Identification of Host Factors Associated with the Development of Equine Herpesvirus Myeloencephalopathy by Transcriptomic Analysis of Peripheral Blood Mononuclear Cells from Horses
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Virus
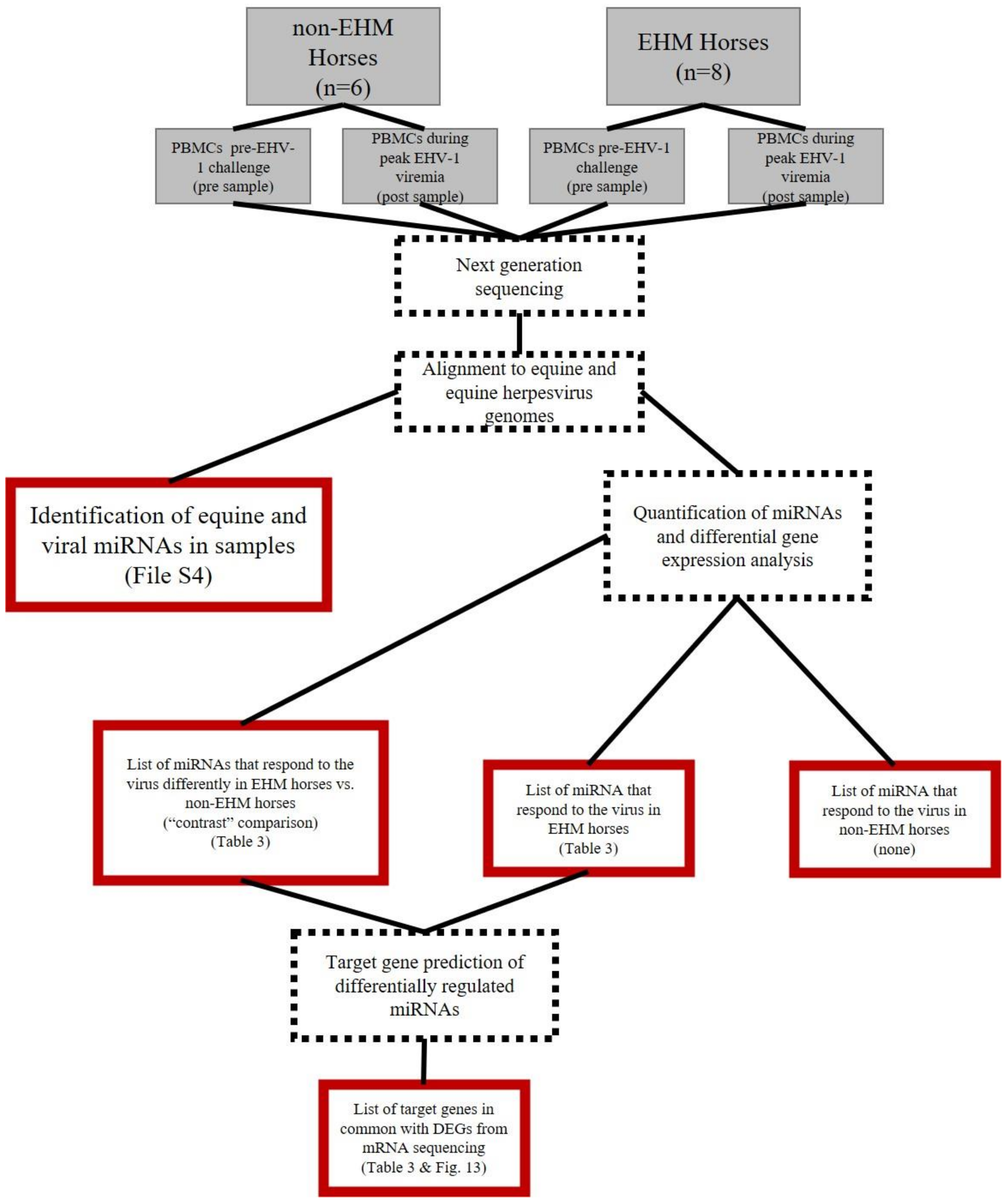
2.3. Experimental Design and Sample Collection
2.4. RNA Isolation, Library Preparation, and Sequencing
2.5. Genome Guided mRNA Alignment
