Abstract
Three human protoparvoviruses, bufavirus (BuV), tusavirus (TuV) and cutavirus (CuV), have recently been discovered in diarrheal stool. BuV has been associated with diarrhea and CuV with cutaneous T-cell lymphoma, but there are hardly any data for TuV or CuV in stool or respiratory samples. Hence, using qPCR and IgG enzyme immunoassays, we analyzed 1072 stool, 316 respiratory and 445 serum or plasma samples from 1098 patients with and without gastroenteritis (GE) or respiratory-tract infections (RTI) from Finland, Latvia and Malawi. The overall CuV-DNA prevalences in stool samples ranged between 0–6.1% among our six patient cohorts. In Finland, CuV DNA was significantly more prevalent in GE patients above rather than below 60 years of age (5.1% vs 0.2%). CuV DNA was more prevalent in stools among Latvian and Malawian children compared with Finnish children. In 10/11 CuV DNA-positive adults and 4/6 CuV DNA-positive children with GE, no known causal pathogens were detected. Interestingly, for the first time, CuV DNA was observed in two nasopharyngeal aspirates from children with RTI and the rare TuV in diarrheal stools of two adults. Our results provide new insights on the occurrence of human protoparvoviruses in GE and RTI in different countries.
Keywords:
parvovirus; bufavirus; tusavirus; cutavirus; gastroenteritis; respiratory-tract infection; leukemia; PCR; serology 1. Introduction
Parvoviridae is a family of small nonenveloped single-stranded DNA viruses that infect host-specifically many diverse animal species. There are two known human pathogens: parvovirus B19 (B19V), causing erythema infectiosum, arthritis, anemias and fetal death; and human bocavirus (HBoV) 1, causing pediatric respiratory-tract infections (RTI) and, infrequently, encephalitis. Furthermore, parvovirus 4 and HBoV2-4 infect humans with unclear disease associations [1]. In 2012–2016, metagenomic studies revealed, in human diarrheal stool samples, three more parvoviruses belonging to the Protoparvovirus genus: bufavirus (BuV), tusavirus (TuV) and cutavirus (CuV) [2,3,4]. Currently there are three known genotypes of BuV [5,6]. However, the clinical impact of the newly discovered protoparvoviruses is largely unknown.
BuV DNA has worldwide been detected by PCR in low prevalence (0–4%) in diarrheal stool samples, whereas non-diarrheal stools have mainly been BuV-DNA negative [5,6,7,8,9,10,11,12,13,14,15,16,17,18,19]. However, the causative role of BuV in gastroenteritis (GE) remains unclear. Currently, TuV DNA has only been reported in the stool of a single child with unexplained diarrhea from Tunisia [3]. No other studies of TuV DNA have to our knowledge been published and further investigation of TuV in human samples is thus warranted. CuV is the newest parvovirus discovered in humans. Its prevalence in stool has hitherto not been reported since the original discovery, which conveyed a low prevalence (1–2%) in Brazilian and Botswanan diarrheic children [4]. Excitingly, dermal CuV DNA is documented to be associated with cutaneous T-cell lymphoma (CTCL) [20,21,22]. It has also been detected in skin biopsies of melanoma and organ transplant patients, but not in those of healthy subjects [21,23,24]. As most CuV studies have focused on screening malignant skin tissues, the prevalence of virus in stool and respiratory samples from respective GE and RTI patients has, unlike for BuV, not been investigated since its discovery, so nothing is known of acute CuV infections.
The aim of the current study was to elucidate how common acute human protoparvovirus infections are in pediatric and adult patients with and without GE or RTI, to identify age- or gender-related and geographic distributions, and to find potential disease associations to GE or RTI. We analyzed in total 1072 stool samples and 316 respiratory samples from patients from six cohorts in three countries, Finland, Latvia and Malawi, for CuV, TuV and BuV DNA, as well as 445 plasma or serum samples for the respective seroprevalences in children from Latvia and Finland. For all children with protoparvovirus IgG-positive serum or plasma, we analyzed also their specific IgG by competition enzyme immunoassay (EIA) [25].
2. Materials and Methods
2.1. Study Cohorts
The Helsinki-B cohort from Finland comprises stool samples originally sent for bacterial diagnosis from 212 patients (age 1–94 years, median 36 years) with GE (Table 1). The samples had been analyzed during October 2012–March 2013 mainly for Salmonella spp., Shigella spp., Campylobacter spp., Yersinia spp., Vibrio cholerae and pathogenic Escherichia coli subtypes by culture or PCR. A bacterial pathogen was detected in 73/212 patient samples.

Table 1.
Characteristics of cohorts used in the current study.
The Helsinki-V cohort from Finland comprises stool samples originally sent for viral diagnosis from 285 patients (age 0–99 years, median 74 years) with GE (Table 1). The samples had been tested for either norovirus alone, or for norovirus, adenovirus, rotavirus and astrovirus, during April–June 2013, by RT-PCR or antigen detection assay (Diarlex MB, Orion Diagnostica, Espoo, Finland). A viral pathogen was discovered in 99/285 samples. All the Helsinki-B and Helsinki-V samples had been sent for routine testing to the Helsinki University Hospital Laboratory (HUSLAB) from diverse locations in Finland, and thus were not from a few isolated outbreaks. Further, all the samples from these two cohorts have been previously studied for BuV [12]. The Ethics Committee of the Hospital District of Helsinki and Uusimaa approved the study of the deidentified samples.
