An Overview of Current Knowledge of Deadly CoVs and Their Interface with Innate Immunity
Abstract
1. Human CoVs
2. SARS-CoV-1
3. MERS-CoV
4. SARS-CoV-2
5. Name, Classification and Viral Structure
6. Epidemiology and Transmission
6.1. SARS-CoV-1
6.2. MERS-CoV
6.3. SARS-CoV-2
7. Viral Cell Entry and Life Cycle
8. Innate Immune Response to Coronaviruses
9. TLR and RLR Signalling
10. Anti-Viral IFNs and the JAK-STAT Pathway
11. SARS-CoV-1 Immune Modulatory Mechanisms
12. MERS-CoV Immune Evasion
13. SARS-CoV-2 Antagonism of IFN
14. Treatment
14.1. SARS-CoV-1
14.2. MERS-CoV
14.3. SARS-CoV-2
15. Conclusions and Future Considerations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Su, S.; Wong, G.; Shi, W.; Liu, J.; Lai, A.C.; Zhou, J.; Liu, W.; Bi, Y.; Gao, G.F. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016, 24, 490–502. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.X.; Ng, Y.L.; Tam, J.P.; Liu, D.X. Human Coronaviruses: A Review of Virus–Host Interactions. Diseases 2016, 4, 26. [Google Scholar] [CrossRef]
- Fehr, A.R.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. Methods Mol. Biol. 2015, 1282, 1–23. [Google Scholar] [PubMed]
- Woo, P.C.Y.; Lau, S.K.P.; Chu, C.-M.; Chan, K.-H.; Tsoi, H.-W.; Huang, Y.; Wong, B.H.L.; Poon, R.W.S.; Cai, J.J.; Luk, W.-K.; et al. Characterization and Complete Genome Sequence of a Novel Coronavirus, Coronavirus HKU1, from Patients with Pneumonia. J. Virol. 2005, 79, 884–895. [Google Scholar] [CrossRef]
- Walsh, E.E.; Shin, J.H.; Falsey, A.R. Clinical Impact of Human Coronaviruses 229E and OC43 Infection in Diverse Adult Populations. J. Infect. Dis. 2013, 208, 1634–1642. [Google Scholar] [CrossRef]
- Meo, S.A.; Alhowikan, A.M.; Al-Khlaiwi, T.; Meo, I.M.; Halepoto, D.M.; Iqbal, M.; Usmani, A.M.; Hajjar, W.; Ahmed, N. Novel coronavirus 2019-nCoV: Prevalence, biological and clinical characteristics comparison with SARS-CoV and MERS-CoV. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 2012–2019. [Google Scholar]
- Xu, R.H.; He, J.F.; Evans, M.R.; Peng, G.W.; Field, H.E.; Yu, D.W.; Lee, C.K.; Luo, H.M.; Lin, W.S.; Lin, P.; et al. Epidemiologic Clues to SARS Origin in China. Emerg. Infect. Dis. 2004, 10, 1030–1037. [Google Scholar] [CrossRef]
- WHO. Summary of Probable SARS Cases with Onset of Illness from 1 November 2002 to 31 July 2003. 2003. Available online: http://www.who.int/csr/sars/country/table2004_04_21/en/index.html (accessed on 26 March 2021).
- Manocha, S.; Walley, K.R.; Russell, J.A. Severe acute respiratory distress syndrome (SARS): A critical care perspective. Crit. Care Med. 2003, 31, 2684–2692. [Google Scholar] [CrossRef]
- Stockman, L.J.; Bellamy, R.; Garner, P. SARS: Systematic Review of Treatment Effects. PLoS Med. 2006, 3, e343. [Google Scholar] [CrossRef]
- Organisation, W.H. SARS Outbreak Contained Worldwide. 2003. Available online: https://www.who.int/mediacentre/news/releases/2003/pr56/en/ (accessed on 6 March 2021).
- Letko, M.; Marzi, A.; Munster, V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569. [Google Scholar] [CrossRef]
- Woo, P.C.; Lau, S.K.; Li, K.S.; Tsang, A.K.; Yuen, K.Y. Genetic relatedness of the novel human group C betacoronavirus to Tylonycteris bat coronavirus HKU4 and Pipistrellus bat coronavirus HKU5. Emerg. Microbes Infect. 2012, 1, 1–5. [Google Scholar] [CrossRef]
- Li, W. Bats Are Natural Reservoirs of SARS-Like Coronaviruses. Science 2005, 310, 676–679. [Google Scholar] [CrossRef]
- Lu, L.; Liu, Q.; Du, L.; Jiang, S. Middle East respiratory syndrome coronavirus (MERS-CoV): Challenges in identifying its source and controlling its spread. Microbes Infect. 2013, 15, 625–629. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Zhang, T.; Wu, Q.; Zhang, Z. Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak. Curr. Biol. 2020, 30, 1346–1351.e2. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yan, M.; Xu, H.; Liang, W.; Kan, B.; Zheng, B.; Chen, H.; Zheng, H.; Xu, Y.; Zhang, E.; et al. SARS-CoV Infection in a Restaurant from Palm Civet. Emerg. Infect. Dis. 2005, 11, 1860–1865. [Google Scholar] [CrossRef] [PubMed]
- Azhar, E.I.; El-Kafrawy, S.A.; Farraj, S.A.; Hassan, A.M.; Al-Saeed, M.S.; Hashem, A.M.; Madani, T.A. Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med. 2014, 370, 2499–2505. [Google Scholar] [CrossRef]
- Byrne, P.; Cullinan, J.; Murphy, C.; Smith, S.M. Cross-sectional analysis of the prevalence and predictors of statin utilisation in Ireland with a focus on primary prevention of cardiovascular disease. BMJ Open 2018, 8, e018524. [Google Scholar] [CrossRef] [PubMed]
- Back, H.; Weld, J.; Walsh, C.; Cullinane, A. Equine Rhinitis A Virus Infection in Thoroughbred Racehorses-A Putative Role in Poor Performance? Viruses 2019, 11, 963. [Google Scholar] [CrossRef]
- Wei, X.; Xiao, Y.T.; Wang, J.; Chen, R.; Zhang, W.; Yang, Y.; Lv, D.; Qin, C.; Gu, D.; Zhang, B.; et al. Sex Differences in Severity and Mortality Among Patients With COVID-19: Evidence from Pooled Literature Analysis and Insights from Integrated Bioinformatic Analysis. arXiv 2020, arXiv:2003.13547. [Google Scholar]
- Baig, A.M.; Khaleeq, A.; Ali, U.; Syeda, H. Evidence of the COVID-19 virus targeting the CNS: Tissue distribution, host–virus interaction, and proposed neurotropic mechanisms. ACS Chem. Neurosci. 2020, 11, 995–998. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Wohlford-Lenane, C.; Perlman, S.; Zhao, J.; Jewell, A.K.; Reznikov, L.R.; Gibson-Corley, K.N.; Meyerholz, D.K.; McCray, P.B., Jr. Middle East respiratory syndrome coronavirus causes multiple organ damage and lethal disease in mice transgenic for human dipeptidyl peptidase 4. J. Infect. Dis. 2016, 213, 712–722. [Google Scholar] [CrossRef] [PubMed]
- Hamming, I.; Timens, W.; Bulthuis, M.L.C.; Lely, A.T.; Navis, G.V.; van Goor, H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004, 203, 631–637. [Google Scholar] [CrossRef]
- van Doremalen, N.; Miazgowicz, K.L.; Milne-Price, S.; Bushmaker, T.; Robertson, S.; Scott, D.; Kinne, J.; McLellan, J.S.; Zhu, J.; Munster, V.J. Host species restriction of Middle East respiratory syndrome coronavirus through its receptor, dipeptidyl peptidase 4. J. Virol. 2014, 88, 9220–9232. [Google Scholar] [CrossRef]
- Klemann, C.; Wagner, L.; Stephan, M.; von Hörsten, S. Cut to the chase: A review of CD26/dipeptidyl peptidase-4′s (DPP4) entanglement in the immune system. Clin. Exp. Immunol. 2016, 185, 1–21. [Google Scholar] [CrossRef]
- Cho, S.Y.; Kang, J.-M.; Ha, Y.E.; Park, G.E.; Lee, J.Y.; Ko, J.-H.; Kim, J.M.; Kang, C.-I.; Jo, I.J.; Ryu, J.G.; et al. MERS-CoV outbreak following a single patient exposure in an emergency room in South Korea: An epidemiological outbreak study. Lancet 2016, 388, 994–1001. [Google Scholar] [CrossRef]
- Zaki, A.M.; Van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.; Fouchier, R.A. Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef]
- Racelis, S.; de los Reyes, V.C.; Sucaldito, M.N.; Deveraturda, I.; Roca, J.B.; Tayag, E. Contact tracing the first Middle East respiratory syndrome case in the Philippines, February 2015. West. Pac. Surveill. Response J. WPSAR 2015, 6, 3. [Google Scholar] [CrossRef]
- WHO: Covid-19 Situation in China 2021. Available online: https://covid19.who.int/region/wpro/country/cn (accessed on 6 March 2021).
- Wu, J.T.; Leung, K.; Leung, G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: A modelling study. Lancet 2020, 395, 689–697. [Google Scholar] [CrossRef]
- Carlos, W.G.; Dela Cruz, C.S.; Cao, B.; Pasnick, S.; Jamil, S. Novel Wuhan (2019-nCoV) Coronavirus. Am. J. Respir. Crit. Care Med. 2020, 201, P7–P8. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Jung, S.M.; Akhmetzhanov, A.R.; Hayashi, K.; Linton, N.M.; Yang, Y.; Yuan, B.; Kobayashi, T.; Kinoshita, R.; Nishiura, H. Real-Time Estimation of the Risk of Death from Novel Coronavirus (COVID-19) Infection: Inference Using Exported Cases. J. Clin. Med. 2020, 9, 523. [Google Scholar] [CrossRef]
- Guan, W.J.; Liang, W.H.; Zhao, Y.; Liang, H.R.; Chen, Z.S.; Li, Y.M.; Liu, X.Q.; Chen, R.C.; Tang, C.L.; Wang, T.; et al. Comorbidity and its impact on 1590 patients with Covid-19 in China: A Nationwide Analysis. Eur. Respir. J. 2020, 55, 2000547. [Google Scholar] [CrossRef] [PubMed]
- Verity, R.; Okell, L.C.; Dorigatti, I.; Winskill, P.; Whittaker, C.; Imai, N.; Cuomo-Dannenburg, G.; Thompson, H.; Walker, P.G.; Fu, H.; et al. Estimates of the severity of coronavirus disease 2019: A model-based analysis. Lancet Infect. Dis. 2020, 20, 669–677. [Google Scholar] [CrossRef]
- Macnaughton, M.R. Occurrence and frequency of coronavirus infections in humans as determined by enzyme-linked immunosorbent assay. Infect. Immun. 1982, 38, 419–423. [Google Scholar] [CrossRef]
- Enserink, M. Amid Heightened Concerns, New Name for Novel Coronavirus Emerges. Science 2013, 340, 673. [Google Scholar] [CrossRef]
- Ren, Z.; Yan, L.; Zhang, N.; Guo, Y.; Yang, C.; Lou, Z.; Rao, Z. The newly emerged SARS-like coronavirus HCoV-EMC also has an” Achilles’ heel”: Current effective inhibitor targeting a 3C-like protease. Protein Cell 2013, 4, 248. [Google Scholar] [CrossRef]
- De Groot, R.J.; Baker, S.C.; Baric, R.S.; Brown, C.S.; Drosten, C.; Enjuanes, L.; Fouchier, R.A.M.; Galiano, M.; Gorbalenya, A.E.; Memish, Z.A.; et al. Middle East Respiratory Syndrome Coronavirus (MERS-CoV): Announcement of the Coronavirus Study Group. J. Virol. 2013, 87, 7790–7792. [Google Scholar] [CrossRef]
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W.; et al. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef]
- Song, Z.; Xu, Y.; Bao, L.; Zhang, L.; Yu, P.; Qu, Y.; Zhu, H.; Zhao, W.; Han, Y.; Qin, C. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses 2019, 11, 59. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.W.; Kok, K.H.; Zhu, Z.; Chu, H.; To, K.K.W.; Yuan, S.; Yuen, K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef] [PubMed]
- Khailany, R.A.; Safdar, M.; Ozaslan, M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020, 19, 100682. [Google Scholar] [CrossRef]
- Mielech, A.M.; Kilianski, A.; Baez-Santos, Y.M.; Mesecar, A.D.; Baker, S.C. MERS-CoV papain-like protease has deISGylating and deubiquitinating activities. Virology 2014, 450–451, 64–70. [Google Scholar] [CrossRef]
- Horner, G.; Daddona, J.; Burke, D.J.; Cullinane, J.; Skeer, M.; Wurcel, A.G. “You’re kind of at war with yourself as a nurse”: Perspectives of inpatient nurses on treating people who present with a comorbid opioid use disorder. PLoS ONE 2019, 14, e0224335. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Peng, L.; Park, J.J.; Hu, Y.; Devarkar, S.C.; Dong, M.B.; Shen, Q.; Wu, S.; Chen, S.; Lomakin, I.B.; et al. Nonstructural Protein 1 of SARS-CoV-2 Is a Potent Pathogenicity Factor Redirecting Host Protein Synthesis Machinery toward Viral RNA. Mol. Cell 2020, 80, 1055–1066.e6. [Google Scholar] [CrossRef]
- Oudshoorn, D.; Rijs, K.; Limpens, R.W.; Groen, K.; Koster, A.J.; Snijder, E.J.; Kikkert, M.; Bárcena, M. Expression and cleavage of Middle East respiratory syndrome coronavirus nsp3-4 polyprotein induce the formation of double-membrane vesicles that mimic those associated with coronaviral RNA replication. MBio 2017, 8, e01658-17. [Google Scholar] [CrossRef]
- Cottam, E.M.; Whelband, M.C.; Wileman, T. Coronavirus NSP6 restricts autophagosome expansion. Autophagy 2014, 10, 1426–1441. [Google Scholar] [CrossRef]
- Zhai, Y.; Sun, F.; Li, X.; Pang, H.; Xu, X.; Bartlam, M.; Rao, Z. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat. Struct. Mol. Biol. 2005, 12, 980–986. [Google Scholar] [CrossRef]
- Clasman, J.R.; Báez-Santos, Y.M.; Mettelman, R.C.; O’Brien, A.; Baker, S.C.; Mesecar, A.D. X-ray Structure and Enzymatic Activity Profile of a Core Papain-like Protease of MERS Coronavirus with utility for structure-based drug design. Sci. Rep. 2017, 7, 40292. [Google Scholar] [CrossRef]
- Miknis, Z.J.; Donaldson, E.F.; Umland, T.C.; Rimmer, R.A.; Baric, R.S.; Schultz, L.W. Severe Acute Respiratory Syndrome Coronavirus nsp9 Dimerization Is Essential for Efficient Viral Growth. J. Virol. 2009, 83, 3007–3018. [Google Scholar] [CrossRef]
- Bouvet, M.; Lugari, A.; Posthuma, C.C.; Zevenhoven, J.C.; Bernard, S.; Betzi, S.; Imbert, I.; Canard, B.; Guillemot, J.-C.; Lécine, P.; et al. Coronavirus Nsp10, a Critical Co-factor for Activation of Multiple Replicative Enzymes. J. Biol. Chem. 2014, 289, 25783–25796. [Google Scholar] [CrossRef]
- Snijder, E.; Decroly, E.; Ziebuhr, J. The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing. In Advances in Clinical Chemistry; Elsevier: Amsterdam, The Netherlands, 2016; Volume 96, pp. 59–126. [Google Scholar]
