendenhall, I. H. (2021). Detection of Tioman Virus in Pteropus vampyrus Near Flores, Indonesia. Viruses, 13(4), 563. https://doi.org/10.3390/v13040563