The Tampere cohort includes 228 stool, nasal swab, and serum samples from 228 children (age 0–15 years, median 1.3 years) with GE (n = 42), RTI (n = 104), or both (n = 82) (Table 1). The patients were recruited between September 2009 and August 2011 in Tampere, Finland, and the samples were collected throughout the year [26]. Further, all the stool and swab samples from this cohort have been previously studied for BuV DNA and the sera for BuV and TuV IgG [13]. A written informed consent was obtained from the parents of all the children enrolled. The study was approved by the Ethics Committee of Pirkanmaa Hospital District, and it was conducted in accordance with the relevant guidelines and regulations.
The Latvia cohort consists of 44 nasopharyngeal aspirate (NPA), 115 stool and 102 plasma samples from 159 children (age 0–5 years, median 1.75 years), with GE (n = 62), RTI (n = 80), or both (n = 17) (Table 1). The study protocol was approved by the Ethics Committee of the Rīga Stradiņš University [27]. Written informed consent was received from all parents or guardians of the participating children.
The Malawi cohort includes 168 stool samples collected from 164 healthy or diseased children (age 6–12 months) from rural Malawi for a Child Nutrition Intervention Study (LCNI-5) between January 2008 and November 2009 (Table 1). The study adhered to the principles of the Declaration of Helsinki and regulatory guidelines in Malawi. Written informed consent was obtained from the participants’ guardians and the trial protocol was approved by the research ethics committees of the University of Malawi College of Medicine and of the Pirkanmaa Hospital District, Finland.
The leukemia cohort comprises 44 nasal swab, 115 serum and 64 stool samples from 50 children (age 0.4–15.3 years, median 5.7 y) with acute leukemia undergoing anticancer treatment, collected between April 2000 and October 2005 at 4 Finnish university hospitals (Table 1) [28]. The study was approved by the Ethical Committees of the Medical faculties of Turku, Oulu, Kuopio, and Helsinki Universities. Written informed consent was obtained from the patients or from their parents.
2.2. CuV-BuV-TuV qPCR Assay
To detect and quantify CuV, BuV, and TuV DNA, a multiplex real-time qPCR was performed with primers and hydrolysis probes located in the VP2 regions of CuV and TuV and the NS1 region of BuV, as previously described [21]. The samples from the Helsinki and Tampere cohorts [12,13], with previously published BuV qPCR results, were analyzed by a duplex qPCR assay for CuV and TuV DNA. For most of the samples, an initial screening was done in multiplex assays and positives were repeated in singleplex. For two CuV DNA-positive samples (from the Helsinki-V and Tampere cohorts each), the qPCR assay could, however, not be repeated due to insufficient amounts of sample. Further, sufficient volumes were not available for 49 Helsinki-V samples, including a TuV DNA-positive one, for the initial qPCR screening and water was added to make up to 5 µl for PCR. All positive samples were confirmed by cloning and sequencing (Figure S1).
2.3. CuV-BuV1-3-TuV IgG EIA
A total of 115 serum and 102 plasma samples from 29 leukemic children and 102 Latvian children, respectively, were screened for CuV-BuV1-3-TuV IgG, and the 228 serum samples from the Tampere children were screened for BuV2 and CuV. BuV and TuV IgG EIA results for these Tampere children have been previously published, but due to frequent cross-reactivity between BuV2 and CuV IgG, learned since then, these samples were here screened besides for CuV, also for BuV2 IgG, with cross blocking [13]. The samples were analyzed by an in-house IgG EIA, with biotinylated VP2 virus-like particles (VLP) as antigen, and all samples with an OD > 0.1 were confirmed using a competition assay as previously described [25], but with a 40-min incubation at room temperature with substrate 3,3′,5,5′-tetramethylbenzidine (BD OptEIA™, Franklin Lakes, NJ, USA). Optical densities (ODs) were measured at 450 nm (Multiskan EX; Thermo Fischer Scientific, Pittsburgh, PA, USA). Samples with OD between 0.1–0.24 were considered to be borderline positives.
2.4. Statistical Analysis
Statistics were calculated with IBM SPSS Statistics v25 (IBM Corp, NY, USA) for Pearson’s χ2. A p value <0.05 was considered statistically significant.
3. Results
3.1. CuV DNA in Stool and Respiratory Samples
In all, CuV DNA was detected in stool samples from 26/1039 (2.5%) adults and children in the current study (Table 2). The prevalence of CuV DNA in stool samples among the six cohorts was between 0–6.1%. The viral loads of CuV DNA-positive stool samples varied between 1.24 × 102 and 1.05 × 104 copies/mL stool supernatant (Table 3). No statistical differences in the CuV viral loads were observed between the different cohorts and between patients with GE or RTI.

Table 2.
Prevalence of CuV, BuV and TuV DNA in stool and respiratory samples in different population cohorts.

Table 3.
CuV, BuV and TuV DNA-positive samples from children and adults in the current study.