- Amraei, R.; Napoleon, M.; Yin, W.; Berrigan, J.; Suder, E.; Zhao, G.; Olejnik, J.; Chandler, K.; Xia, C.; Gummuluru, S.; et al. CD209L/L-SIGN and CD209/DC-SIGN act as receptors for SARS-CoV-2 and are differentially expressed in lung and kidney epithelial and endothelial cells. bioRxiv 2020. [Google Scholar] [CrossRef]
- Guarino, L.A.; Bhardwaj, K.; Dong, W.; Sun, J.; Holzenburg, A.; Kao, C. Mutational Analysis of the SARS Virus Nsp15 Endoribonuclease: Identification of Residues Affecting Hexamer Formation. J. Mol. Biol. 2005, 353, 1106–1117. [Google Scholar] [CrossRef]
- Guan, Y.J.; Zheng, B.J.; He, Y.Q.; Liu, X.L.; Zhuang, Z.X.; Cheung, C.L.; Luo, S.W.; Li, P.H.; Zhang, L.J.; Butt, K.M.; et al. Isolation and Characterization of Viruses Related to the SARS Coronavirus from Animals in Southern China. Science 2003, 302, 276–278. [Google Scholar] [CrossRef]
- Kan, B.; Wang, M.; Jing, H.; Xu, H.; Jiang, X.; Yan, M.; Liang, W.; Zheng, H.; Wan, K.; Liu, Q.; et al. Molecular Evolution Analysis and Geographic Investigation of Severe Acute Respiratory Syndrome Coronavirus-Like Virus in Palm Civets at an Animal Market and on Farms. J. Virol. 2005, 79, 11892–11900. [Google Scholar] [CrossRef] [PubMed]
- Poon, L.L.M.; Chu, D.K.W.; Chan, K.H.; Wong, O.K.; Ellis, T.M.; Leung, Y.H.C.; Lau, S.K.P.; Woo, P.C.Y.; Suen, K.Y.; Guan, Y.; et al. Identification of a Novel Coronavirus in Bats. J. Virol. 2005, 79, 2001–2009. [Google Scholar] [CrossRef]
- Wang, L.-F.; Shi, Z.; Zhang, S.; Field, H.; Daszak, P.; Eaton, B.T. Review of Bats and SARS. Emerg. Infect. Dis. 2006, 12, 1834–1840. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Hui, D.; Wu, A.; Chan, P.; Cameron, P.; Joynt, G.M.; Ahuja, A.; Yung, M.Y.; Leung, C.; To, K.; et al. A Major Outbreak of Severe Acute Respiratory Syndrome in Hong Kong. N. Engl. J. Med. 2003, 348, 1986–1994. [Google Scholar] [CrossRef] [PubMed]
- Gormley, M.; Aspray, T.J.; Kelly, D.A.; Rodriguez-Gil, C. Pathogen cross-transmission via building sanitary plumbing systems in a full scale pilot test-rig. PLoS ONE 2017, 12, e0171556. [Google Scholar] [CrossRef]
- Bassetti, S.; Bischoff, W.E.; Sherertz, R.J. Are SARS Superspreaders Cloud Adults? Emerg. Infect. Dis. 2005, 11, 637–638. [Google Scholar] [CrossRef]
- Booth, T.F.; Kournikakis, B.; Bastien, N.; Ho, J.; Kobasa, D.; Stadnyk, L.; Li, Y.; Spence, M.; Paton, S.; Henry, B.; et al. Detection of Airborne Severe Acute Respiratory Syndrome (SARS) Coronavirus and Environmental Contamination in SARS Outbreak Units. J. Infect. Dis. 2005, 191, 1472–1477. [Google Scholar] [CrossRef]
- Yu, I.T.S.; Wong, T.W.; Chiu, Y.L.; Lee, N.; Li, Y. Temporal-Spatial Analysis of Severe Acute Respiratory Syndrome among Hospital Inpatients. Clin. Infect. Dis. 2005, 40, 1237–1243. [Google Scholar] [CrossRef]
- Lessler, J.; Reich, N.G.; Brookmeyer, R.; Perl, T.M.; Nelson, K.E.; Cummings, D.A. Incubation periods of acute respiratory viral infections: A systematic review. Lancet Infect. Dis. 2009, 9, 291–300. [Google Scholar] [CrossRef]
- Wilder-Smith, A.; Teleman, M.D.; Heng, B.H.; Earnest, A.; Ling, A.E.; Leo, Y.S. Asymptomatic SARS Coronavirus Infection among Healthcare Workers, Singapore. Emerg. Infect. Dis. 2005, 11, 1142–1145. [Google Scholar] [CrossRef]
- Yang, G.-G.; Lin, S.-Z.; Liao, K.-W.; Lee, J.-J.; Wang, L.-S. SARS-associated Coronavirus Infection in Teenagers. Emerg. Infect. Dis. 2004, 10, 382–383. [Google Scholar] [CrossRef]
- Chan-Yeung, M.; Xu, R.H. SARS: Epidemiology. Respirology 2003, 8, S9–S14. [Google Scholar] [CrossRef]
- Memish, Z.A.; Mishra, N.; Olival, K.J.; Fagbo, S.F.; Kapoor, V.; Epstein, J.H.; AlHakeem, R.; Durosinloun, A.; Al Asmari, M.; Islam, A.; et al. Middle East Respiratory Syndrome Coronavirus in Bats, Saudi Arabia. Emerg. Infect. Dis. 2013, 19, 1819–1823. [Google Scholar] [CrossRef]
- Raj, V.S.; Osterhaus, A.D.; Fouchier, R.A.; Haagmans, B.L. MERS: Emergence of a novel human coronavirus. Curr. Opin. Virol. 2014, 5, 58–62. [Google Scholar] [CrossRef]
- Reusken, C.B.; Haagmans, B.L.; Müller, M.A.; Gutierrez, C.; Godeke, G.J.; Meyer, B.; Muth, D.; Raj, V.S.; Smits-De Vries, L.; Corman, V.M.; et al. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: A comparative serological study. Lancet Infect. Dis. 2013, 13, 859–866. [Google Scholar] [CrossRef]
- Zumla, A.; Hui, D.S.; Perlman, S. Middle East respiratory syndrome. Lancet 2015, 386, 995–1007. [Google Scholar] [CrossRef]
- Al Muhairi, S.; Al Hosani, F.; Eltahir, Y.M.; Al Mulla, M.; Yusof, M.F.; Serhan, W.S.; Hashem, F.M.; Elsayed, E.A.; Marzoug, B.A.; Abdelazim, A.S. Epidemiological investigation of Middle East respiratory syndrome coronavirus in dromedary camel farms linked with human infection in Abu Dhabi Emirate, United Arab Emirates. Virus Genes 2016, 52, 848–854. [Google Scholar] [CrossRef]
- Hemida, M.G.; Chu, D.K.; Poon, L.L.; Perera, R.A.; Alhammadi, M.A.; Ng, H.-Y.; Siu, L.Y.; Guan, Y.; Alnaeem, A.; Peiris, M. MERS Coronavirus in Dromedary Camel Herd, Saudi Arabia. Emerg. Infect. Dis. 2014, 20, 1231–1234. [Google Scholar] [CrossRef] [PubMed]
- Alagaili, A.N.; Briese, T.; Mishra, N.; Kapoor, V.; Sameroff, S.C.; De Wit, E.; Munster, V.J.; Hensley, L.E.; Zalmout, I.S.; Kapoor, A.; et al. Middle East Respiratory Syndrome Coronavirus Infection in Dromedary Camels in Saudi Arabia. mBio 2014, 5, e00884-14. [Google Scholar] [CrossRef]
- Reusken, C.B.; Farag, E.A.; Jonges, M.; Godeke, G.J.; El-Sayed, A.M.; Pas, S.D.; Raj, V.S.; Mohran, K.A.; Moussa, H.A.; Ghobashy, H.; et al. Middle East respiratory syndrome coronavirus (MERS-CoV) RNA and neutralising antibodies in milk collected according to local customs from dromedary camels, Qatar, April 2014. Eurosurveillance 2014, 19, 20829. [Google Scholar] [CrossRef] [PubMed]
- Van Doremalen, N.; Bushmaker, T.; Karesh, W.B.; Munster, V.J. Stability of Middle East Respiratory Syndrome Coronavirus in Milk. Emerg. Infect. Dis. 2014, 20, 1263–1264. [Google Scholar] [CrossRef] [PubMed]
- Lee, J. Better Understanding on MERS Corona Virus Outbreak in Korea. J. Korean Med. Sci. 2015, 30, 835–836. [Google Scholar] [CrossRef]
- Ng, D.L.; Al Hosani, F.; Keating, M.K.; Gerber, S.I.; Jones, T.L.; Metcalfe, M.G.; Tong, S.; Tao, Y.; Alami, N.N.; Haynes, L.M.; et al. Clinicopathologic, Immunohistochemical, and Ultrastructural Findings of a Fatal Case of Middle East Respiratory Syndrome Coronavirus Infection in the United Arab Emirates, April 2014. Am. J. Pathol. 2016, 186, 652–658. [Google Scholar] [CrossRef] [PubMed]
- Omrani, A.S.; Matin, M.A.; Haddad, Q.; Al-Nakhli, D.; Memish, Z.A.; Albarrak, A.M. A family cluster of Middle East Respiratory Syndrome Coronavirus infections related to a likely unrecognized asymptomatic or mild case. Int. J. Infect. Dis. 2013, 17, e668–e672. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Li, X.; Zai, J.; Wang, X.; Li, Y. Potential of large “first generation” human-to-human transmission of 2019-nCoV. J. Med Virol. 2020, 92, 448–454. [Google Scholar] [CrossRef]
- Li, X.; Wang, W.; Zhao, X.; Zai, J.; Zhao, Q.; Li, Y.; Chaillon, A. Transmission dynamics and evolutionary history of 2019-nCoV. J. Med. Virol. 2020, 92, 501–511. [Google Scholar] [CrossRef]
- Lam, T.T.Y.; Shum, M.H.H.; Zhu, H.C.; Tong, Y.G.; Ni, X.B.; Liao, Y.S.; Wei, W.; Cheung, W.Y.M.; Li, W.J.; Li, L.F.; et al. Identification of 2019-nCoV related coronaviruses in Malayan pangolins in southern China. bioRxiv 2020. [Google Scholar] [CrossRef]
- Liu, P.; Jiang, J.-Z.; Wan, X.-F.; Hua, Y.; Li, L.; Zhou, J.; Wang, X.; Hou, F.; Chen, J.; Zou, J.; et al. Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)? PLoS Pathog. 2020, 16, e1008421. [Google Scholar] [CrossRef]
- Frutos, R.; Serra-Cobo, J.; Chen, T.; Devaux, C.A. COVID-19: Time to exonerate the pangolin from the transmission of SARS-CoV-2 to humans. Infect. Genet. Evol. 2020, 84, 104493. [Google Scholar] [CrossRef]
- Organisation, W.H. WHO Timeline—COVID-19. 2020. Available online: https://www.who.int/news-room/detail/08-04-2020-who-timeline---covid-19 (accessed on 6 March 2021).
- Apolone, G.; Montomoli, E.; Manenti, A.; Boeri, M.; Sabia, F.; Hyseni, I.; Mazzini, L.; Martinuzzi, D.; Cantone, L.; Milanese, G.; et al. Unexpected detection of SARS-CoV-2 antibodies in the prepandemic period in Italy. Tumor. J. 2020. [Google Scholar] [CrossRef]
- Fongaro, G.; Stoco, P.H.; Souza, D.S.M.; Grisard, E.C.; Magri, M.E.; Rogovski, P.; Schorner, M.A.; Barazzetti, F.H.; Christoff, A.P.; de Oliveira, L.F.V.; et al. SARS-CoV-2 in human sewage in Santa Catalina, Brazil, November 2019. medRxiv 2020. [Google Scholar] [CrossRef]
- Basavaraju, S.V.; Patton, M.E.; Grimm, K.; Rasheed, M.A.U.; Lester, S.; Mills, L.; Stumpf, M.; Freeman, B.; Tamin, A.; Harcourt, J.; et al. Serologic testing of US blood donations to identify SARS-CoV-2-reactive antibodies: December 2019–January 2020. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Zou, L.; Ruan, F.; Huang, M.; Liang, L.; Huang, H.; Hong, Z.; Yu, J.; Kang, M.; Song, Y.; Xia, J.; et al. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N. Engl. J. Med. 2020, 382, 1177–1179. [Google Scholar] [CrossRef]
- Almadhi, M.A.; Abdulrahman, A.; Sharaf, S.A.; AlSaad, D.; Stevenson, N.J.; Atkin, S.L.; AlQahtani, M.M. The high prevalence of asymptomatic SARS-CoV-2 infection reveals the silent spread of COVID-19. Int. J. Infect. Dis. 2021. [Google Scholar] [CrossRef]
- Van Doremalen, N.; Bushmaker, T.; Lloyd-Smith, J.O.; De Wit, E.; Munster, V.J.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; et al. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567. [Google Scholar] [CrossRef]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.; Lau, E.H.; Wong, J.Y.; et al. Early transmission dynamics in Wuhan, China, of Novel Coronavirus—Infected pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef]
- Kong, W.H.; Li, Y.; Peng, M.W.; Kong, D.G.; Yang, X.B.; Wang, L.; Liu, M.Q. SARS-CoV-2 detection in patients with influen-za-like illness. Nat. Microbiol. 2020, 5, 675–678. [Google Scholar] [CrossRef]
- Müller, M.A.; Corman, V.M.; Jores, J.; Meyer, B.; Younan, M.; Liljander, A.; Bosch, B.-J.; Lattwein, E.; Hilali, M.; Musa, B.E.; et al. MERS Coronavirus Neutralizing Antibodies in Camels, Eastern Africa, 1983–1997. Emerg. Infect. Dis. 2014, 20, 2093–2095. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Li, Y.; Wang, L.; Zhao, G.; Tao, X.; Tseng, C.-T.K.; Zhou, Y.; Du, L.; Jiang, S. Intranasal vaccination with recombinant receptor-binding domain of MERS-CoV spike protein induces much stronger local mucosal immune responses than subcutaneous immunization: Implication for designing novel mucosal MERS vaccines. Vaccine 2014, 32, 2100–2108. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef]
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; Van Der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, eabd2985. [Google Scholar] [CrossRef]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef] [PubMed]
- Faria, N.R.; Claro, I.M.; Candido, D.; Moyses Franco, L.A.; Andrade, P.S.; Coletti, T.M.; Silva, C.A.; Sales, F.C.; Manuli, E.R.; Aguiar, R.S. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: Preliminary findings. Virological 2021, in press. [Google Scholar]
- Bayarri-Olmos, R.; Rosbjerg, A.; Johnsen, L.B.; Helgstrand, C.; Bak-Thomsen, T.; Garred, P.; Skjoedt, M.-O. The SARS-CoV-2 Y453F mink variant displays a pronounced increase in ACE-2 affinity but does not challenge antibody neutralization. J. Biol. Chem. 2021. [Google Scholar] [CrossRef] [PubMed]
- Thomson, E.C.; Rosen, L.E.; Shepherd, J.G.; Spreafico, R.; Filipe, A.D.S.; Wojcechowskyj, J.A.; Davis, C.; Piccoli, L.; Pascall, D.J.; Dillen, J.; et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell 2021, 184, 1171–1187.e20. [Google Scholar] [CrossRef]
- Liu, L.; Wei, Q.; Alvarez, X.; Wang, H.; Du, Y.; Zhu, H.; Jiang, H.; Zhou, J.; Lam, P.; Zhang, L.; et al. Epithelial Cells Lining Salivary Gland Ducts Are Early Target Cells of Severe Acute Respiratory Syndrome Coronavirus Infection in the Upper Respiratory Tracts of Rhesus Macaques. J. Virol. 2011, 85, 4025–4030. [Google Scholar] [CrossRef]
- Gu, J.; Korteweg, C. Pathology and Pathogenesis of Severe Acute Respiratory Syndrome. Am. J. Pathol. 2007, 170, 1136–1147. [Google Scholar] [CrossRef] [PubMed]
- Jeffers, S.A.; Tusell, S.M.; Gillim-Ross, L.; Hemmila, E.M.; Achenbach, J.E.; Babcock, G.J.; Thomas, J.W.D.; Thackray, L.B.; Young, M.D.; Mason, R.J.; et al. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA 2004, 101, 15748–15753. [Google Scholar] [CrossRef]
- Zhang, H.; Penninger, J.M.; Li, Y.; Zhong, N.; Slutsky, A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: Molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020, 46, 586–590. [Google Scholar] [CrossRef]