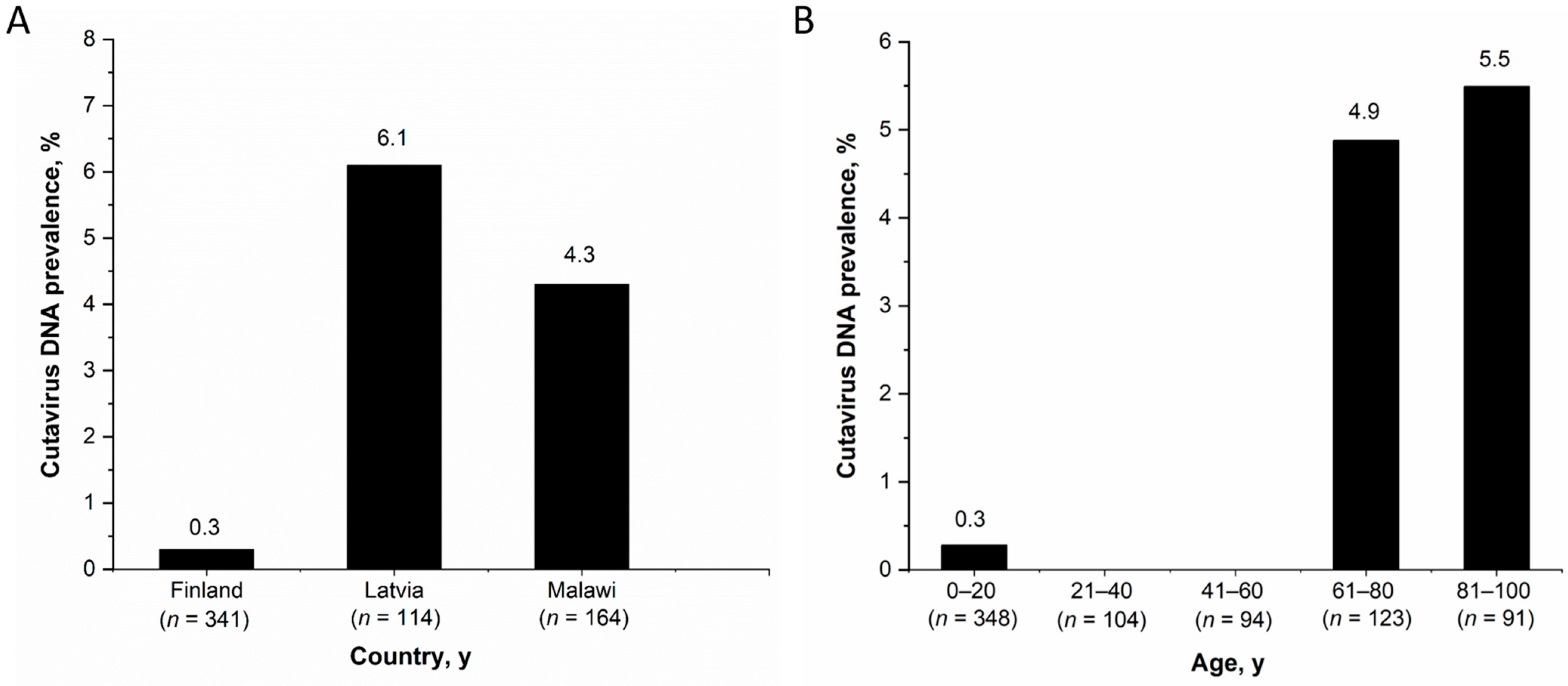
Among the observed country-specific CuV-DNA prevalences in stool samples of children <15 years, Latvia had the highest prevalence (7/115, 6.1%) followed by Malawi (7/164, 4.3%) and Finland (1/341, 0.3%) (Figure 1A). The CuV-DNA prevalence in Latvian and Malawian children was significantly higher than in the Finnish children (p < 0.05). However, there was no significant differences in CuV DNA prevalence between children in different age groups of all the three cohorts; 0–1 years (10/291, 3.4%), >1–5 years (4/192, 2.1%), >5–15 years (1/24, 4.2%) (p ≥ 0.5). Nevertheless, when considering both adults and children from all cohorts in Finland (n= 761) (including Helsinki-B, Helsinki-V, Tampere and Leukemia cohorts), the CuV DNA prevalence among individuals above 60 years of age (5.1%, 11/214) was significantly higher (p < 0.001) than that of individuals below 60 years of age (0.2%, 1/546) (Figure 1B). This was also significant when including adults and children from only the 2 Helsinki cohorts.
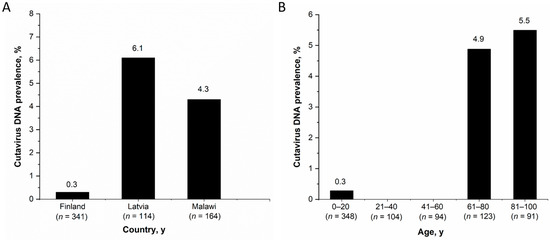
Figure 1.
CuV-DNA stool prevalence (A) Among children from Finland, Latvia and Malawi, (B) Among all patients from Finland by age.
Among children (<15 years) from the Helsinki-B, Helsinki-V, Tampere and Latvia cohorts, CuV DNA was detected in stools of 4/180 (2.2%) with GE alone, of 2/183 (1.1%) with RTI alone, of 2/99 (2%) with symptoms of both. The difference in CuV-DNA prevalence between the GE and RTI groups was not significant (p > 0.5). Further, stool samples from 4 healthy children from Malawi were CuV-DNA positive (Table 3).
Among the respiratory samples, CuV DNA was detected in 2/44 (4.5%) NPAs from children from Latvia but not in any of 272 pediatric nasal swabs from the Leukemia or Tampere cohorts from Finland. The two CuV DNA-positive NPAs from the Latvian cohort were from children 45- and 39-months of age, both with RTI and fever at the time of sampling. The viral loads of the CuV DNA-positive NPA samples were 8.62 ×102 and 2.04 × 103 copies/mL of NPA respectively (Table 3). No stool samples were available from these two individuals and the corresponding plasma samples were CuV-DNA and -IgG negative. In addition, they were both positive for HBoV1 DNA in NPA, but not in serum, and one of the children was also positive for human rhinovirus RNA.
The 91-nt long CuV amplicons from all CuV DNA-positive samples in the current study, had 1 to 5 nucleotide mismatches in the non-primer-binding region, compared with the CuV sequence NC_039050.1, serving as positive control in qPCR (Figure S1).