- Kang, Y.-L.; Chou, Y.-Y.; Rothlauf, P.W.; Liu, Z.; Soh, T.K.; Cureton, D.; Case, J.B.; Chen, R.E.; Diamond, M.S.; Whelan, S.P.J.; et al. Inhibition of PIKfyve kinase prevents infection by Zaire ebolavirus and SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 20803–20813. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Chen, W.; Zhou, Y.-S.; Lian, J.-Q.; Zhang, Z.; Du, P.; Gong, L.; Zhang, Y.; Cui, H.-Y.; Geng, J.-J.; et al. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ahmetaj-Shala, B.; Vaja, R.; Atanur, S.S.; George, P.M.; Kirkby, N.S.; Mitchell, J.A. Cardiorenal Tissues Express SARS-CoV-2 Entry Genes and Basigin (BSG/CD147) Increases With Age in Endothelial Cells. JACC: Basic Transl. Sci. 2020, 5, 1111–1123. [Google Scholar] [CrossRef]
- Radzikowska, U.; Ding, M.; Tan, G.; Zhakparov, D.; Peng, Y.; Wawrzyniak, P.; Wang, M.; Li, S.; Morita, H.; Altunbulakli, C.; et al. Distribution of ACE2, CD147, CD26, and other SARS-CoV-2 associated molecules in tissues and immune cells in health and in asthma, COPD, obesity, hypertension, and COVID-19 risk factors. Allergy 2020, 75. [Google Scholar] [CrossRef]
- Wei, J.; Alfajaro, M.M.; Deweirdt, P.C.; Hanna, R.E.; Lu-Culligan, W.J.; Cai, W.L.; Strine, M.S.; Zhang, S.-M.; Graziano, V.R.; Schmitz, C.O.; et al. Genome-wide CRISPR Screens Reveal Host Factors Critical for SARS-CoV-2 Infection. Cell 2021, 184, 76–91.e13. [Google Scholar] [CrossRef]
- Hoffmann, H.-H.; Schneider, W.M.; Rozen-Gagnon, K.; Miles, L.A.; Schuster, F.; Razooky, B.; Jacobson, E.; Wu, X.; Yi, S.; Rudin, C.M.; et al. TMEM41B Is a Pan-flavivirus Host Factor. Cell 2021, 184, 133–148.e20. [Google Scholar] [CrossRef]
- Daniloski, Z.; Jordan, T.X.; Wessels, H.-H.; Hoagland, D.A.; Kasela, S.; Legut, M.; Maniatis, S.; Mimitou, E.P.; Lu, L.; Geller, E.; et al. Identification of Required Host Factors for SARS-CoV-2 Infection in Human Cells. Cell 2021, 184, 92–105.e16. [Google Scholar] [CrossRef]
- Wang, R.; Simoneau, C.R.; Kulsuptrakul, J.; Bouhaddou, M.; Travisano, K.A.; Hayashi, J.M.; Carlson-Stevermer, J.; Zengel, J.R.; Richards, C.M.; Fozouni, P.; et al. Genetic Screens Identify Host Factors for SARS-CoV-2 and Common Cold Coronaviruses. Cell 2021, 184, 106–119.e14. [Google Scholar] [CrossRef]
- Schneider, W.M.; Luna, J.M.; Hoffmann, H.-H.; Sánchez-Rivera, F.J.; Leal, A.A.; Ashbrook, A.W.; Le Pen, J.; Ricardo-Lax, I.; Michailidis, E.; Peace, A.; et al. Genome-Scale Identification of SARS-CoV-2 and Pan-coronavirus Host Factor Networks. Cell 2021, 184, 120–132.e14. [Google Scholar] [CrossRef]
- Raj, V.S.; Mou, H.; Smits, S.L.; Dekkers, D.H.W.; Müller, M.A.; Dijkman, R.; Muth, D.; Demmers, J.A.A.; Zaki, A.; Fouchier, R.A.M.; et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013, 495, 251–254. [Google Scholar] [CrossRef]
- Raj, V.S.; Farag, E.A.; Reusken, C.B.; Lamers, M.M.; Pas, S.D.; Voermans, J.; Smits, S.L.; Osterhaus, A.D.; Al-Mawlawi, N.; Al-Romaihi, H.E.; et al. Isolation of MERS Coronavirus from a Dromedary Camel, Qatar, 2014. Emerg. Infect. Dis. 2014, 20, 1339–1342. [Google Scholar] [CrossRef]
- Lu, G.; Hu, Y.; Wang, Q.; Qi, J.; Gao, F.; Li, Y.; Zhang, Y.; Zhang, W.; Yuan, Y.; Bao, J.; et al. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature 2013, 500, 227–231. [Google Scholar] [CrossRef] [PubMed]
- Needle, D.; Lountos, G.T.; Waugh, D.S. Structures of theMiddle East respiratory syndrome coronavirus3C-like protease reveal insights into substrate specificity. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015, 71, 1102–1111. [Google Scholar] [CrossRef] [PubMed]
- De Wit, E.; Van Doremalen, N.; Falzarano, D.; Munster, V.J. SARS and MERS: Recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016, 14, 523. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. Innate immune recognition of viral infection. Nat. Immunol. 2006, 7, 131–137. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. TLR signaling. Cell Death Differ. 2006, 13, 816–825. [Google Scholar] [CrossRef]
- Reikine, S.; Nguyen, J.B.; Modis, Y. Pattern Recognition and Signaling Mechanisms of RIG-I and MDA5. Front. Immunol. 2014, 5, 342. [Google Scholar] [CrossRef]
- Bego, M.G.; Mercier, J.; Cohen, É.A. Virus-Activated Interferon Regulatory Factor 7 Upregulates Expression of the Interferon-Regulated BST2 Gene Independently of Interferon Signaling. J. Virol. 2012, 86, 3513–3527. [Google Scholar] [CrossRef]
- Lin, R.; Noyce, R.S.; Collins, S.E.; Everett, R.D.; Mossman, K.L. The Herpes Simplex Virus ICP0 RING Finger Domain Inhibits IRF3- and IRF7-Mediated Activation of Interferon-Stimulated Genes. J. Virol. 2004, 78, 1675–1684. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Reddy, P. Intracellular sensors of immunity and allogeneic hematopoietic stem cell transplantation. In Immune Biology of Allogeneic Hematopoietic Stem Cell Transplantation; Elsevier: Amsterdam, The Netherlands, 2013; pp. 425–447. [Google Scholar]
- Mazaleuskaya, L.; Veltrop, R.; Ikpeze, N.; Martin-Garcia, J.; Navas-Martín, S. Protective Role of Toll-like Receptor 3-Induced Type I Interferon in Murine Coronavirus Infection of Macrophages. Viruses 2012, 4, 901–923. [Google Scholar] [CrossRef]
- Zhao, J.; Li, K.; Wohlford-Lenane, C.; Agnihothram, S.S.; Fett, C.; Zhao, J.; Gale, J.M.J.; Baric, R.S.; Enjuanes, L.; Gallagher, T.; et al. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc. Natl. Acad. Sci. USA 2014, 111, 4970–4975. [Google Scholar] [CrossRef] [PubMed]
- Totura, A.L.; Whitmore, A.C.; Agnihothram, S.; Schäfer, A.; Katze, M.G.; Heise, M.T.; Baric, R.S. Toll-Like Receptor 3 Signaling via TRIF Contributes to a Protective Innate Immune Response to Severe Acute Respiratory Syndrome Coronavirus Infection. mBio 2015, 6, e00638-15. [Google Scholar] [CrossRef] [PubMed]
- Sheahan, T.; Morrison, T.E.; Funkhouser, W.; Uematsu, S.; Akira, S.; Baric, R.S.; Heise, M.T. MyD88 Is Required for Protection from Lethal Infection with a Mouse-Adapted SARS-CoV. PLOS Pathog. 2008, 4, e1000240. [Google Scholar] [CrossRef]
- Al-Qahtani, A.A.; Lyroni, K.; Aznaourova, M.; Tseliou, M.; Al-Anazi, M.R.; Al-Ahdal, M.N.; Alkahtani, S.; Sourvinos, G.; Tsatsanis, C. Middle east respiratory syndrome corona virus spike glycoprotein suppresses macrophage responses via DPP4-mediated induction of IRAK-M and PPARγ. Oncotarget 2017, 8, 9053–9066. [Google Scholar] [CrossRef]
- Cervantes-Barragan, L.; Züst, R.; Weber, F.; Spiegel, M.; Lang, K.S.; Akira, S.; Thiel, V.; Ludewig, B. Control of coronavirus infection through plasmacytoid dendritic-cell–derived type I interferon. Blood 2006, 109, 1131–1137. [Google Scholar] [CrossRef]
- Channappanavar, R.; Fehr, A.R.; Zheng, J.; Wohlford-Lenane, C.; Abrahante, J.E.; Mack, M.; Sompallae, R.; McCray, P.B.; Meyerholz, D.K.; Perlman, S. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Investig. 2019, 129, 3625–3639. [Google Scholar] [CrossRef]
- Bowie, A.G.; Unterholzner, L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat. Rev. Immunol. 2008, 8, 911–922. [Google Scholar] [CrossRef]
- Wilkins, C.; Gale, M. Recognition of viruses by cytoplasmic sensors. Curr. Opin. Immunol. 2010, 22, 41–47. [Google Scholar] [CrossRef]
- Roth-Cross, J.K.; Bender, S.J.; Weiss, S.R. Murine Coronavirus Mouse Hepatitis Virus Is Recognized by MDA5 and Induces Type I Interferon in Brain Macrophages/Microglia. J. Virol. 2008, 82, 9829–9838. [Google Scholar] [CrossRef]
- Züst, R.; Cervantes-Barragan, L.; Habjan, M.; Maier, R.; Neuman, B.W.; Ziebuhr, J.; Szretter, K.J.; Baker, S.C.; Barchet, W.; Diamond, M.S.; et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011, 12, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Riva, L.; Pu, Y.; Martin-Sancho, L.; Kanamune, J.; Yamamoto, Y.; Sakai, K.; Gotoh, S.; Miorin, L.; De Jesus, P.D.; et al. MDA5 Governs the Innate Immune Response to SARS-CoV-2 in Lung Epithelial Cells. Cell Rep. 2021, 34, 108628. [Google Scholar] [CrossRef]
- Rebendenne, A.; Valadão, A.L.C.; Tauziet, M.; Maarifi, G.; Bonaventure, B.; McKellar, J.; Planès, R.; Nisole, S.; Arnaud-Arnould, M.; Moncorgé, O.; et al. SARS-CoV-2 triggers an MDA-5-dependent interferon response which is unable to control replication in lung epithelial cells. J. Virol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Barber, G.N. Host defense, viruses and apoptosis. Cell Death Differ. 2001, 8, 113–126. [Google Scholar] [CrossRef] [PubMed]
- Coccia, E.M.; Severa, M.; Giacomini, E.; Monneron, D.; Remoli, M.E.; Julkunen, I.; Cella, M.; Lande, R.; Uzé, G. Viral infection and Toll-like receptor agonists induce a differential expression of type I and λ interferons in human plasmacytoid and monocyte-derived dendritic cells. Eur. J. Immunol. 2004, 34, 796–805. [Google Scholar] [CrossRef] [PubMed]
- Levy, D.E.; Marié, I.J.; Durbin, J.E. Induction and function of type I and III interferon in response to viral infection. Curr. Opin. Virol. 2011, 1, 476–486. [Google Scholar] [CrossRef]
- Platanias, L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005, 5, 375–386. [Google Scholar] [CrossRef]
- Andreakos, E.; Salagianni, M.; Galani, I.E.; Koltsida, O. Interferon-λs: Front-Line Guardians of Immunity and Homeostasis in the Respiratory Tract. Front. Immunol. 2017, 8, 1232. [Google Scholar] [CrossRef]
- Trinchieri, G. Type I interferon: Friend or foe? J. Exp. Med. 2010, 207, 2053–2063. [Google Scholar] [CrossRef] [PubMed]
- Ank, N.; West, H.; Paludan, S.R. IFN-λ: Novel Antiviral Cytokines. J. Interf. Cytokine Res. 2006, 26, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Wathelet, M.G.; Lin, C.H.; Parekh, B.S.; Ronco, L.V.; Howley, P.M.; Maniatis, T. Virus Infection Induces the Assembly of Coordinately Activated Transcription Factors on the IFN-β Enhancer In Vivo. Mol. Cell 1998, 1, 507–518. [Google Scholar] [CrossRef]
- Mahony, R.; Gargan, S.; Roberts, K.L.; Bourke, N.; Keating, S.E.; Bowie, A.G.; O’Farrelly, C.; Stevenson, N.J. A novel anti-viral role for STAT3 in IFN-α signalling responses. Cell. Mol. Life Sci. 2017, 74, 1755–1764. [Google Scholar] [CrossRef]
- Darnell, J.E.; Kerr, I.M.; Stark, G.R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 1994, 264, 1415–1421. [Google Scholar] [CrossRef] [PubMed]
- Morales, D.J.; Lenschow, D.J. The antiviral activities of ISG15. J. Mol. Biol. 2013, 425, 4995–5008. [Google Scholar] [CrossRef] [PubMed]
- Akerström, S.; Mousavi-Jazi, M.; Klingström, J.; Leijon, M.; Lundkvist, A.; Mirazimi, A. Nitric Oxide Inhibits the Replication Cycle of Severe Acute Respiratory Syndrome Coronavirus. J. Virol. 2005, 79, 1966–1969. [Google Scholar] [CrossRef]
- Huang, I.-C.; Bailey, C.C.; Weyer, J.L.; Radoshitzky, S.R.; Becker, M.M.; Chiang, J.J.; Brass, A.L.; Ahmed, A.A.; Chi, X.; Dong, L.; et al. Distinct Patterns of IFITM-Mediated Restriction of Filoviruses, SARS Coronavirus, and Influenza A Virus. PLoS Pathog. 2011, 7, e1001258. [Google Scholar] [CrossRef]
- Zhao, X.; Zheng, S.; Chen, D.; Zheng, M.; Li, X.; Li, G.; Lin, H.; Chang, J.; Zeng, H.; Guo, J.-T. LY6E Restricts Entry of Human Coronaviruses, Including Currently Pandemic SARS-CoV-2. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [PubMed]
- Busnadiego, I.; Fernbach, S.; Pohl, M.O.; Karakus, U.; Huber, M.; Trkola, A.; Stertz, S.; Hale, B.G. Antiviral Activity of Type I, II, and III Interferons Counterbalances ACE2 Inducibility and Restricts SARS-CoV-2. mBio 2020, 11. [Google Scholar] [CrossRef]
- Boudewijns, R.; Thibaut, H.J.; Kaptein, S.J.F.; Li, R.; Vergote, V.; Seldeslachts, L.; Van Weyenbergh, J.; De Keyzer, C.; Bervoets, L.; Sharma, S.; et al. STAT2 signaling restricts viral dissemination but drives severe pneumonia in SARS-CoV-2 infected hamsters. Nat. Commun. 2020, 11, 5838. [Google Scholar] [CrossRef] [PubMed]
- Porritt, R.A.; Hertzog, P.J. Dynamic control of type I IFN signalling by an integrated network of negative regulators. Trends Immunol. 2015, 36, 150–160. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, C.; Xue, M.; Fu, F.; Zhang, X.; Li, L.; Yin, L.; Xu, W.; Feng, L.; Liu, P. The Coronavirus Transmissible Gastroenteritis Virus Evades the Type I Interferon Response through IRE1α-Mediated Manipulation of the MicroRNA miR-30a-5p/SOCS1/3 Axis. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [PubMed]
- Chiang, S.-F.; Lin, T.-Y.; Chow, K.-C.; Chiou, S.-H. SARS spike protein induces phenotypic conversion of human B cells to macrophage-like cells. Mol. Immunol. 2010, 47, 2575–2586. [Google Scholar] [CrossRef]
- Johnson, H.M.; Lewin, A.S.; Ahmed, C.M. SOCS, Intrinsic Virulence Factors, and Treatment of COVID-19. Front. Immunol. 2020, 11, 582102. [Google Scholar] [CrossRef]
- Knoops, K.; Kikkert, M.; Worm, S.H.E.V.D.; Zevenhoven-Dobbe, J.C.; Van Der Meer, Y.; Koster, A.J.; Mommaas, A.M.; Snijder, E.J. SARS-Coronavirus Replication Is Supported by a Reticulovesicular Network of Modified Endoplasmic Reticulum. PLoS Biol. 2008, 6, e226. [Google Scholar] [CrossRef]
- Bouvet, M.; Debarnot, C.; Imbert, I.; Selisko, B.; Snijder, E.J.; Canard, B.; Decroly, E. In Vitro Reconstitution of SARS-Coronavirus mRNA Cap Methylation. PLOS Pathog. 2010, 6, e1000863. [Google Scholar] [CrossRef]
- Spiegel, M.; Pichlmair, A.; Martínez-Sobrido, L.; Cros, J.; García-Sastre, A.; Haller, O.; Weber, F. Inhibition of Beta Interferon Induction by Severe Acute Respiratory Syndrome Coronavirus Suggests a Two-Step Model for Activation of Interferon Regulatory Factor. J. Virol. 2005, 79, 2079–2086. [Google Scholar] [CrossRef]
- Kopecky-Bromberg, S.A.; Martínez-Sobrido, L.; Frieman, M.; Baric, R.A.; Palese, P. Severe Acute Respiratory Syndrome Coronavirus Open Reading Frame (ORF) 3b, ORF 6, and Nucleocapsid Proteins Function as Interferon Antagonists. J. Virol. 2006, 81, 548–557. [Google Scholar] [CrossRef]
- Hu, Y.; Li, W.; Gao, T.; Cui, Y.; Jin, Y.; Li, P.; Ma, Q.; Liu, X.; Cao, C. The Severe Acute Respiratory Syndrome Coronavirus Nucleocapsid Inhibits Type I Interferon Production by Interfering with TRIM25-Mediated RIG-I Ubiquitination. J. Virol. 2017, 91, e02143-16. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, K.; Huang, C.; Lokugamage, K.; Kamitani, W.; Ikegami, T.; Tseng, C.-T.K.; Makino, S. Severe Acute Respiratory Syndrome Coronavirus nsp1 Suppresses Host Gene Expression, Including That of Type I Interferon, in Infected Cells. J. Virol. 2008, 82, 4471–4479. [Google Scholar] [CrossRef]
- Kamitani, W.; Narayanan, K.; Huang, C.; Lokugamage, K.; Ikegami, T.; Ito, N.; Kubo, H.; Makino, S. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl. Acad. Sci. USA 2006, 103, 12885–12890. [Google Scholar] [CrossRef] [PubMed]
- Ratia, K.; Saikatendu, K.S.; Santarsiero, B.D.; Barretto, N.; Baker, S.C.; Stevens, R.C.; Mesecar, A.D. Severe acute respiratory syndrome coronavirus papain-like protease: Structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. USA 2006, 103, 5717–5722. [Google Scholar] [CrossRef] [PubMed]
- Lindner, H.A.; Lytvyn, V.; Qi, H.; Lachance, P.; Ziomek, E.; Ménard, R. Selectivity in ISG15 and ubiquitin recognition by the SARS coronavirus papain-like protease. Arch. Biochem. Biophys. 2007, 466, 8–14. [Google Scholar] [CrossRef]
- Zevini, A.; Olagnier, D.; Hiscott, J. Crosstalk between Cytoplasmic RIG-I and STING Sensing Pathways. Trends Immunol. 2017, 38, 194–205. [Google Scholar] [CrossRef]
- Castanier, C.; Garcin, D.; Vazquez, A.; Arnoult, D. Mitochondrial dynamics regulate the RIG-I-like receptor antiviral pathway. EMBO Rep. 2009, 11, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Zhong, B.; Yang, Y.; Li, S.; Wang, Y.-Y.; Li, Y.; Diao, F.; Lei, C.; He, X.; Zhang, L.; Tien, P.; et al. The Adaptor Protein MITA Links Virus-Sensing Receptors to IRF3 Transcription Factor Activation. Immunity 2008, 29, 538–550. [Google Scholar] [CrossRef]
- Liu, S.; Cai, X.; Wu, J.; Cong, Q.; Chen, X.; Li, T.; Du, F.; Ren, J.; Wu, Y.-T.; Grishin, N.V.; et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 2015, 347, aaa2630. [Google Scholar] [CrossRef]
- Chen, X.; Yang, X.; Zheng, Y.; Yang, Y.; Xing, Y.; Chen, Z. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell 2014, 5, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Matthews, K.; Schäfer, A.; Pham, A.; Frieman, M.B. The SARS coronavirus papain like protease can inhibit IRF3 at a post activation step that requires deubiquitination activity. Virol. J. 2014, 11, 1–12. [Google Scholar] [CrossRef]
- Sun, L.; Xing, Y.; Chen, X.; Zheng, Y.; Yang, Y.; Nichols, D.B.; Clementz, M.A.; Banach, B.S.; Li, K.; Baker, S.C.; et al. Coronavirus Papain-like Proteases Negatively Regulate Antiviral Innate Immune Response through Disruption of STING-Mediated Signaling. PLoS ONE 2012, 7, e30802. [Google Scholar] [CrossRef] [PubMed]
- Minakshi, R.; Padhan, K.; Rani, M.; Khan, N.; Ahmad, F.; Jameel, S. The SARS Coronavirus 3a Protein Causes Endoplasmic Reticulum Stress and Induces Ligand-Independent Downregulation of the Type 1 Interferon Receptor. PLoS ONE 2009, 4, e8342. [Google Scholar] [CrossRef]
- Frieman, M.; Yount, B.; Heise, M.; Kopecky-Bromberg, S.A.; Palese, P.; Baric, R.S. Severe Acute Respiratory Syndrome Coronavirus ORF6 Antagonizes STAT1 Function by Sequestering Nuclear Import Factors on the Rough Endoplasmic Reticulum/Golgi Membrane. J. Virol. 2007, 81, 9812–9824. [Google Scholar] [CrossRef]
- Wathelet, M.G.; Orr, M.; Frieman, M.B.; Baric, R.S. Severe Acute Respiratory Syndrome Coronavirus Evades Antiviral Signaling: Role of nsp1 and Rational Design of an Attenuated Strain. J. Virol. 2007, 81, 11620–11633. [Google Scholar] [CrossRef]
- Mizutani, T.; Fukushi, S.; Murakami, M.; Hirano, T.; Saijo, M.; Kurane, I.; Morikawa, S. Tyrosine dephosphorylation of STAT3 in SARS coronavirus-infected Vero E6 cells. FEBS Lett. 2004, 577, 187–192. [Google Scholar] [CrossRef]
- Liu, Y.-J. IPC: Professional Type 1 Interferon-Producing Cells and Plasmacytoid Dendritic Cell Precursors. Annu. Rev. Immunol. 2005, 23, 275–306. [Google Scholar] [CrossRef]
- Cheung, C.Y.; Poon, L.L.M.; Ng, I.H.Y.; Luk, W.; Sia, S.-F.; Wu, M.H.S.; Chan, K.-H.; Yuen, K.-Y.; Gordon, S.; Guan, Y.; et al. Cytokine Responses in Severe Acute Respiratory Syndrome Coronavirus-Infected Macrophages In Vitro: Possible Relevance to Pathogenesis. J. Virol. 2005, 79, 7819–7826. [Google Scholar] [CrossRef]
- Spiegel, M.; Weber, F. Inhibition of cytokine gene expression and induction of chemokine genes in non-lymphatic cells infected with SARS coronavirus. Virol. J. 2006, 3, 17. [Google Scholar] [CrossRef] [PubMed]
- Cinatl, J.; Hoever, G.; Morgenstern, B.; Preiser, W.; Vogel, J.-U.; Hofmann, W.-K.; Bauer, G.; Michaelis, M.; Rabenau, H.F.; Doerr, H.W. Infection of cultured intestinal epithelial cells with severe acute respiratory syndrome coronavirus. Cell. Mol. Life Sci. 2004, 61, 2100–2112. [Google Scholar] [CrossRef]
- Cinatl, J.; Michaelis, M.; Morgenstern, B.; Doerr, H.W. High-dose hydrocortisone reduces expression of the pro-inflammatory chemokines CXCL8 and CXCL10 in SARS coronavirus-infected intestinal cells. Int. J. Mol. Med. 2005, 15, 323–327. [Google Scholar] [CrossRef]
- He, R.; Leeson, A.; Andonov, A.; Li, Y.; Bastien, N.; Cao, J.; Osiowy, C.; Dobie, F.; Cutts, T.; Ballantine, M.; et al. Activation of AP-1 signal transduction pathway by SARS coronavirus nucleocapsid protein. Biochem. Biophys. Res. Commun. 2003, 311, 870–876. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-J.; Liu, C.Y.-Y.; Chiang, B.-L.; Chao, Y.-C.; Chen, C.-C. Induction of IL-8 Release in Lung Cells via Activator Protein-1 by Recombinant Baculovirus Displaying Severe Acute Respiratory Syndrome-Coronavirus Spike Proteins: Identification of Two Functional Regions. J. Immunol. 2004, 173, 7602–7614. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Xu, J.; Zhou, C.; Wu, Z.; Zhong, S.; Liu, J.; Luo, W.; Chen, T.; Qin, Q.; Deng, P. Characterization of Cytokine/Chemokine Profiles of Severe Acute Respiratory Syndrome. Am. J. Respir. Crit. Care Med. 2005, 171, 850–857. [Google Scholar] [CrossRef]
- Tang, N.L.-S.; Chan, P.K.-S.; Wong, C.-K.; To, K.-F.; Wu, A.K.-L.; Sung, Y.-M.; Hui, D.S.-C.; Sung, J.J.-Y.; Lam, C.W.-K. Early Enhanced Expression of Interferon-Inducible Protein-10 (CXCL-10) and Other Chemokines Predicts Adverse Outcome in Severe Acute Respiratory Syndrome. Clin. Chem. 2005, 51, 2333–2340. [Google Scholar] [CrossRef]
- Huang, K.-J.; Su, I.-J.; Theron, M.; Wu, Y.-C.; Lai, S.-K.; Liu, C.-C.; Lei, H.-Y. An interferon-γ-related cytokine storm in SARS patients. J. Med Virol. 2004, 75, 185–194. [Google Scholar] [CrossRef]
- Josset, L.; Menachery, V.D.; Gralinski, L.E.; Agnihothram, S.; Sova, P.; Carter, V.S.; Yount, B.L.; Graham, R.L.; Baric, R.S.; Katze, M.G. Cell Host Response to Infection with Novel Human Coronavirus EMC Predicts Potential Antivirals and Important Differences with SARS Coronavirus. mBio 2013, 4, e00165-13. [Google Scholar] [CrossRef]
- Hui, K.P.Y.; Cheung, M.-C.; Perera, R.A.P.M.; Ng, K.-C.; Bui, C.H.T.; Ho, J.C.W.; Ng, M.M.T.; Kuok, D.I.T.; Shih, K.C.; Tsao, S.-W.; et al. Tropism, replication competence, and innate immune responses of the coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva: An analysis in ex-vivo and in-vitro cultures. Lancet Respir. Med. 2020, 8, 687–695. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, L.; Geng, H.; Deng, Y.; Huang, B.; Guo, Y.; Zhao, Z.; Tan, W. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell 2013, 4, 951–961. [Google Scholar] [CrossRef] [PubMed]
- Siu, K.-L.; Yeung, M.L.; Kok, K.-H.; Yuen, K.-Y.; Kew, C.; Lui, P.-Y.; Chan, C.-P.; Tse, H.; Woo, P.C.Y.; Jin, D.-Y. Middle East Respiratory Syndrome Coronavirus 4a Protein Is a Double-Stranded RNA-Binding Protein That Suppresses PACT-Induced Activation of RIG-I and MDA5 in the Innate Antiviral Response. J. Virol. 2014, 88, 4866–4876. [Google Scholar] [CrossRef] [PubMed]
- Menachery, V.D.; Mitchell, H.D.; Cockrell, A.S.; Gralinski, L.E.; Yount, B.L.; Graham, R.L.; McAnarney, E.T.; Douglas, M.G.; Scobey, T.; Beall, A.; et al. MERS-CoV Accessory ORFs Play Key Role for Infection and Pathogenesis. mBio 2017, 8, e00665-17. [Google Scholar] [CrossRef]
- Matthews, K.L.; Coleman, C.M.; Van Der Meer, Y.; Snijder, E.J.; Frieman, M.B. The ORF4b-encoded accessory proteins of Middle East respiratory syndrome coronavirus and two related bat coronaviruses localize to the nucleus and inhibit innate immune signalling. J. Gen. Virol. 2014, 95, 874–882. [Google Scholar] [CrossRef]
- Yang, Y.; Ye, F.; Zhu, N.; Wang, W.; Deng, Y.; Zhao, Z.; Tan, W. Middle East respiratory syndrome coronavirus ORF4b protein inhibits type I interferon production through both cytoplasmic and nuclear targets. Sci. Rep. 2015, 5, 17554. [Google Scholar] [CrossRef] [PubMed]
- Canton, J.; Fehr, A.R.; Fernandez-Delgado, R.; Gutierrez-Alvarez, F.J.; Sanchez-Aparicio, M.T.; García-Sastre, A.; Perlman, S.; Enjuanes, L.; Sola, I. MERS-CoV 4b protein interferes with the NF-κB-dependent innate immune response during infection. PLoS Pathog. 2018, 14, e1006838. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.K.W.; Hui, K.P.Y.; Perera, R.A.P.M.; Miguel, E.; Niemeyer, D.; Zhao, J.; Channappanavar, R.; Dudas, G.; Oladipo, J.O.; Traoré, A.; et al. MERS coronaviruses from camels in Africa exhibit region-dependent genetic diversity. Proc. Natl. Acad. Sci. USA 2018, 115, 3144–3149. [Google Scholar] [CrossRef]
- Wong, L.-Y.R.; Ye, Z.-W.; Lui, P.-Y.; Zheng, X.; Yuan, S.; Zhu, L.; Fung, S.-Y.; Yuen, K.-S.; Siu, K.-L.; Yeung, M.-L.; et al. Middle East Respiratory Syndrome Coronavirus ORF8b Accessory Protein Suppresses Type I IFN Expression by Impeding HSP70-Dependent Activation of IRF3 Kinase IKKε. J. Immunol. 2020, 205, 1564–1579. [Google Scholar] [CrossRef]
- Lee, J.Y.; Bae, S.; Myoung, J. Middle East Respiratory Syndrome Coronavirus-Encoded Accessory Proteins Impair MDA5-and TBK1-Mediated Activation of NF-κB. J. Microbiol. Biotechnol. 2019, 29, 1316–1323. [Google Scholar] [CrossRef]
- Lee, J.Y.; Kim, S.-J.; Myoung, J. Middle East Respiratory Syndrome Coronavirus-Encoded ORF8b Inhibits RIG-I-Like Receptors in a Differential Mechanism. J. Microbiol. Biotechnol. 2019, 29, 2014–2021. [Google Scholar] [CrossRef] [PubMed]
- Menachery, V.D.; Gralinski, L.E.; Mitchell, H.D.; Dinnon, K.H.; Leist, S.R.; Yount, B.L.; Graham, R.L.; McAnarney, E.T.; Stratton, K.G.; Cockrell, A.S.; et al. Middle East Respiratory Syndrome Coronavirus Nonstructural Protein 16 Is Necessary for Interferon Resistance and Viral Pathogenesis. mSphere 2017, 2, e00346-17. [Google Scholar] [CrossRef] [PubMed]
- Alosaimi, B.; Hamed, M.E.; Naeem, A.; Alsharef, A.A.; Alqahtani, S.Y.; Aldosari, K.M.; Alamri, A.A.; Al-Eisa, K.; Khojah, T.; Assiri, A.M.; et al. MERS-CoV infection is associated with downregulation of genes encoding Th1 and Th2 cytokines/chemokines and elevated inflammatory innate immune response in the lower respiratory tract. Cytokine 2020, 126, 154895. [Google Scholar] [CrossRef]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Péré, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.-C.; Uhl, S.; Hoagland, D.; Møller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045.e9. [Google Scholar] [CrossRef]