3.2. BuV and TuV DNA in Stool and Respiratory Samples
BuV DNA in stools was analyzed only from the Latvia, Malawi and Leukemia cohorts, where 1/314 (0.3%) children were PCR positive. The single BuV DNA-positive stool sample had a virus load of 1.23 × 103 copies/mL and was from a healthy child, 6 months of age, from Malawi (Table 3). Of note, we did not analyze for BuV DNA in the samples from the Helsinki and Tampere cohorts because they had already been analyzed in our previous study, where it was detected in stools of the Helsinki-V (3/386, 0.7%), Helsinki-B (4/243, 1.6%), and Tampere cohorts (3/955, 0.3%), and in a nasal swab (1/955, 0.1%) of a child from Tampere [12,13]. Furthermore, none of the current respiratory samples from 76 children in the Latvia or Leukemia cohorts contained BuV DNA (Table 2).
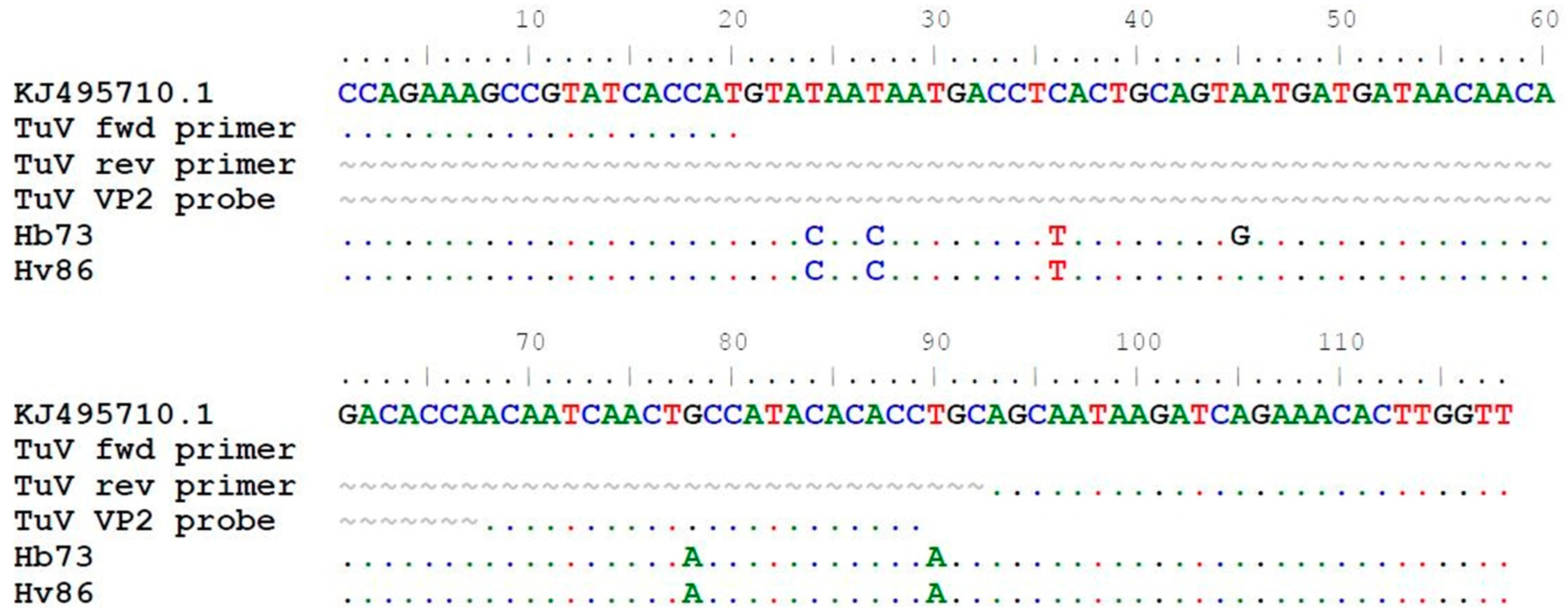
TuV DNA was detected in stool samples from 2/1039 (0.2%) individuals but in none of the 316 respiratory samples (Table 2). The TuV DNA-positive stool samples were from 22- and 27-year-old women in the Helsinki-V and -B cohorts, respectively, from Finland. The viral loads of the samples were 8.90 × 101 and 4.42 × 101 copies/mL stool suspension, respectively (Table 3). The young adults had gastrointestinal symptoms, but no other causative pathogens were found. Further, the 27-year-old woman had recently travelled to Jamaica and Turkey (Istanbul). In the sequence alignment, the 118-nt long TuV amplicons had 5 and 6 nucleotide mismatches in the non-primer-binding target region, respectively, compared to the only known TuV sequence KJ495710.1, also serving as positive control in qPCR (Figure 2). There were no more stool materials left for further sequencing of longer amplicons.
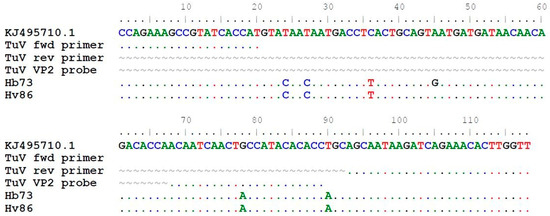
Figure 2.
Alignment of the 118 nt sequenced qPCR amplicons from TuV DNA-positive stool samples in the current study to the only published TuV reference sequence KJ495710.1 (3085–3202 bp). Dot (.) indicates identical nucleotides and tilde (~) lacking nucleotides.
3.3. Protoparvovirus IgG in Children
We screened 228 serum samples from the Tampere children for CuV IgG and the seroprevalence was 2/228 (0.9%). For the two CuV IgG-positive samples, the corresponding nasal swab and stool samples from the same patients were CuV-DNA negative (Table 4). Further, from one child whose stool was CuV DNA-positive, we could not detect CuV DNA in the swab or CuV DNA or IgG in serum. For the 102 Latvian children, the CuV seroprevalence was 2.9% (3/102). All three CuV-seropositive children (La00, La78 and La34) had GE symptoms, but we did not detect CuV DNA in their stool samples, and NPA samples were not available from these three children. Further, three other children (La82, La03 and La14) with CuV DNA-positive stools and two children (La49 and La50) with CuV DNA-positive NPA samples were seronegative. No follow-up samples were available.

Table 4.
Comparison of CuV DNA in stool and respiratory samples with CuV IgG from CuV DNA- or IgG-positive individuals.
No BuV1, 2, 3, or TuV IgG antibodies were detected in 102 plasma samples from the Latvian children. BuV1, 2, 3 and TuV IgG EIA results for 228 serum samples from the Tampere children have been previously published and seven children (7/228, 3.1%) had BuV IgG and one child (1/228, 0.4%) had TuV IgG [13]. Of the BuV IgG-positive cases, one had BuV1, three had BuV2, two had BuV3 and one had both BuV1 and 2. However, due to the known cross-reactivity of BuV2 and CuV IgG, we re-analyzed the four previously reported BuV2 IgG-positive samples by competition EIAs and proved two of them to actually be CuV IgG and not BuV2 IgG, whereas the 2 remaining positives were truly BuV2 IgG. Therefore, the correct BuV IgG seroprevalence among the Tampere children is 2.6% (6/228).
A total of 115 serum samples from 29 leukemic children were screened for CuV, BuV1-3, and TuV IgG. One child (1/29, 3.4%) had BuV2 IgG and four children (4/29, 13.8%) had slightly raised CuV IgG absorbances. Multiple serum samples were available from three CuV IgG-positive children and the observed OD values remained similar for the whole follow-up period, up to one month for 2 children and one year for one child. Overall, the CuV absorbance values were low (0.14–0.27), but specific by competition EIA, and the corresponding stool and swab samples from these children were CuV-DNA negative. No BuV1, BuV3, or TuV IgG antibodies were detected in this cohort.
4. Discussion
CuV has already gained much interest due to its association with cutaneous T-cell lymphoma, despite being discovered only in 2016. It has been predominantly searched for in skin cancers [4,20,21,22], however, nothing is known of its acute infections. No respiratory or stool samples have been analyzed for its prevalence or to characterize its possible symptoms in acute infections. Likewise, with TuV, no PCR results have been reported in human samples apart from the initial paper where it was discovered in the stool of a single Tunisian child with unexplained diarrhea [3]. Hence in the current study, we analyzed CuV and TuV DNA prevalences and their age- or gender-related and geographic distributions in human stool and respiratory samples of patients, with and without GE or RTI, and compared the findings with those of the more studied BuV.
The highest CuV DNA prevalence in stool samples was detected among the Latvian children as compared to all other cohorts analyzed in both this and the original discovery study [4]. The observed CuV viral loads in stool samples, in this study, were similar to those of BuV from previous studies of stool samples from GE and RTI patients, including both adults and children [12,13]. Low viral loads in stool samples of these protoparvoviruses may be remnants from a previous infection. Nevertheless, no known pathogens were detected in the majority of the CuV DNA-positive adults and children with GE, allowing speculations for a possible causal role of CuV. However, further evidence from serological studies together with viral DNA detection in paired samples is required to provide additional clues about CuV acute infections in humans and its possible role in GE.
CuV was significantly more prevalent in stool samples from Finnish GE patients above 60 years than below. By contrast, the BuV-DNA stool-positive samples were from a broader age group of Finnish adults, of 21–89 years, with a median of 53 years [12]. Unfortunately, stool samples from adults from Latvia and Malawi were not available for similar prevalence comparisons in these two countries. Further, we did not have serum samples from the adults for CuV IgG prevalence comparisons in different age groups.
Interestingly, we observed for the first time CuV DNA in NPAs of children with RTI, indicating that CuV is shedding into the airways. However, due to the low prevalence, we did not observe a significant difference (p > 0.5) in the CuV airway genoprevalences between the GE and RTI groups, which in our association studies served as controls for each other. Due to the lack of association, the low DNA prevalence and copy numbers, and the presence of other RTI-causing pathogens, we did not see a causal role for CuV in RTI.
CuV has been more widely studied in skin samples than in stool or respiratory samples. So far, CuV DNA has been detected in skin biopsies of CTCL, melanoma and transplant patients with prevalences between 1.1–16% [4,21,22,23], but not in those of healthy subjects [4,21]. However, viral DNA has surprisingly been found in skin surface swabs of healthy adults (9/237, 3.8%) and of HIV-positive men (35/205, 17.1%) [24]. Recently, CuV DNA was found in the skin biopsy of an acute lymphoblastic leukemia patient [29]. The overall occurrence of CuV DNA in skin seems to be more common than in stool or respiratory samples, which may be because the virus persists in skin tissues for years [21], whereas shedding may be limited to the acute phase of infection.
Previously, the CuV-IgG prevalence was observed to be low, ranging from 0–6% among healthy individuals in the USA, Iran, Iraq and Finland, with the exception of 9.5% in CTCL patients [21,25]. The observed CuV seroprevalences in the Tampere (0.9%) and Latvian (2.9%) children are thus in line with previous studies. CuV DNA was detected relatively often in skin and stool when taking into consideration the observed lower seroprevalence in any population worldwide. This could be interpreted in three ways: (i) B-cell immunity is not induced due to the infections being local; (ii) some antibody responses wane with time below the detection limit, as seen for human bocavirus [30]; or (iii) a prior BuV infection could inhibit the induction of CuV IgG due to an immunological phenomenon called original antigenic sin or imprinting, also seen among the related human bocaviruses [31]. However, the first scenario seems logical only for stool positivity, since CuV persists in malignant skin tissue and lymph nodes [21], the middle scenario seems the most logical unless the whole virion persists or viral VP2 is expressed, which would rather boost immunity, and the latter would be more expected in Asia and Africa, where the BuV seroprevalence is much higher [25]. In contrast to our previous skin study, we observed no correlation between CuV DNA positivity in stool or respiratory samples and CuV IgG positivity in the same individuals [21]. This could be due to scenario (i) above, or more likely, as for other human parvoviruses, to acute systemic infections occurring before the antibody responses become measurable. Unfortunately, we did not have follow-up sera to observe possible seroconversions.