- Lokugamage, K.G.; Hage, A.; De Vries, M.; Valero-Jimenez, A.M.; Schindewolf, C.; Dittmann, M.; Rajsbaum, R.; Menachery, V.D. Type I Interferon Susceptibility Distinguishes SARS-CoV-2 from SARS-CoV. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.-X.; Yang, K.; Liu, X.; Liu, X.-Y.; Wei, B.; Shan, Y.-F.; Zhu, L.-H.; Wang, C. Positive Regulation of Interferon Regulatory Factor 3 Activation by Herc5 via ISG15 Modification. Mol. Cell. Biol. 2010, 30, 2424–2436. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.; Mukherjee, R.; Grewe, D.; Bojkova, D.; Baek, K.; Bhattacharya, A.; Schulz, L.; Widera, M.; Mehdipour, A.R.; Tascher, G.; et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature 2020, 1–10. [Google Scholar] [CrossRef]
- Lei, X.; Dong, X.; Ma, R.; Wang, W.; Xiao, X.; Tian, Z.; Wang, C.; Wang, Y.; Li, L.; Ren, L.; et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020, 11, 1–12. [Google Scholar] [CrossRef]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef]
- Miorin, L.; Kehrer, T.; Sanchez-Aparicio, M.T.; Zhang, K.; Cohen, P.; Patel, R.S.; Cupic, A.; Makio, T.; Mei, M.; Moreno, E.; et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 28344–28354. [Google Scholar] [CrossRef]
- Li, J.-Y.; Liao, C.-H.; Wang, Q.; Tan, Y.-J.; Luo, R.; Qiu, Y.; Ge, X.-Y. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020, 286, 198074. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, J.; Chen, Y.; Luo, B.; Yuan, Y.; Huang, F.; Yang, T.; Yu, F.; Liu, J.; Liu, B.; et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through potently downregulating MHC-I. bioRxiv 2020. [Google Scholar] [CrossRef]
- Young, B.E.; Fong, S.W.; Chan, Y.H.; Mak, T.M.; Ang, L.W.; Anderson, D.E.; Lee, C.Y.P.; Amrun, S.N.; Lee, B.; Goh, Y.S.; et al. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: An observational cohort study. Lancet 2020, 396, 603–611. [Google Scholar] [CrossRef]
- Yuen, C.-K.; Lam, J.-Y.; Wong, W.-M.; Mak, L.-F.; Wang, X.; Chu, H.; Cai, J.-P.; Jin, D.-Y.; To, K.K.-W.; Chan, J.F.-W.; et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg. Microbes Infect. 2020, 9, 1418–1428. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Cao, Z.; Xie, X.; Zhang, X.; Chen, J.Y.C.; Wang, H.; Menachery, V.D.; Rajsbaum, R.; Shi, P.Y. Evasion of Type I Interferon by SARS-CoV-2. Cell Rep. 2020, 33, 108234. [Google Scholar] [CrossRef]
- Ren, Y.; Shu, T.; Wu, D.; Mu, J.; Wang, C.; Huang, M.; Han, Y.; Zhang, X.-Y.; Zhou, W.; Qiu, Y.; et al. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell. Mol. Immunol. 2020, 17, 881–883. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, P.; Niyogi, S. ORF3a mutation associated higher mortality rate in SARS-CoV-2 infection. Epidemiol. Infect. 2020, 148, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Konno, Y.; Kimura, I.; Uriu, K.; Fukushi, M.; Irie, T.; Koyanagi, Y.; Sauter, D.; Gifford, R.J.; Nakagawa, S.; Sato, K. SARS-CoV-2 ORF3b Is a Potent Interferon Antagonist Whose Activity Is Increased by a Naturally Occurring Elongation Variant. Cell Rep. 2020, 32, 108185. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhuang, M.-W.; Han, L.; Zhang, J.; Nan, M.-L.; Zhan, P.; Kang, D.; Liu, X.; Gao, C.; Wang, P.-H. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) membrane (M) protein inhibits type I and III interferon production by targeting RIG-I/MDA-5 signaling. Signal Transduct. Target. Ther. 2020, 5, 1–13. [Google Scholar] [CrossRef]
- Bastard, P.; Rosen, L.B.; Zhang, Q.; Michailidis, E.; Hoffmann, H.-H.; Zhang, Y.; Dorgham, K.; Philippot, Q.; Rosain, J.; Béziat, V.; et al. Auto-antibodies against type I IFNs in patients with life-threatening COVID-19. Sci. 2020, 370, eabd4585. [Google Scholar] [CrossRef]
- Knowles, S.R.; Phillips, E.J.; Dresser, L.; Matukas, L. Common Adverse Events Associated with the Use of Ribavirin for Severe Acute Respiratory Syndrome in Canada. Clin. Infect. Dis. 2003, 37, 1139–1142. [Google Scholar] [CrossRef]
- Van Vonderen, M.G.; Bos, J.C.; Prins, J.M.; Dillen, P.W.-V.; Speelman, P. Ribavirin in the treatment of severe acute respiratory syndrome (SARS). Neth. J. Med. 2003, 61, 238–241. [Google Scholar] [PubMed]
- Wang, H.; Ding, Y.; Li, X.; Yang, L.; Zhang, W.; Kang, W. Fatal Aspergillosis in a Patient with SARS Who Was Treated with Corticosteroids. N. Engl. J. Med. 2003, 349, 507–508. [Google Scholar] [CrossRef]
- Hwang, D.M.; Chamberlain, D.W.; Poutanen, S.M.; Low, D.E.; Asa, S.L.; Butany, J. Pulmonary pathology of severe acute respiratory syndrome in Toronto. Mod. Pathol. 2004, 18, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Chan, K.A.; Hui, D.S.; Ng, E.K.; Wu, A.; Chiu, R.W.; Wong, V.W.; Chan, P.K.; Wong, K.; Wong, E.; et al. Effects of early corticosteroid treatment on plasma SARS-associated Coronavirus RNA concentrations in adult patients. J. Clin. Virol. 2004, 31, 304–309. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, F.; Xu, M.; Huang, K.; Zhong, W.; Cai, W.; Yin, Z.; Huang, S.; Deng, Z.; Wei, M.; et al. Description and clinical treatment of an early outbreak of severe acute respiratory syndrome (SARS) in Guangzhou, PR China. J. Med. Microbiol. 2003, 52, 715–720. [Google Scholar] [CrossRef]
- Loutfy, M.R.; Blatt, L.M.; Siminovitch, K.A.; Ward, S.; Wolff, B.; Lho, H.; Pham, D.H.; Deif, H.; LaMere, E.A.; Chang, M.; et al. Interferon Alfacon-1 Plus Corticosteroids in Severe Acute Respiratory Syndrome. JAMA 2003, 290, 3222–3228. [Google Scholar] [CrossRef] [PubMed]
- Sung, J.J.Y.; Wu, A.; Joynt, G.M.; Yuen, K.Y.; Lee, N.; Chan, P.K.S.; Cockram, C.S.; Ahuja, A.T.; Yu, L.M.; Wong, V.W.; et al. Severe acute respiratory syndrome: Report of treatment and outcome after a major outbreak. Thorax 2004, 59, 414–420. [Google Scholar] [CrossRef]
- Chu, C.M.; Cheng, V.C.C.; Hung, I.F.N.; Wong, M.M.L.; Chan, K.H.; Kao, R.Y.T.; Poon, L.L.M.; Wong, C.L.P.; Guan, Y.; Peiris, J.S.M.; et al. Role of lopinavir/ritonavir in the treatment of SARS: Initial virological and clinical findings. Thorax 2004, 59, 252–256. [Google Scholar] [CrossRef]
- Chan, K.S.; Lai, S.T.; Chu, C.M.; Tsui, E.; Tam, C.Y.; Wong, M.M.L.; Tse, M.W.; Que, T.L.; Peiris, J.S.M.; Sung, J.; et al. Treatment of severe acute respiratory syndrome with lopinavir/ritonavir: A multicentre retrospective matched cohort study. Hong Kong Med. J. 2003, 9, 399–406. [Google Scholar] [PubMed]
- Mair-Jenkins, J.; Saavedra-Campos, M.; Baillie, J.K.; Cleary, P.; Khaw, F.-M.; Lim, W.S.; Makki, S.; Rooney, K.D.; Nguyen-Van-Tam, J.S.; Beck, C.R.; et al. The Effectiveness of Convalescent Plasma and Hyperimmune Immunoglobulin for the Treatment of Severe Acute Respiratory Infections of Viral Etiology: A Systematic Review and Exploratory Meta-analysis. J. Infect. Dis. 2015, 211, 80–90. [Google Scholar] [CrossRef]
- Bolles, M.; Deming, D.; Long, K.; Agnihothram, S.; Whitmore, A.; Ferris, M.; Funkhouser, W.; Gralinski, L.; Totura, A.; Heise, M.; et al. A Double-Inactivated Severe Acute Respiratory Syndrome Coronavirus Vaccine Provides Incomplete Protection in Mice and Induces Increased Eosinophilic Proinflammatory Pulmonary Response upon Challenge. J. Virol. 2011, 85, 12201–12215. [Google Scholar] [CrossRef] [PubMed]
- See, R.H.; Zakhartchouk, A.N.; Petric, M.; Lawrence, D.J.; Mok, C.P.Y.; Hogan, R.J.; Rowe, T.; Zitzow, L.A.; Karunakaran, K.P.; Hitt, M.M.; et al. Comparative evaluation of two severe acute respiratory syndrome (SARS) vaccine candidates in mice challenged with SARS coronavirus. J. Gen. Virol. 2006, 87, 641–650. [Google Scholar] [CrossRef]
- Zhou, Z.; Post, P.; Chubet, R.; Holtz, K.; McPherson, C.; Petric, M.; Cox, M. A recombinant baculovirus-expressed S glycoprotein vaccine elicits high titers of SARS-associated coronavirus (SARS-CoV) neutralizing antibodies in mice. Vaccine 2006, 24, 3624–3631. [Google Scholar] [CrossRef]
- Deming, D.; Sheahan, T.; Heise, M.; Yount, B.; Davis, N.; Sims, A.; Suthar, M.; Harkema, J.; Whitmore, A.; Pickles, R.; et al. Vaccine Efficacy in Senescent Mice Challenged with Recombinant SARS-CoV Bearing Epidemic and Zoonotic Spike Variants. PLoS Med. 2006, 3, e525. [Google Scholar] [CrossRef]
- Lokugamage, K.G.; Yoshikawa-Iwata, N.; Ito, N.; Watts, D.M.; Wyde, P.R.; Wang, N.; Newman, P.; Tseng, C.-T.K.; Peters, C.; Makino, S. Chimeric coronavirus-like particles carrying severe acute respiratory syndrome coronavirus (SCoV) S protein protect mice against challenge with SCoV. Vaccine 2008, 26, 797–808. [Google Scholar] [CrossRef]
- See, R.H.; Petric, M.; Lawrence, D.J.; Mok, C.P.Y.; Rowe, T.; Zitzow, L.A.; Karunakaran, K.P.; Voss, T.G.; Brunham, R.C.; Gauldie, J.; et al. Severe acute respiratory syndrome vaccine efficacy in ferrets: Whole killed virus and adenovirus-vectored vaccines. J. Gen. Virol. 2008, 89, 2136–2146. [Google Scholar] [CrossRef]
- Bisht, H.; Roberts, A.; Vogel, L.; Bukreyev, A.; Collins, P.L.; Murphy, B.R.; Subbarao, K.; Moss, B. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc. Natl. Acad. Sci. USA 2004, 101, 6641–6646. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.-T.; Zhang, J.-S.; Su, N.; Xu, J.-G.; Wang, N.; Chen, J.-T.; Chen, X.; Liu, Y.-X.; Gao, H.; Jia, Y.-P.; et al. Safety and immunogenicity from a phase I trial of inactivated severe acute respiratory syndrome coronavirus vaccine. Antivir. Ther. 2007, 12, 1107. [Google Scholar] [PubMed]
- Martin, J.E.; Louder, M.K.; Holman, L.A.; Gordon, I.J.; Enama, M.E.; Larkin, B.D.; Andrews, C.A.; Vogel, L.; Koup, R.A.; Roederer, M.; et al. A SARS DNA vaccine induces neutralizing antibody and cellular immune responses in healthy adults in a Phase I clinical trial. Vaccine 2008, 26, 6338–6343. [Google Scholar] [CrossRef] [PubMed]
- Falzarano, D.; De Wit, E.; Rasmussen, A.L.; Feldmann, F.; Okumura, A.; Scott, D.P.; Brining, D.; Bushmaker, T.; Martellaro, C.; Baseler, L.; et al. Treatment with interferon-α2b and ribavirin improves outcome in MERS-CoV–infected rhesus macaques. Nat. Med. 2013, 19, 1313–1317. [Google Scholar] [CrossRef]
- Chan, J.F.-W.; Yao, Y.; Yeung, M.-L.; Deng, W.; Bao, L.; Jia, L.; Li, F.; Xiao, C.; Gao, H.; Yu, P.; et al. Treatment with Lopinavir/Ritonavir or Interferon-β1b Improves Outcome of MERS-CoV Infection in a Nonhuman Primate Model of Common Marmoset. J. Infect. Dis. 2015, 212, 1904–1913. [Google Scholar] [CrossRef] [PubMed]
- de Wilde, A.H.; Raj, V.S.; Oudshoorn, D.; Bestebroer, T.M.; van Nieuwkoop, S.; Limpens, R.W.; Posthuma, C.C.; van der Meer, Y.; Bárcena, M.; Haagmans, B.L.; et al. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-α treatment. J. Gen. Virol. 2013, 94, 1749–1760. [Google Scholar] [CrossRef]
- Reinhold, D.; Brocke, S. DPP4-directed therapeutic strategies for MERS-CoV. Lancet Infect. Dis. 2014, 14, 100–101. [Google Scholar] [CrossRef]
- Zhao, J.; Alshukairi, A.N.; Baharoon, S.A.; Ahmed, W.A.; Bokhari, A.A.; Nehdi, A.M.; Layqah, L.A.; Alghamdi, M.G.; Al Gethamy, M.M.; Dada, A.M.; et al. Recovery from the Middle East respiratory syndrome is associated with antibody and T-cell responses. Sci. Immunol. 2017, 2, eaan5393. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.-C.; Agnihothram, S.S.; Jiao, Y.; Stanhope, J.; Graham, R.L.; Peterson, E.C.; Avnir, Y.; Tallarico, A.S.C.; Sheehan, J.; Zhu, Q.; et al. Identification of human neutralizing antibodies against MERS-CoV and their role in virus adaptive evolution. Proc. Natl. Acad. Sci. USA 2014, 111, E2018–E2026. [Google Scholar] [CrossRef]
- Haagmans, B.L.; Brand, J.M.A.V.D.; Raj, V.S.; Volz, A.; Wohlsein, P.; Smits, S.L.; Schipper, D.; Bestebroer, T.M.; Okba, N.; Fux, R.; et al. An orthopoxvirus-based vaccine reduces virus excretion after MERS-CoV infection in dromedary camels. Science 2016, 351, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Koch, T.; Dahlke, C.; Fathi, A.; Kupke, A.; Krähling, V.; Okba, N.M.; Halwe, S.; Rohde, C.; Eickmann, M.; Volz, A.; et al. Safety and immunogenicity of a modified vaccinia virus Ankara vector vaccine candidate for Middle East respiratory syndrome: An open-label, phase 1 trial. Lancet Infect. Dis. 2020, 20, 827–838. [Google Scholar] [CrossRef]
- Lokugamage, K.G.; Hage, A.; Schindewolf, C.; Rajsbaum, R.; Menachery, V.D. SARS-CoV-2 sensitive to type I interferon pretreatment. bioRxiv 2020. [Google Scholar] [CrossRef]
- Mantlo, E.; Bukreyeva, N.; Maruyama, J.; Paessler, S.; Huang, C. Potent Antiviral Activities of Type I Interferons to SARS-CoV-2 Infection. bioRxiv 2020. [Google Scholar] [CrossRef]
- Grein, J.; Ohmagari, N.; Shin, D.; Diaz, G.; Asperges, E.; Castagna, A.; Feldt, T.; Green, G.; Green, M.L.; Lescure, F.-X.; et al. Compassionate Use of Remdesivir for Patients with Severe Covid-19. N. Engl. J. Med. 2020, 382, 2327–2336. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration Approves Gilead’s Antiviral Veklury® (Remdesivir) for Treatment of COVID-19. 2020. Available online: https://www.gilead.com/news-and-press/press-room/press-releases/2020/10/us-food-and-drug-administration-approves-gileads-antiviral-veklury-remdesivir-for-treatment-of-covid19 (accessed on 26 March 2021).
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef]
- Cavalcanti, A.B.; Zampieri, F.G.; Rosa, R.G.; Azevedo, L.C.; Veiga, V.C.; Avezum, A.; Damiani, L.P.; Marcadenti, A.; Kawano-Dourado, L.; Lisboa, T.; et al. Hydroxychloroquine with or without Azithromycin in Mild-to-Moderate Covid-19. N. Engl. J. Med. 2020, 383, 2041–2052. [Google Scholar] [CrossRef]
- Smiley, A.; Cullin, J.; Kaschalk, E.; He, K.; Smiley, A. Comments on “Soy isoflavone intake and its association with depressive symptoms during pregnancy”: Consider sleep and physical activity as possible confounders. Eur. J. Nutr. 2017, 56, 1793–1794. [Google Scholar] [CrossRef]
- Zhang, W.; Zhao, Y.; Zhang, F.; Wang, Q.; Li, T.; Liu, Z.; Wang, J.; Qin, Y.; Zhang, X.; Yan, X.; et al. The use of anti-inflammatory drugs in the treatment of people with severe coronavirus disease 2019 (COVID-19): The Perspectives of clinical immunologists from China. Clin. Immunol. 2020, 214, 108393. [Google Scholar] [CrossRef]
- Duan, K.; Liu, B.; Li, C.; Zhang, H.; Yu, T.; Qu, J.; Zhou, M.; Chen, L.; Meng, S.; Hu, Y.; et al. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. USA 2020, 117, 9490–9496. [Google Scholar] [CrossRef]
- Mejía-Salazar, M.F.; Goldizen, A.W.; Menz, C.S.; Dwyer, R.G.; Blomberg, S.P.; Waldner, C.L.; Cullingham, C.I.; Bollinger, T.K. Mule deer spatial association patterns and potential implications for transmission of an epizootic disease. PLoS ONE 2017, 12, e0175385. [Google Scholar] [CrossRef]
- Kawase, M.; Shirato, K.; Van Der Hoek, L.; Taguchi, F.; Matsuyama, S. Simultaneous Treatment of Human Bronchial Epithelial Cells with Serine and Cysteine Protease Inhibitors Prevents Severe Acute Respiratory Syndrome Coronavirus Entry. J. Virol. 2012, 86, 6537–6545. [Google Scholar] [CrossRef] [PubMed]
- Caly, L.; Druce, J.D.; Catton, M.G.; Jans, D.A.; Wagstaff, K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020, 178, 104787. [Google Scholar] [CrossRef] [PubMed]
- Gursel, M.; Gursel, I. WITHDRAWN: Is global BCG vaccination coverage relevant to the progression of SARS-CoV-2 pandemic? Med. Hypotheses 2020, 6, 109707. [Google Scholar] [CrossRef]
- Mullard, A. COVID-19 vaccine development pipeline gears up. Lancet 2020, 395, 1751–1752. [Google Scholar] [CrossRef]
- Breining, P.; Frølund, A.L.; Højen, J.F.; Gunst, J.D.; Staerke, N.B.; Saedder, E.; Cases-Thomas, M.; Little, P.; Nielsen, L.P.; Søgaard, O.S.; et al. Camostat mesylate against SARS-CoV-2 and COVID-19—Rationale, dosing and safety. Basic Clin. Pharmacol. Toxicol. 2021, 128, 204–212. [Google Scholar] [CrossRef]
- Sidwell, R.W.; Huffman, J.H.; Khare, L.G.P.; Allen, L.B.; Witkowski, R.J.T.; Robins, R.K. Broad-Spectrum Antiviral Activity of Virazole: 1-f8- D-Ribofuranosyl- 1,2,4-triazole- 3-carboxamide. Science 1972, 177, 705–706. [Google Scholar] [CrossRef]
- Falzarano, D.; De Wit, E.; Martellaro, C.; Callison, J.; Munster, V.J.; Feldmann, H. Inhibition of novel β coronavirus replication by a combination of interferon-α2b and ribavirin. Sci. Rep. 2013, 3, srep01686. [Google Scholar] [CrossRef] [PubMed]
- Cronin, R.S.; Li, M.; Wise, M.; Bradford, B.; Culling, V.; Zuccollo, J.; Thompson, J.M.D.; Mitchell, E.A.; McCowan, L.M.E. Late stillbirth post mortem examination in New Zealand: Maternal decision-making. Aust. N. Z. J. Obstet. Gynaecol. 2018, 58, 667–673. [Google Scholar] [CrossRef]
- Gong, W.-J.; Zhou, T.; Wu, S.-L.; Ye, J.-L.; Xu, J.-Q.; Zeng, F.; Su, Y.-Y.; Han, Y.; Lv, Y.-N.; Zhang, Y.; et al. A retrospective analysis of clinical efficacy of ribavirin in adults hospitalized with severe COVID-19. J. Infect. Chemother. 2021. [Google Scholar] [CrossRef] [PubMed]
- Agostini, M.L.; Andres, E.L.; Sims, A.C.; Graham, R.L.; Sheahan, T.P.; Lu, X.; Smith, E.C.; Case, J.B.; Feng, J.Y.; Jordan, R.; et al. Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. mBio 2018, 9, e00221-18. [Google Scholar] [CrossRef]
- European Medicines Agency. Risks Associated with Remdesivir; Veklury|European Medicines Agency: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Min, J.S.; Kim, G.-W.; Kwon, S.; Jin, Y.-H. A Cell-Based Reporter Assay for Screening Inhibitors of MERS Coronavirus RNA-Dependent RNA Polymerase Activity. J. Clin. Med. 2020, 9, 2399. [Google Scholar] [CrossRef]
- Chen, C.; Huang, J.; Cheng, Z.; Wu, J.; Chen, S.; Zhang, Y.; Chen, B.; Lu, M.; Luo, Y.; Zhang, J.; et al. Favipiravir versus arbidol for COVID-19: A randomized clinical trial. MedRxiv 2020. [Google Scholar] [CrossRef]
- Cao, B.; Wang, Y.; Wen, D.; Liu, W.; Wang, J.; Fan, G.; Ruan, L.; Song, B.; Cai, Y.; Wei, M.; et al. A Trial of Lopinavir–Ritonavir in Adults Hospitalized with Severe Covid-19. N. Engl. J. Med. 2020, 382, 1787–1799. [Google Scholar] [CrossRef]
- Yamamoto, N.; Yang, R.; Yoshinaka, Y.; Amari, S.; Nakano, T.; Cinatl, J.; Rabenau, H.; Doerr, H.W.; Hunsmann, G.; Otaka, A.; et al. HIV protease inhibitor nelfinavir inhibits replication of SARS-associated coronavirus. Biochem. Biophys. Res. Commun. 2004, 318, 719–725. [Google Scholar] [CrossRef]
- Chan, J.F.; Chan, K.-H.; Kao, R.Y.; To, K.K.; Zheng, B.-J.; Li, C.P.; Li, P.T.; Dai, J.; Mok, F.K.; Chen, H.; et al. Broad-spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. J. Infect. 2013, 67, 606–616. [Google Scholar] [CrossRef]
- Xu, Z.; Peng, C.; Shi, Y.; Zhu, Z.; Mu, K.; Wang, X.; Zhu, W. Nelfinavir was predicted to be a potential inhibitor of 2019-nCov main protease by an integrative approach combining homology modelling, molecular docking and binding free energy calculation. bioRxiv 2020. [Google Scholar] [CrossRef]
- Foo, C.S.; Abdelnabi, R.; Kaptein, S.J.; Zhang, X.; ter Horst, S.; Mols, R.; Delang, L.; Rocha-Pereira, J.; Coelmont, L.; Leyssen, P.; et al. Nelfinavir markedly improves lung pathology in SARS-CoV-2-infected Syrian hamsters despite a lack of an antiviral effect. bioRxiv 2021. [Google Scholar] [CrossRef]
- Zhang, L.; Lin, D.; Kusov, Y.; Nian, Y.; Ma, Q.; Wang, J.; Von Brunn, A.; Leyssen, P.; Lanko, K.; Neyts, J.; et al. α-Ketoamides as Broad-Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, Synthesis, and Activity Assessment. J. Med. Chem. 2020, 63, 4562–4578. [Google Scholar] [CrossRef]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Becker, S.; Rox, K.; Hilgenfeld, R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 2020, 368, 409–412. [Google Scholar] [CrossRef]
- Riva, L.; Yuan, S.; Yin, X.; Martin-Sancho, L.; Matsunaga, N.; Pache, L.; Burgstaller-Muehlbacher, S.; De Jesus, P.D.; Teriete, P.; Hull, M.V.; et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature 2020, 586, 113–119. [Google Scholar] [CrossRef]
- Khamitov, R.A.; SIa, L.; Shchukina, V.N.; Borisevich, S.V.; Maksimov, V.A.; Shuster, A.M. Antiviral activity of arbidol and its derivatives against the pathogen of severe acute respiratory syndrome in the cell cultures. Vopr. Virusol. 2008, 53, 9–13. [Google Scholar] [PubMed]
- Zhu, Z.; Lu, Z.; Xu, T.; Chen, C.; Yang, G.; Zha, T.; Lu, J.; Xue, Y. Arbidol monotherapy is superior to lopinavir/ritonavir in treating COVID-19. J. Infect. 2020, 81, e21–e23. [Google Scholar] [CrossRef] [PubMed]
- French, B.; Funamoto, S.; Sugiyama, H.; Sakata, R.; Cologne, J.; Cullings, H.M.; Mabuchi, K.; Preston, D.L. Population Density in Hiroshima and Nagasaki Before the Bombings in 1945: Its Measurement and Impact on Radiation Risk Estimates in the Life Span Study of Atomic Bomb Survivors. Am. J. Epidemiol. 2018, 187, 1623–1629. [Google Scholar] [CrossRef] [PubMed]
- Chaccour, C.; Casellas, A.; Matteo, A.B.-D.; Pineda, I.; Fernandez-Montero, A.; Ruiz-Castillo, P.; Richardson, M.-A.; Rodríguez-Mateos, M.; Jordán-Iborra, C.; Brew, J.; et al. The effect of early treatment with ivermectin on viral load, symptoms and humoral response in patients with non-severe COVID-19: A pilot, double-blind, placebo-controlled, randomized clinical trial. EClinicalMedicine 2021, 32, 100720. [Google Scholar] [CrossRef]
- Alam, M.T.; Murshed, R.; Bhiuyan, E.; Saber, S.; Alam, R.F.; Robin, R.C. A Case Series of 100 COVID-19 Positive Patients Treated with Combination of Ivermectin and Doxycycline. J. Bangladesh Coll. Physicians Surg. 2020, 10–15. [Google Scholar] [CrossRef]
- Liu, D.; Ahmet, A.; Ward, L.; Krishnamoorthy, P.; Mandelcorn, E.D.; Leigh, R.; Brown, J.P.; Cohen, A.; Kim, H. A practical guide to the monitoring and management of the complications of systemic corticosteroid therapy. Allergy Asthma Clin. Immunol. 2013, 9, 30. [Google Scholar] [CrossRef]
- Arabi, Y.M.; Mandourah, Y.; Al-Hameed, F.; Sindi, A.A.; Almekhlafi, G.A.; Hussein, M.A.; Jose, J.; Pinto, R.; Al-Omari, A.; Kharaba, A.; et al. Corticosteroid Therapy for Critically Ill Patients with Middle East Respiratory Syndrome. Am. J. Respir. Crit. Care Med. 2018, 197, 757–767. [Google Scholar] [CrossRef]
- Westrupp, E.M.; Bennett, C.; Cullinane, M.; Hackworth, N.J.; Berthelsen, D.; Reilly, S.; Mensah, F.K.; Gold, L.; Bennetts, S.K.; Levickis, P.; et al. EHLS at School: School-age follow-up of the Early Home Learning Study cluster randomized controlled trial. BMC Pediatr. 2018, 18, 148. [Google Scholar] [CrossRef] [PubMed]
- Jang, C.-H.; Choi, J.-H.; Byun, M.-S.; Jue, D.-M. Chloroquine inhibits production of TNF-α, IL-1β and IL-6 from lipopolysaccharide-stimulated human monocytes/macrophages by different modes. Rheumatology 2006, 45, 703–710. [Google Scholar] [CrossRef]
- Vincent, M.J.; Bergeron, E.; Benjannet, S.; Erickson, B.R.; Rollin, P.E.; Ksiazek, T.G.; Seidah, N.G.; Nichol, S.T. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005, 2, 69. [Google Scholar] [CrossRef] [PubMed]