In our earlier study, we revealed a high BuV seroprevalence in Kenyan adults of 72% (compared to 2% in Finland) and in children 21% [25], nevertheless, in our current study, we found only one child from Malawi with BuV DNA in stool. This could be due to the lower age of the children, or to regional differences within Africa. BuV has been associated with GE, as most BuV DNA-positive cases have been diarrheic [6]. However, no such symptoms had been described for the single BuV DNA-positive child in Malawi. The BuV-DNA prevalence in stool was lower than that of CuV in the Malawian children and no BuV DNA was detected in the Latvian children, who had the highest CuV-DNA prevalence. This would suggest that the prevalence of BuV in these countries is much lower than that of CuV. The low BuV-DNA prevalence in children is supported by previous studies where all or a majority of the BuV DNA-positive stool samples were from adults [6,9,10,12,15].
This is the first published study of TuV DNA in human stool and respiratory samples since the initial metagenomics study [3]. TuV DNA was detected, though in low quantities, in stool samples from two adults from Finland with GE. There is currently only one TuV sequence published, on which our qPCR and VP2-VLP EIAs were based on [3]. Further, the observed 5 and 6 nucleotide mutations in the non-primer binding region of the 118-nt-long amplicon sequences from the two TuV DNA-positive stools, indicate that TuV may be more diverse in nature than previously believed. The low prevalence could therefore be attributed to non-optimal primers or probe that affects the sensitivity of the current PCR method. This could also explain the low TuV geno- and seroprevalence in the populations studied so far [13,25]. Unfortunately, there was not enough sample material to sequence more of the genome. More studies are therefore needed to confirm whether there are different strains of TuV circulating in the population and its true prevalence globally.
Unlike for TuV, there are many published sequences for CuV, which differ only by a few nucleotides in the region covering our primer and probe sites [4,21]. Hence with a degenerate CuV forward primer and probe in our qPCR assays, we were able to detect also variant strains of CuV. However, apart from the known mismatches, there could be CuV strains with other mutations at the primer and probe binding sites, resulting in lack of detection with our current qPCR assay.
Children with acute leukemia are more prone to opportunistic infections due to cytotoxic and immunosuppressive treatments [32]. Hence, in our study, we wanted to see the geno- and seroprevalences of protoparvoviruses in leukemic children. However, none of the 64 stool samples and 44 swab samples from 35 and 32 leukemic children, respectively, were CuV, TuV or BuV DNA positive in our current study. Interestingly, the CuV IgG seroprevalence of leukemic children, despite their immunosuppressive state, was 13.8% in our study, one of the highest observed in any cohort so far [21,25]. By competition EIA, we observed that the CuV-specific reactivity was blocked completely only by homotypic CuV antigen and not by heterotypic antigen (BuV1 and 2), thereby confirming specificity. Despite the confirmatory blocking results, the overall absorbance values were unusually low and therefore the results should be interpreted with caution. As these children with acute leukemia had not received blood transfusions, the observed higher CuV IgG prevalence and overall lower absorbance values could be attributed to their immunosuppressive state.
5. Conclusions
Since their discoveries, we observed for the first time CuV DNA in nasal swabs and report the genoprevalences of CuV and TuV in stool and respiratory samples. CuV DNA was detected relatively often when taking into consideration the observed seroprevalence. The overall low geno- and seroprevalences and low DNA loads, together with a lack of association to GE or RTI, indicate that CuV is not at least a frequent cause of GE or RTI. However, the reason for viral shedding in the airways and stool in different populations and the ability of these newly discovered human protoparvoviruses to cause diseases remain to be elucidated.
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4915/13/3/483/s1, Figure S1: qPCR-amplicons of 91 nt from CuV DNA-positive stool and NPA samples.
Author Contributions
Conceptualization, U.M., E.V. and M.S.-V.; methodology, E.V.; software, U.M.; investigation, U.M., M.J., R.R.T. and E.V.; resources, M.P., S.O., Y.-M.F., P.A., T.V., M.L., E.T., Z.N.-K., A.V., K.V. and C.M.; writing—original draft preparation, U.M.; writing—review and editing, M.S.-V.; visualization, U.M.; supervision, M.S-V.; funding acquisition, M.S.-V., U.M., E.V., Z.N.-K. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by grants from the Sigrid Jusélius Foundation, the Life and Health Medical Support Association, the Finnish Norwegian Medical Foundation, the Ida Montin Foundation and Rīga Stradiņš University research project funding (RSU ZP 17/2013). Open access funding provided by University of Helsinki.