- Dyall, J.; Coleman, C.M.; Hart, B.J.; Venkataraman, T.; Holbrook, M.R.; Kindrachuk, J.; Johnson, R.F.; Olinger, G.G.; Jahrling, P.B.; Laidlaw, M.; et al. Repurposing of Clinically Developed Drugs for Treatment of Middle East Respiratory Syndrome Coronavirus Infection. Antimicrob. Agents Chemother. 2014, 58, 4885–4893. [Google Scholar] [CrossRef]
- Gao, J.; Tian, Z.; Yang, X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends 2020, 14, 72–73. [Google Scholar] [CrossRef] [PubMed]
- Goldman, A.; Bomze, D.; Dankner, R.; Hod, H.; Meirson, T.; Boursi, B.; Maor, E. Cardiovascular adverse events associated with hydroxychloroquine and chloroquine: A comprehensive pharmacovigilance analysis of pre-COVID-19 reports. Br. J. Clin. Pharmacol. 2021, 87, 1432–1442. [Google Scholar] [CrossRef] [PubMed]
- Aziz, M.; Haghbin, H.; Abu Sitta, E.; Nawras, Y.; Fatima, R.; Sharma, S.; Lee-Smith, W.; Duggan, J.; Kammeyer, J.A.; Hanarahan, J.; et al. Efficacy of tocilizumab in COVID-19: A systematic review and meta-analysis. J. Med. Virol. 2021, 93, 1620–1630. [Google Scholar] [CrossRef]
- Morena, V.; Milazzo, L.; Oreni, L.; Bestetti, G.; Fossali, T.; Bassoli, C.; Torre, A.; Cossu, M.V.; Minari, C.; Ballone, E.; et al. Off-label use of tocilizumab for the treatment of SARS-CoV-2 pneumonia in Milan, Italy. Eur. J. Intern. Med. 2020, 76, 36–42. [Google Scholar] [CrossRef]
- Balkhair, A.; Al-Zakwani, I.; Al Busaidi, M.; Al-Khirbash, A.; Al Mubaihsi, S.; BaTaher, H.; Al Aghbari, J.; Al Kindi, M.; Baawain, S.; Al Alawi, A.; et al. Anakinra in hospitalized patients with severe COVID-19 pneumonia requiring oxygen therapy: Results of a prospective, open-label, interventional study. Int. J. Infect. Dis. 2021, 103, 288–296. [Google Scholar] [CrossRef]
- Huet, T.; Beaussier, H.; Voisin, O.; Jouveshomme, S.; Dauriat, G.; Lazareth, I.; Sacco, E.; Naccache, J.-M.; Bézie, Y.; Laplanche, S.; et al. Anakinra for severe forms of COVID-19: A cohort study. Lancet Rheumatol. 2020, 2, e393–e400. [Google Scholar] [CrossRef]
- Zhang, X.-J.; Qin, J.-J.; Cheng, X.; Shen, L.; Zhao, Y.-C.; Yuan, Y.; Lei, F.; Chen, M.-M.; Yang, H.; Bai, L.; et al. In-Hospital Use of Statins Is Associated with a Reduced Risk of Mortality among Individuals with COVID-19. Cell Metab. 2020, 32, 176–187.e4. [Google Scholar] [CrossRef]
- Haagmans, B.L.; Kuiken, T.; Martina, B.E.; Fouchier, R.A.; Rimmelzwaan, G.F.; Van Amerongen, G.; Van Riel, D.; De Jong, T.; Itamura, S.; Chan, K.H.; et al. Pegylated interferon-α protects type 1 pneumocytes against SARS coronavirus infection in macaques. Nat. Med. 2004, 10, 290–293. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Zhan, Y.; Zhu, L.; Hou, Z.; Liu, F.; Song, P.; Qiu, F.; Wang, X.; Zou, X.; Wan, D.; et al. Retrospective Multicenter Cohort Study Shows Early Interferon Therapy Is Associated with Favorable Clinical Responses in COVID-19 Patients. Cell Host Microbe 2020, 28, 455–464.e2. [Google Scholar] [CrossRef] [PubMed]
- Cinatl, J.; Morgenstern, B.; Bauer, G.; Chandra, P.; Rabenau, H.; Doerr, H. Treatment of SARS with human interferons. Lancet 2003, 362, 293–294. [Google Scholar] [CrossRef]
- Davoudi-Monfared, E.; Rahmani, H.; Khalili, H.; Hajiabdolbaghi, M.; Salehi, M.; Abbasian, L.; Kazemzadeh, H.; Yekaninejad, M.S. A Randomized Clinical Trial of the Efficacy and Safety of Interferon β-1a in Treatment of Severe COVID-19. Antimicrob. Agents Chemother. 2020, 64. [Google Scholar] [CrossRef] [PubMed]
- Rossignol, J.-F. Nitazoxanide, a new drug candidate for the treatment of Middle East respiratory syndrome coronavirus. J. Infect. Public Health 2016, 9, 227–230. [Google Scholar] [CrossRef]
- Chen, S.; Chan, K.; Jiang, Y.; Kao, R.; Lu, H.; Fan, K.; Cheng, V.; Tsui, W.; Hung, I.; Lee, T. In vitro susceptibility of 10 clinical isolates of SARS coronavirus to selected antiviral compounds. J. Clin. Virol. 2004, 31, 69–75. [Google Scholar] [CrossRef]
- Omrani, A.S.; Saad, M.M.; Baig, K.; Bahloul, A.; Abdul-Matin, M.; Alaidaroos, A.Y.; Almakhlafi, G.A.; Albarrak, M.M.; Memish, Z.A.; Albarrak, A.M. Ribavirin and interferon alfa-2a for severe Middle East respiratory syndrome coronavirus infection: A retrospective cohort study. Lancet Infect. Dis. 2014, 14, 1090–1095. [Google Scholar] [CrossRef]
- Li, H.; Xiong, N.; Li, C.; Gong, Y.; Liu, L.; Yang, H.; Tan, X.; Jiang, N.; Zong, Q.; Wang, J.; et al. Efficacy of ribavirin and interferon-α therapy for hospitalized patients with COVID-19: A multicenter, retrospective cohort study. Int. J. Infect. Dis. 2021, 104, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Morgenstern, B.; Michaelis, M.; Baer, P.C.; Doerr, H.W.; Cinatl, J. Ribavirin and interferon-β synergistically inhibit SARS-associated coronavirus replication in animal and human cell lines. Biochem. Biophys. Res. Commun. 2005, 326, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Arabi, Y.M.; Shalhoub, S.; Mandourah, Y.; Al-Hameed, F.; Al-Omari, A.; Al Qasim, E.; Jose, J.; Alraddadi, B.; AlMotairi, A.; Al Khatib, K.; et al. Ribavirin and Interferon Therapy for Critically Ill Patients with Middle East Respiratory Syndrome: A Multicenter Observational Study. Clin. Infect. Dis. 2020, 70, 1837–1844. [Google Scholar] [CrossRef]
- Pereira, M.; Williams, S.; Restrick, L.; Cullinan, P.; Hopkinson, N.S. Barriers to influenza vaccination in healthcare workers. BMJ 2018, 360, k1141. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.M.; Ma, E.S.K.; Peiris, J.S.M.; Wong, P.C.; Ho, J.C.M.; Lam, B.; Lai, K.N.; Tsang, K.W.T. Prolonged disturbances of in vitro cytokine production in patients with severe acute respiratory syndrome (SARS) treated with ribavirin and steroids. Clin. Exp. Immunol. 2004, 135, 467–473. [Google Scholar] [CrossRef]
- Kim, U.J.; Won, E.-J.; Kee, S.-J.; Jung, S.-I.; Jang, H.-C. Combination therapy with lopinavir/ritonavir, ribavirin and interferon-alpha for Middle East respiratory syndrome: A case report. Antivir. Ther. 2015, 21, 455–459. [Google Scholar] [CrossRef]
- Hung, I.F.-N.; Lung, K.-C.; Tso, E.Y.-K.; Liu, R.; Chung, T.W.-H.; Chu, M.-Y.; Ng, Y.-Y.; Lo, J.; Chan, J.; Tam, A.R.; et al. Triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: An open-label, randomised, phase 2 trial. Lancet 2020, 395, 1695–1704. [Google Scholar] [CrossRef]
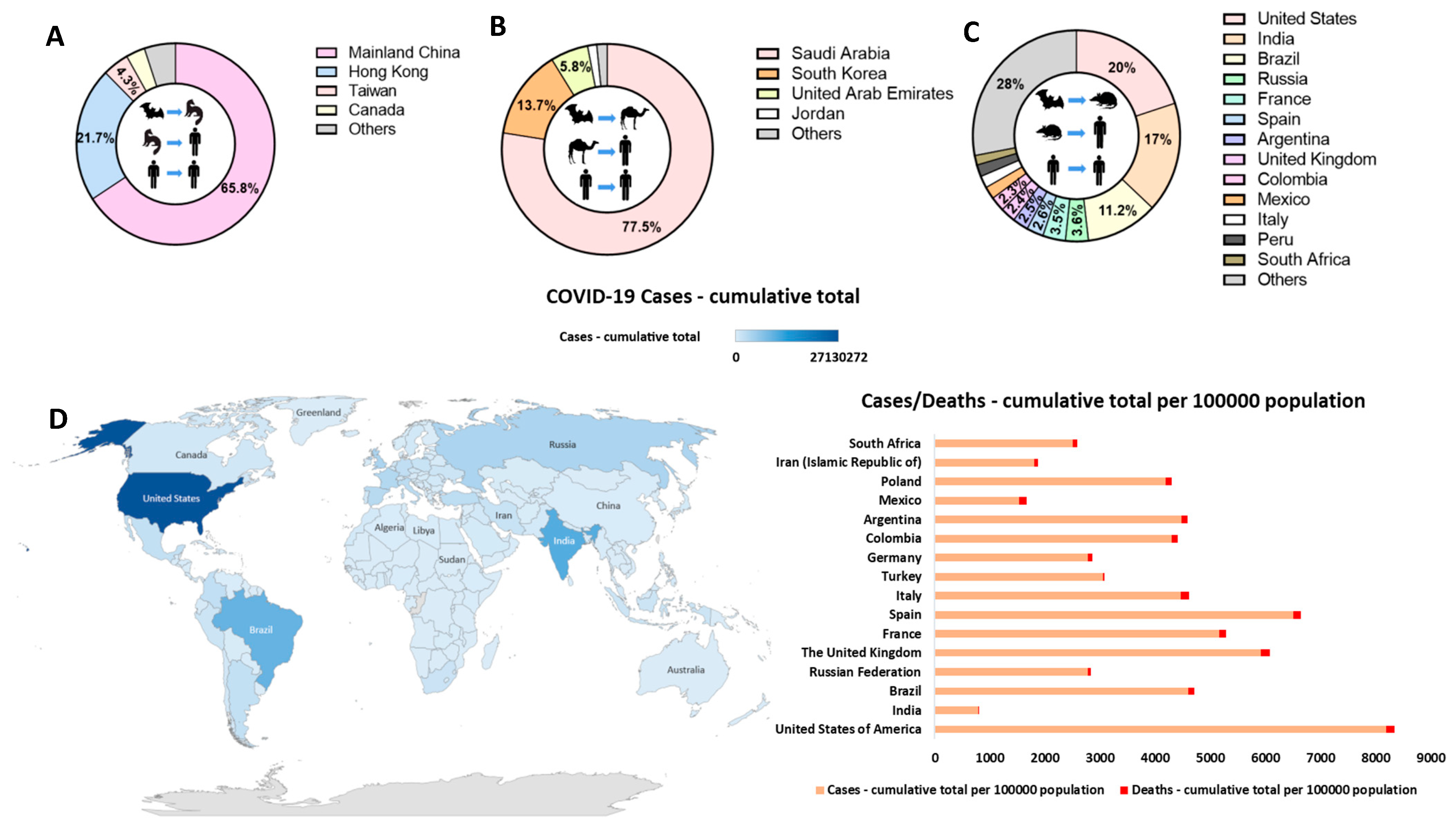
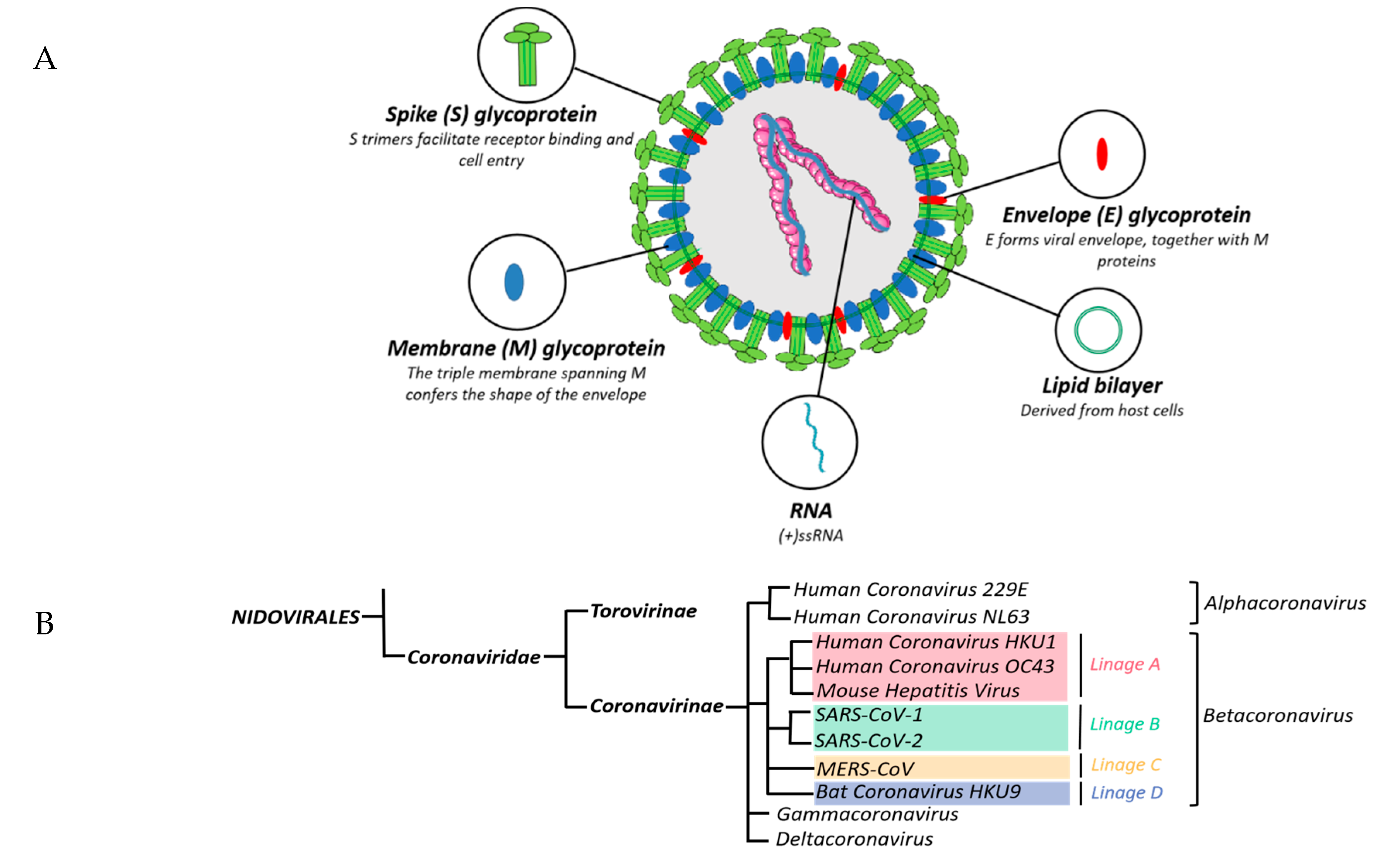
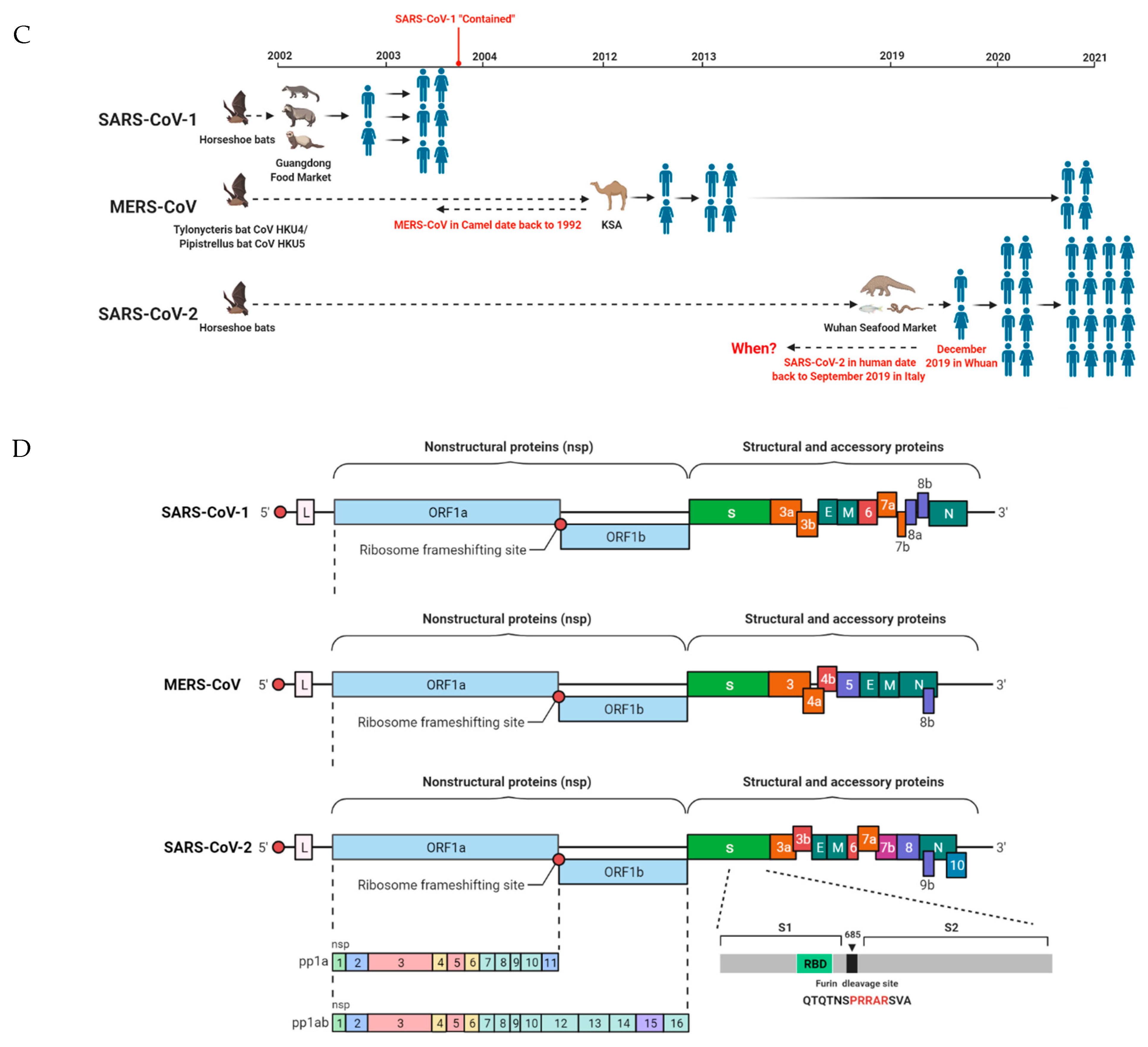
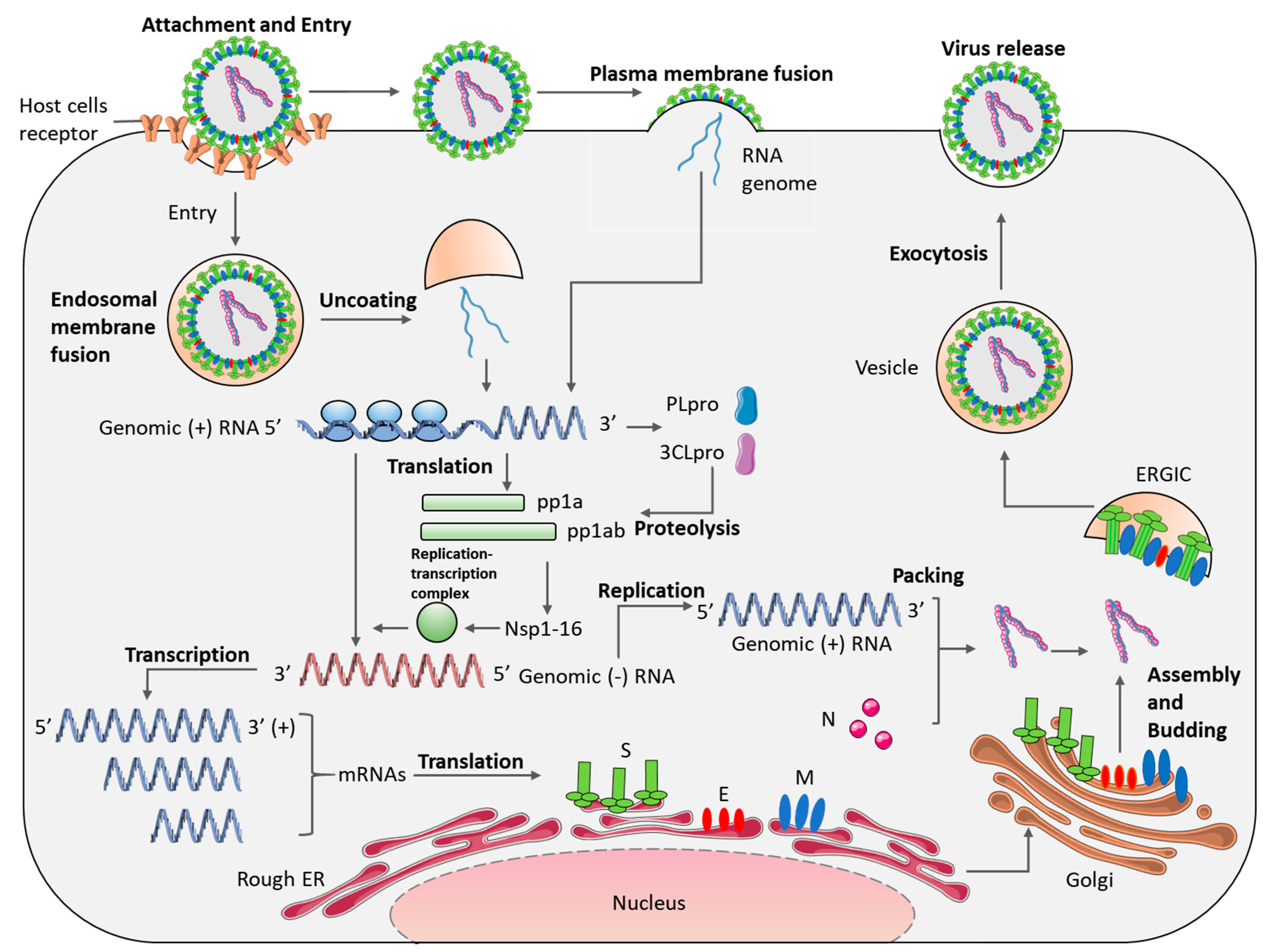
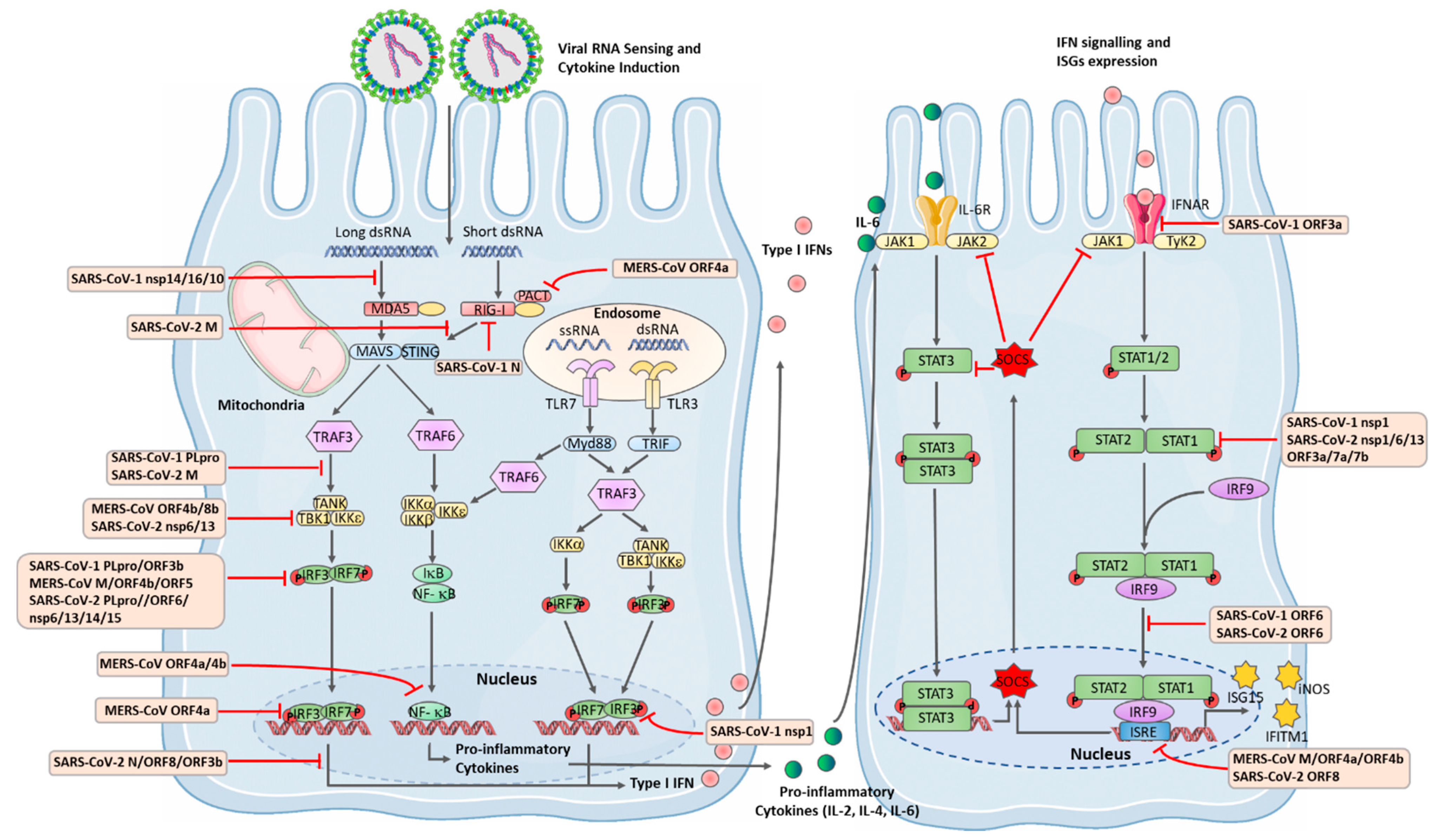
| SARS-CoV-1 | MERS-CoV | SARS-CoV-2 | ||
|---|---|---|---|---|
| Genome type | ssRNA(+) | ssRNA(+) | ssRNA(+) | |
| Genome size | 29.7 kb | 30.1 kb | 29.9 kb | |
| Genus | Beta-CoV, linage B [12] | Beta-CoV, linage C [13] | Beta-CoV, linage B [12] | |
| Origin | Guangdong, China | Saudi Arabia | Wuhan, China | |
| Possible natural reservoir | Horseshoe bats [14] | Tylonycteris and Pipistrellus bats [15] | Horseshoe bats or Pangolin [16,17] | |
| Possible intermediate host | Civet cat [18] | Dromedary camel [19] | Pangolin [17] | |
| Epidemiological Characteristics | Total laboratory-confirmed cases | 8089 | 2566 | 117,764,619 |
| Infected countries | 29 | 27 | 215 | |
| Gender (% of male) | 47% [20] | 78% [21] | 53% [22] | |
| Median age of patients | 43 [20] | 55 [21] | 61 [22] | |
| Reported death number | 774 | 881 | 2,613,747 | |
| Mortality | 9.6% | 34.4% | 0.8–10.8% | |
| Latency period | 2–10 | 5–14 | 12–20 | |
| Clinical Symptoms | Acute respiratory distress syndrome (ARDS) | Acute respiratory distress syndrome (ARDS) | Fever, cough, loss of taste, shortness of breath, sore throat and dyspnoea | |
| Functional receptor | Human angiotensin-converting enzyme 2 (ACE2), CD209 L | Human dipeptidyl peptidase 4 (DPP4 or CD26) | Human angiotensin-converting enzyme 2 (ACE2), CD209 L, NRP1, PIKfyve, BSG | |
| Receptor localised organ | Lungs, intestines, kidneys, heart, brain liver and testicles [23] | Brain, heart, lung, kidney, spleen, intestine, and liver [24] | Lungs, intestines, kidneys, heart, brain liver and testicles [23] | |
| Receptor distributed cell lines | Lung alveolar epithelial cells, enterocytes of the small intestine, arterial and venous endothelial cells and arterial smooth muscle cells [25] | Bronchiolar epithelium cells, alveolar interstitium cells, kidney vascular smooth muscle cells and immune cells [26,27] | Lung alveolar epithelial cells, enterocytes of the small intestine, arterial and venous endothelial cells and arterial smooth muscle cells [25] | |
| Strategy | Treatment | Mechanism | Efficacy | Side Effect/Disadvantage | ||
|---|---|---|---|---|---|---|
| SARS-CoV-1 | MERS-CoV | SARS-CoV-2 | ||||
| Serine protease inhibitor | Camostat mesylate | Block the entry of virus into TMPRSS2-expressing cells | Significantly reduced the entry of pseudotyped SARS-1-S (1–5 μM) [101] | Significantly reduced the entry of pseudotyped MERS-S (1–5 μM) [101] | Significantly reduced the entry of pseudotyped SARS-2-S and authentic infection (1–5 μM) [101] | The drug has few and mild side effects even at high dosages for other diseases [268]. Currently in many phase I and phase 2 clinical trials |
| Nucleoside inhibitor | Ribavirin | Stops viral RNA synthesis and viral mRNA capping [269] | No demonstrable anti-viral activity | Reduced virus replication in susceptible Vero cell lines [270] | Ribavirin was recommended by Chinese National Health Commission because of its in vitro effect [271], but no significant benefit was observed with ribavirin treatment for COVID-19 patients later [272] | Haemolytic anaemia, hypocalcemia and hypomagnesmia in SARS-CoV-1 infection [226] MERS-CoV infected Vero cells displayed a high level of resistance to the activity of ribavirin [270] |
| RNA polymerase inhibitor | Remdesivir | Inserts into viral RNA chains, causing their premature termination | Highly inhibited viral titres [273] | Highly inhibited viral titres [273] | Significantly improved time to recovery and reduced disease progression in patients needing oxygen, compared to placebo, and was approved by FDA [257] for emergency use | Approximately 10% healthy volunteers had raised blood levels of liver enzymes. Another common effect is nausea [274] |
| Favipiravir | Inhibits the RNA-dependent RNA polymerase (RdRp) of RNA viruses | Weak inhibitory effects on MERS-CoV RdRp activity [275] | No reports | Significantly improved the latency to cough relief and decreased the duration of pyrexia in moderate COVID-19 patients [276] | Increased serum uric acid and Psychiatric symptom reactions in COVID-19 patients [276] and clinical trials are ongoing | |
| HIV Protease inhibitors | Lopinavir/ ritonavir | Inhibits viral replication | Reduced use of pulse methylprednisolone, milder disease course and reduced viral load [234] | Improved clinical, radiological, and pathological outcomes and lower mean viral loads in lung tissues [247] | No benefit was observed with lopinavir–ritonavir treatment [277] | Gastrointestinal adverse events including nausea, vomiting, and diarrhoea were more common in lopinavir–ritonavir group [277] |
| Nelfinavir | Inhibits viral replication | Strongly inhibited viral replication in Vero E6 cells [278] | No activity found in vitro [279] | Potential inhibitor against main protease (Mpro) [280] Did not reduce viral load in the lungs of SARS-CoV-2-infected hamsters, but markedly improved lung pathology despite a massive infiltration of neutrophils [281] | Limited clinical data | |
| 3 CLpro inhibiotr | α-ketoamide inhibitors | Inhibit viral replication | Exhibited excellent anti-MERS-CoV activity in virus-infected Vero E6 cells [282] | Exhibited excellent anti-MERS-CoV activity in virus-infected Huh7 cells [282] | Inhibited SARS-CoV-2 replication in human Calu-3 lung cells [283] | No human proteases with a similar cleavage specificity are known; such inhibitors are unlikely to be toxic [283] |
| PIKfyve kinase inhibitor | Apilimod | Inhibit viral replication during entry | Inhibited MERS pseudotyped particles entry and replication in Vero E6 cell [284] | No reports | Strongly blocked SARS-CoV-2 infection in Vero E6 cells [111] | Phase I and phase II clinical trials of other diseases have shown that apilimod is safe and well tolerated [111] |
| Anti-viral compound | Arbidol | Prevents viral entry into the target cells | Reduced virus reproduction in vitro [285] | No reports | Patients had a shorter duration of infection (positive RNA test results), compared to those in the lopinavir/ritonavir group [286] | A clinical trial in patients with COVID-19 has been initiated in China [287] |
| Anti-parasitic agent | Ivermectin | Inhibits viral replication | No reports | No reports | 5000-fold reduction in SARS-CoV-2 RNA levels in vitro [265] Patients treated with ivermectin recovered earlier from hyposmia/anosmia [288] | No noticeable side effects [289] and larger clinical trials are needed |
| Anti-inflammatory agents | Corticosteroids | Blocks the action of inflammatory mediators and induce anti-inflammatory mediators [290] | Delayed viral clearance in blood [230] | Delayed viral clearance in respiratory tract [291] | Routine corticosteroids should be avoided unless they are being used to treat another condition (WHO guideline) [292] | Secondary bacterial/fungal infections, hyperglycemia, electrolyte imbalance and psychosis in SARS-CoV-1 infection treatment [230] |
| Chloroquine/hydroxychloroquine | Inhibits the production of inflammatory cytokines [293] and interferes with terminal glycosylation of the cellular receptor [294] | Ameliorated the hyperinflammatory response induced by viral infection [293] Prevented the spread of SARS-CoV-1 in cell culture [294] | Effectively blocked viral infection in vitro [295] | Several randomised controlled trials showed potential effects in reducing respiratory symptoms and pulmonary inflammation in COVID-19 patients [296] | High doses of CQ/HCQ in COVID-19 patients can be associated with increased cardiac adverse events [297] | |
| Tocilizumab | IL-6 receptor antagonist | No reports | No reports | Decreased the mortality rate in severe COVID-19 patients [298] | Some reports showed an increase in hepatic enzymes (29%), thrombocytopenia (14%), and serious bacterial and fungal infections (27%) [299] | |
| Anakinra | IL-1 receptor antagonist | No reports | No reports | Averted the need for mechanical ventilation in patients with severe COVID-19 pneumonia. Significantly reduced biomarkers of inflammation [300] | A three times increased level of aminotransferase in liver in 13% patients [301] | |
| Statins | Decreases inflammation and proinflammatory cytokines production | No reports | No reports | Statins can reduce inflammation and the progression of lung injury in experimental models. Statin use was associated with improved survival l [302] | Statins are first-line lipid-lowering therapies, with well-tolerated side effects. Limited clinical data [302] | |
| Anti-viral cytokine (type I interferon) | Interferon-α | Anti-viral ISGs induction inhibits viral replication and production | IFN alfacon-1 (recombinant synthetic type I IFN) was well tolerated by patients [232] PEG-IFN-α-2 b inhibited symptoms in macaques [303] | Inhibited viral induced cytopathic effect and RNA levels in vitro [248] | Early usage of IFN-α2 b decreased in-hospital death, but late usage raised mortality [304] | Mild neutropenia and some elevation of serum transaminase levels in SARS-CoV-1 infection [232] |
| Interferon-β | Anti-viral ISG induction inhibits viral replication and production | IFN-β-1 b inhibited viral replication more effectively than IFN-α-2 b in vitro and showed prophylactic protection and anti-viral ability [305] | Improved clinical, radiological, and pathological outcomes and lower mean viral loads in lung tissues [247] | Early administration significantly reduced mortality in severe COVID-19 patients [306] | Injection-related side effects (fever, chills, myalgia, and headache) a few hours after injection of IFN, happened in 19% of COVID-19 patients [306] | |
| Broad-spectrum anti-viral drug | Nitazoxanide | Broadly amplifying cytoplasmic RNA sensing and type I IFN pathways | Inhibited MERS-CoV infection in LLC-MK2 cells and reduced production of pro-inflammatory cytokines [307] | No reports | Inhibited the viral infection at a low micromolar concentration [258] | Clinical trials are ongoing with limited data |
| Combination treatment | Interferon-α and corticosteroids | Combined anti-viral and anti-inflammatory effects | More rapid resolution of radiographic lung abnormalities and better oxygen saturation levels [232] | No reports | No reports | Lack of randomisation and limited sample size [232] |
| Interferon-α and ribavirin | Anti-viral effects and inhibition of viral RNA replication | Highly synergistic effect and more effective than either single treatment [308] | Reduced viral replication and levels of proinflammatory cytokines in infected rhesus macaques [246] The survival rate of patients was improved significantly at 14 days [309] | RBV/IFN-α therapy was not observed to improve clinical outcomes in COVID-19 [310] | Not beneficial for severe and late MERS-CoV infection patients [309]. RBV/IFN-α therapy was associated with a higher probability of hospital stay [310] | |
| Interferon-β and ribavirin | Anti-viral effects and inhibition of RNA replication | Inhibited viral replication drastically compared to either single treatment in vitro [311] | No mortality reduction and fast viral RNA clearance in patients [312] | Clinical trial combined with lopinavir/ritonavir is ongoing [313] | Clinical data are limited | |
| Corticosteroids and ribavirin | Combined anti-inflammatory effects and inhibition of viral replication | Evaluated clinical outcomes, including recovered normal production of cytokines for establishing both cell-mediated and humoral immunity in patients [314] | No reports | No reports | Clinical data are limited | |
| Lopinavir/ritonavir and ribavirin | Combined inhibition of viral replication | Showed stronger suppression on viral load and the adverse clinical outcomes (ARDS or mortality) was significantly lower [234] | Showed rapid viral RNA clearance and patient recovery when combined with IFN-α [315] | Lopinavir/ritonavir in combination with ribavirin and IFNβ-1 b was safe and superior to lopinavir–ritonavir alone in alleviating symptoms and shortening the duration of viral shedding and hospital stay in patients with mild to moderate COVID-19 [316] | Anaemia and fall of haemoglobin in SARS-CoV-1 infection [234] Self-limited nausea and diarrhoea [316] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Gargan, S.; Lu, Y.; Stevenson, N.J. An Overview of Current Knowledge of Deadly CoVs and Their Interface with Innate Immunity. Viruses 2021, 13, 560. https://doi.org/10.3390/v13040560
Zhang Y, Gargan S, Lu Y, Stevenson NJ. An Overview of Current Knowledge of Deadly CoVs and Their Interface with Innate Immunity. Viruses. 2021; 13(4):560. https://doi.org/10.3390/v13040560
Chicago/Turabian StyleZhang, Yamei, Siobhan Gargan, Yongxu Lu, and Nigel J. Stevenson. 2021. "An Overview of Current Knowledge of Deadly CoVs and Their Interface with Innate Immunity" Viruses 13, no. 4: 560. https://doi.org/10.3390/v13040560
APA StyleZhang, Y., Gargan, S., Lu, Y., & Stevenson, N. J. (2021). An Overview of Current Knowledge of Deadly CoVs and Their Interface with Innate Immunity. Viruses, 13(4), 560. https://doi.org/10.3390/v13040560