Institutional Review Board Statement
The Helsinki samples were routine stool samples from deidentified patients and informed consents were given from all other patients or their guardians. The study protocols were approved by the Ethics Committees of the local hospital districts of Helsinki, Tampere, Riga and Malawi, and they were conducted in accordance with the relevant guidelines and regulations and followed the principles of the Declaration of Helsinki.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Acknowledgments
We thank Dace Gardovska and Inga Ziemele at the Infectious Diseases Department of the Children’s Clinical University Hospital of Riga, Latvia, for the pediatric sample collection and all patients and the guardians of the children.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Qiu, J.; Söderlund-Venermo, M.; Young, N.S. Human parvoviruses. Clin. Microbiol. Rev. 2017, 30, 43–113. [Google Scholar] [CrossRef]
- Phan, T.G.; Vo, N.P.; Bonkoungou, I.J.; Kapoor, A.; Barro, N.; O’Ryan, M.; Kapusinszky, B.; Wang, C.; Delwart, E. Acute diarrhea in West African children: Diverse enteric viruses and a novel parvovirus genus. J. Virol. 2012, 86, 11024–11030. [Google Scholar] [CrossRef]
- Phan, T.G.; Sdiri-Loulizi, K.; Aouni, M.; Ambert-Balay, K.; Pothier, P.; Deng, X.; Delwart, E. New parvovirus in child with unexplained diarrhea, Tunisia. Emerg. Infect. Dis. 2014, 20, 1911–1913. [Google Scholar] [CrossRef]
- Phan, T.G.; Dreno, B.; da Costa, A.C.; Li, L.; Orlandi, P.; Deng, X.; Kapusinszky, B.; Siqueira, J.; Knol, A.C.; Halary, F.; et al. A new protoparvovirus in human fecal samples and cutaneous T cell lymphomas (mycosis fungoides). Virology 2016, 496, 299–305. [Google Scholar] [CrossRef]
- Yahiro, T.; Wangchuk, S.; Tshering, K.; Bandhari, P.; Zangmo, S.; Dorji, T.; Tshering, K.; Matsumoto, T.; Nishizono, A.; Söderlund-Venermo, M.; et al. Novel human bufavirus genotype 3 in children with severe diarrhea, Bhutan. Emerg. Infect. Dis. 2014, 20, 1037–1039. [Google Scholar] [CrossRef]
- Väisänen, E.; Fu, Y.; Hedman, K.; Söderlund-Venermo, M. Human protoparvoviruses. Viruses 2017, 9, 354. [Google Scholar] [CrossRef]
- Okitsu, S.; Khamrin, P.; Takanashi, S.; Thongprachum, A.; Hoque, S.A.; Takeuchi, H.; Khan, M.A.; Hasan, S.M.T.; Iwata, T.; Shimizu, H.; et al. Molecular detection of enteric viruses in the stool samples of children without diarrhea in Bangladesh. Infect. Genet. Evol. 2020, 77, 104055. [Google Scholar] [CrossRef]
- Mohammad, H.A.; Madi, N.M.; Al-Nakib, W. Analysis of viral diversity in stool samples from infants and children with acute gastroenteritis in Kuwait using Metagenomics approach. Virol. J. 2020, 17, 10. [Google Scholar] [CrossRef]
- Smits, S.L.; Schapendonk, C.M.E.; van Beek, J.; Vennema, H.; Schürch, A.C.; Schipper, D.; Bodewes, R.; Haagmans, B.L.; Osterhaus, A.D.M.E.; Koopmans, M.P. New viruses in idiopathic human diarrhea cases, the Netherlands. Emerg. Infect. Dis. 2014, 20, 1218–1222. [Google Scholar] [CrossRef]
- Chieochansin, T.; Vutithanachot, V.; Theamboonlers, A.; Poovorawan, Y. Bufavirus in fecal specimens of patients with and without diarrhea in Thailand. Arch. Virol. 2015, 160, 1781–1784. [Google Scholar] [CrossRef]
- Ayouni, S.; Estienney, M.; Hammami, S.; Guediche, M.N.; Pothier, P.; Aouni, M.; Belliot, G.; De Rougemont, A. Cosavirus, Salivirus and Bufavirus in diarrheal Tunisian infants. PLoS ONE 2016, 11, e0162255. [Google Scholar] [CrossRef] [PubMed]
- Väisänen, E.; Kuisma, I.; Phan, T.G.; Delwart, E.; Lappalainen, M.; Tarkka, E.; Hedman, K.; Söderlund-Venermo, M. Bufavirus in feces of patients with gastroenteritis, Finland. Emerg. Infect. Dis. 2014, 20, 1077–1079. [Google Scholar] [CrossRef] [PubMed]
- Väisänen, E.; Paloniemi, M.; Kuisma, I.; Lithovius, V.; Kumar, A.; Franssila, R.; Ahmed, K.; Delwart, E.; Vesikari, T.; Hedman, K.; et al. Epidemiology of two human protoparvoviruses, bufavirus and tusavirus. Sci Rep 2016, 6, 39267. [Google Scholar] [CrossRef] [PubMed]
- Altay, A.; Yahiro, T.; Bozdayi, G.; Matsumoto, T.; Sahin, F.; Ozkan, S.; Nishizono, A.; Söderlund-Venermo, M.; Ahmed, K. Bufavirus genotype 3 in Turkish children with severe diarrhoea. Clin. Microbiol. Infect. 2015, 21, 965.e1–965.e4. [Google Scholar] [CrossRef]
- Huang, D.D.; Wang, W.; Lu, Q.B.; Zhao, J.; Guo, C.T.; Wang, H.Y.; Zhang, X.A.; Tong, Y.G.; Liu, W.; Cao, W.C. Identification of Bufavirus-1 and Bufavirus-3 in feces of patients with acute diarrhea, China. Sci. Rep. 2015, 5, 13272. [Google Scholar] [CrossRef] [PubMed]
- Altan, E.; Del Valle Mendoza, J.; Deng, X.; Phan, T.G.; Sadeghi, M.; Delwarta, E.L. Small circular rep-encoding single-stranded DNA genomes in peruvian diarrhea virome. Genome Announc. 2017, 5. [Google Scholar] [CrossRef] [PubMed]
- Altan, E.; Aiemjoy, K.; Phan, T.G.; Deng, X.; Aragie, S.; Tadesse, Z.; Callahan, K.E.; Keenan, J.; Delwart, E. Enteric virome of Ethiopian children participating in a clean water intervention trial. PLoS ONE 2018, 13, e0202054. [Google Scholar] [CrossRef]
- Siqueira, J.D.; Dominguez-Bello, M.G.; Contreras, M.; Lander, O.; Caballero-Arias, H.; Xutao, D.; Noya-Alarcon, O.; Delwart, E. Complex virome in feces from Amerindian children in isolated Amazonian villages. Nat. Commun. 2018, 9, 4270. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Latorre, L.; Romero, B.; Bonifaz, E.; Timoneda, N.; Rusiñol, M.; Girones, R.; Rios-Touma, B. Quito’s virome: Metagenomic analysis of viral diversity in urban streams of Ecuador’s capital city. Sci. Total Environ. 2018, 645, 1334–1343. [Google Scholar] [CrossRef]
- Phan, T.; Nagaro, K. Cutavirus: A newly discovered parvovirus on the rise. Infect. Genet. Evol. 2020, 80, 104175. [Google Scholar] [CrossRef]
- Väisänen, E.; Fu, Y.; Koskenmies, S.; Fyhrquist, N.; Wang, Y.; Keinonen, A.; Mäkisalo, H.; Väkevä, L.; Pitkänen, S.; Ranki, A. Cutavirus DNA in malignant and nonmalignant skin of cutaneous T-cell lymphoma and organ transplant patients but not of healthy adults. Clin. Infect. Dis. 2019, 68, 1904–1910. [Google Scholar] [CrossRef]
- Kreuter, A.; Nasserani, N.; Tigges, C.; Oellig, F.; Silling, S.; Akgul, B.; Wieland, U. Cutavirus infection in primary cutaneous B- and T-Cell lymphoma. JAMA Dermatol. 2018, 154, 965–967. [Google Scholar] [CrossRef]
- Mollerup, S.; Fridholm, H.; Vinner, L.; Kjartansdottir, K.R.; Friis-Nielsen, J.; Asplund, M.; Herrera, J.A.; Steiniche, T.; Mourier, T.; Brunak, S.; et al. Cutavirus in cutaneous malignant melanoma. Emerg. Infect. Dis. 2017, 23, 363–365. [Google Scholar] [CrossRef]
- Wieland, U.; Silling, S.; Hufbauer, M.; Mauch, C.; Zigrino, P.; Oellig, F.; Kreuter, A.; Akgül, B. No evidence for role of cutavirus in malignant melanoma. Emerg. Infect. Dis. 2019, 25, 1600–1610. [Google Scholar] [CrossRef]
- Väisänen, E.; Mohanraj, U.; Kinnunen, P.M.; Jokelainen, P.; Al-Hello, H.; Barakat, A.M.; Sadeghi, M.; Jalilian, F.A.; Majlesi, A.; Masika, M.; et al. Global distribution of human protoparvoviruses. Emerg. Infect. Dis. 2018, 24, 1292–1299. [Google Scholar] [CrossRef] [PubMed]
- Paloniemi, M.; Lappalainen, S.; Salminen, M.; Kätkä, M.; Kantola, K.; Hedman, L.; Hedman, K.; Söderlund-Venermo, M.; Vesikari, T. Human bocaviruses are commonly found in stools of hospitalized children without causal association to acute gastroenteritis. Eur. J. Pediatr. 2014, 173, 1051–1057. [Google Scholar] [CrossRef] [PubMed]
- Nora-Krukle, Z.; Vilmane, A.; Xu, M.; Rasa, S.; Ziemele, I.; Silina, E.; Söderlund-Venermo, M.; Gardovska, D.; Murovska, M. Human bocavirus infection markers in peripheral blood and stool samples of children with acute gastroenteritis. Viruses 2018, 10, 639. [Google Scholar] [CrossRef] [PubMed]
- Koskenvuo, M.; Möttönen, M.; Rahiala, J.; Saarinen-Pihkala, U.M.; Riikonen, P.; Waris, M.; Ziegler, T.; Uhari, M.; Ruuskanen, O.; Salmi, T.T. Mixed bacterial-viral infections in septic children with leukemia. Pediatr. Infect. Dis. J. 2007, 26, 1133–1136. [Google Scholar] [CrossRef]
- Zanella, M.C.; Cordey, S.; Laubscher, F.; Docquier, M.; Vieille, G.; Van Delden, C.; Braunersreuther, V.; TA, M.K.; Lobrinus, J.A.; Masouridi-Levrat, S.; et al. Unmasking viral sequences by metagenomic next-generation sequencing in adult human blood samples during steroid-refractory/dependent graft-versus-host disease. Microbiome 2021, 9, 28. [Google Scholar] [CrossRef] [PubMed]
- Meriluoto, M.; Hedman, L.; Tanner, L.; Simell, V.; Mäkinen, M.; Simell, S.; Mykkänen, J.; Korpelainen, J.; Ruuskanen, O.; Ilonen, J.; et al. Association of human bocavirus 1 infection with respiratory disease in childhood follow-up study Finland. Emerg. Infect. Dis. 2012, 18, 264–271. [Google Scholar] [CrossRef]
- Li, X.; Kantola, K.; Hedman, L.; Arku, B.; Hedman, K.; Söderlund-Venermo, M. Original antigenic sin with human bocaviruses 1–4. J. Gen. Virol. 2015, 96, 3099–3108. [Google Scholar] [CrossRef]
- Inaba, H.; Pei, D.; Wolf, J.; Howard, S.C.; Hayden, R.T.; Go, M.; Varechtchouk, O.; Hahn, T.; Buaboonnam, J.; Metzger, M.L.; et al. Infection-related complications during treatment for childhood acute lymphoblastic leukemia. Ann. Oncol. 2017, 28, 386–392. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).