2.6. Host and Viral miRNA Identification and Quantification
2.7. Differential Gene Expression Analysis
2.8. Gene Ontology (GO) Enrichment Analysis
2.9. In Silico Cell Sorting
2.10. Whole Blood Cytokine RT-qPCR
3. Results
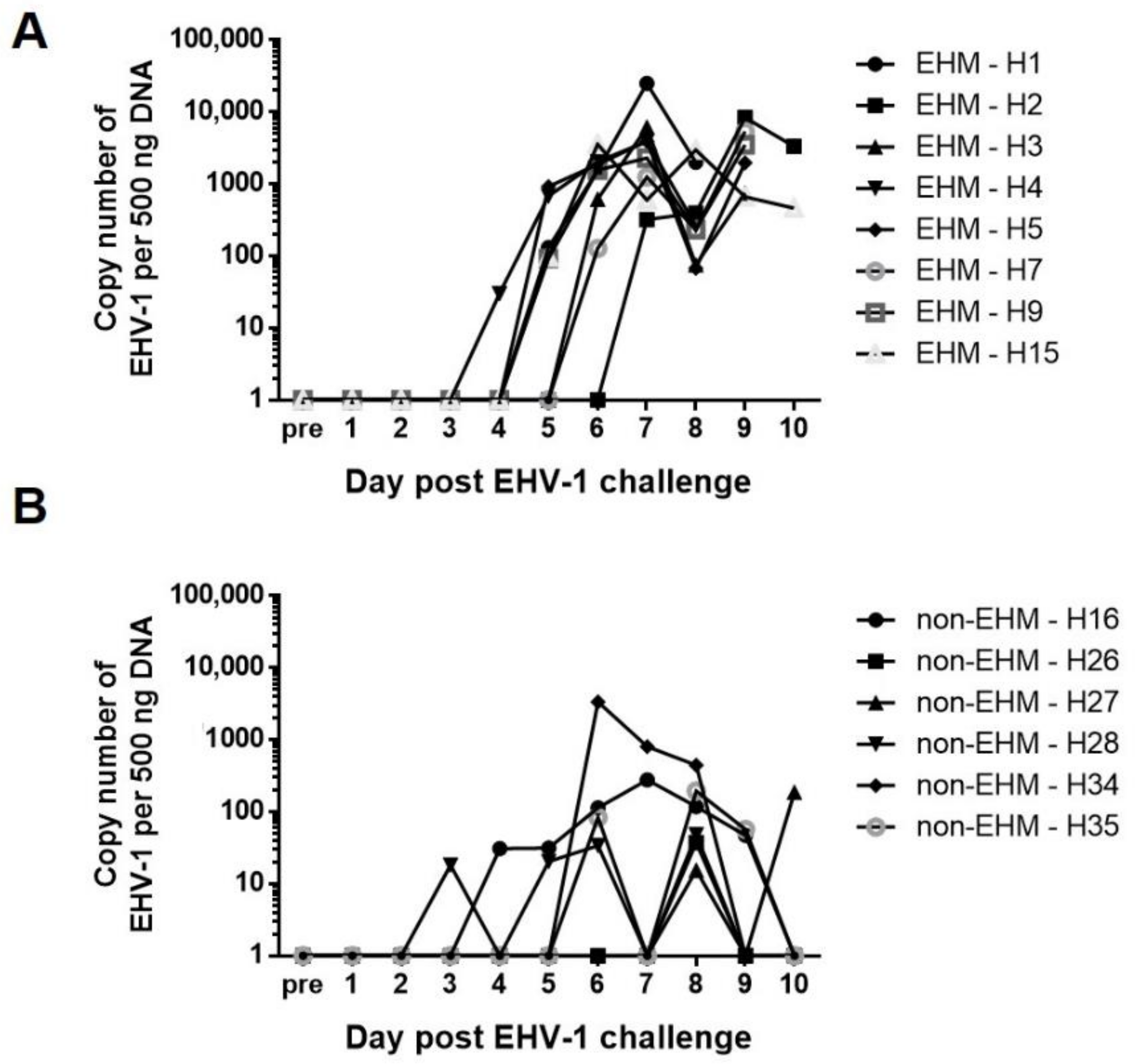
3.1. Clinical Disease and Viremia Differed between EHM and Non-EHM Horses in Response to EHV-1 Challenge Infection
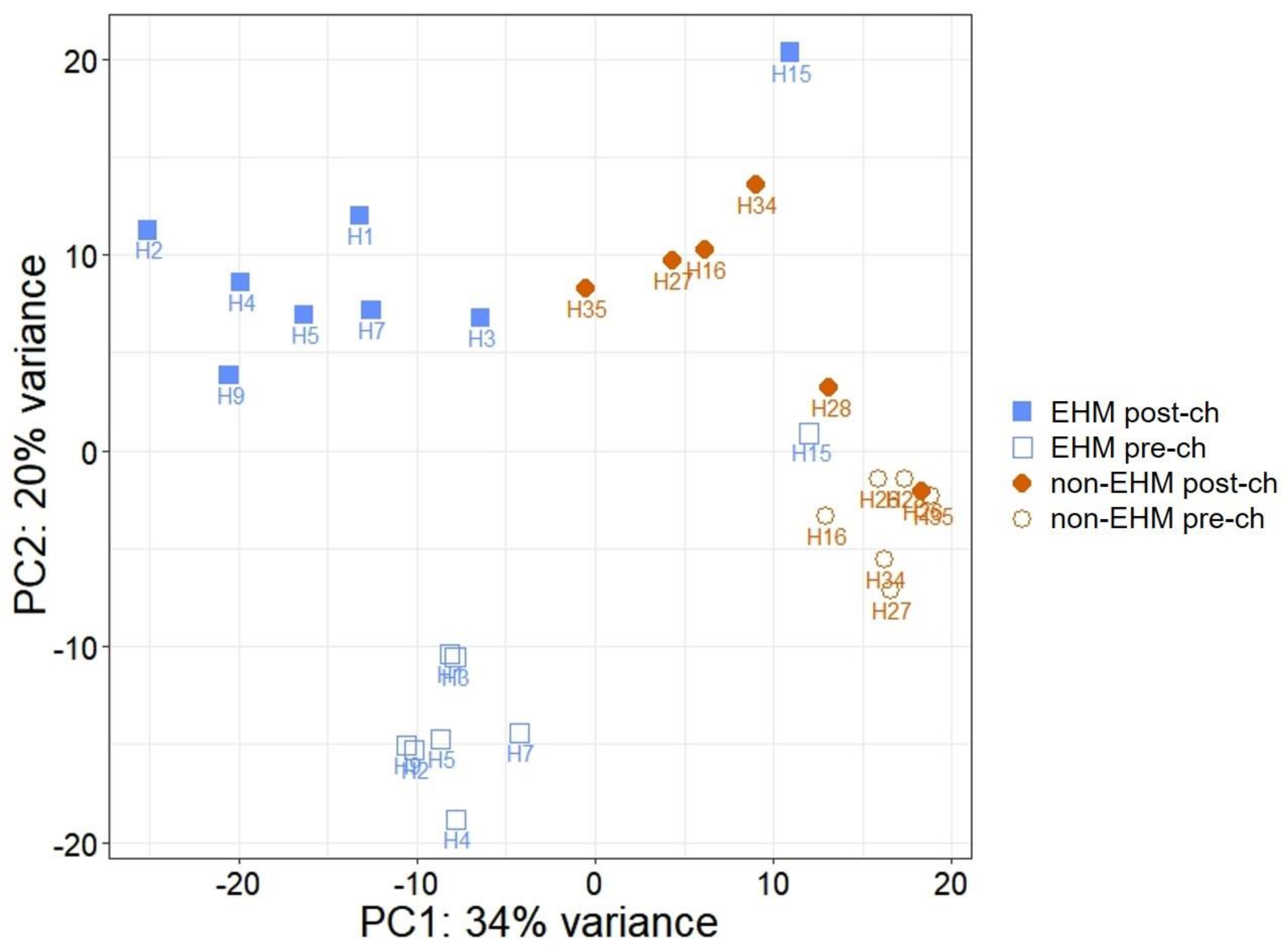
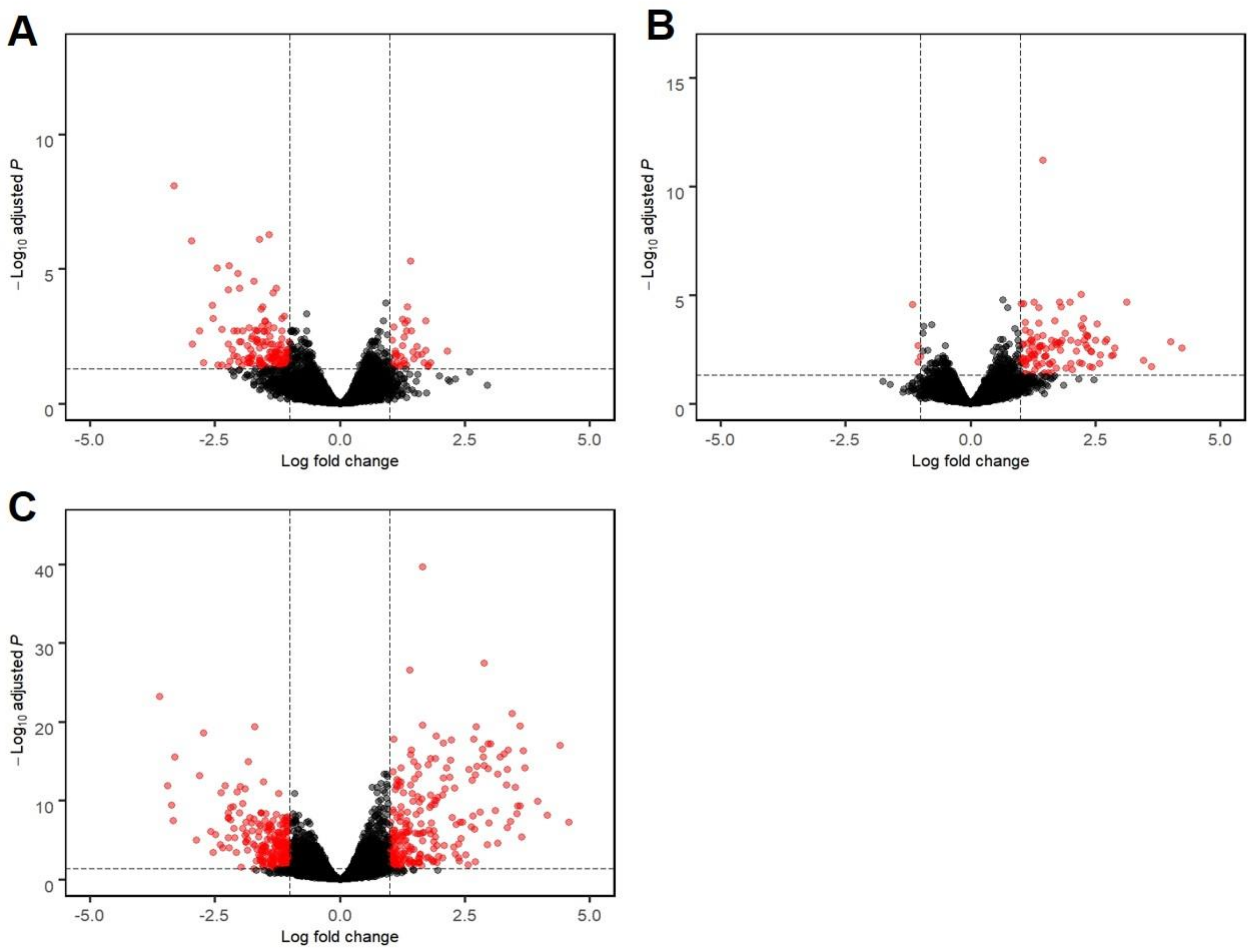
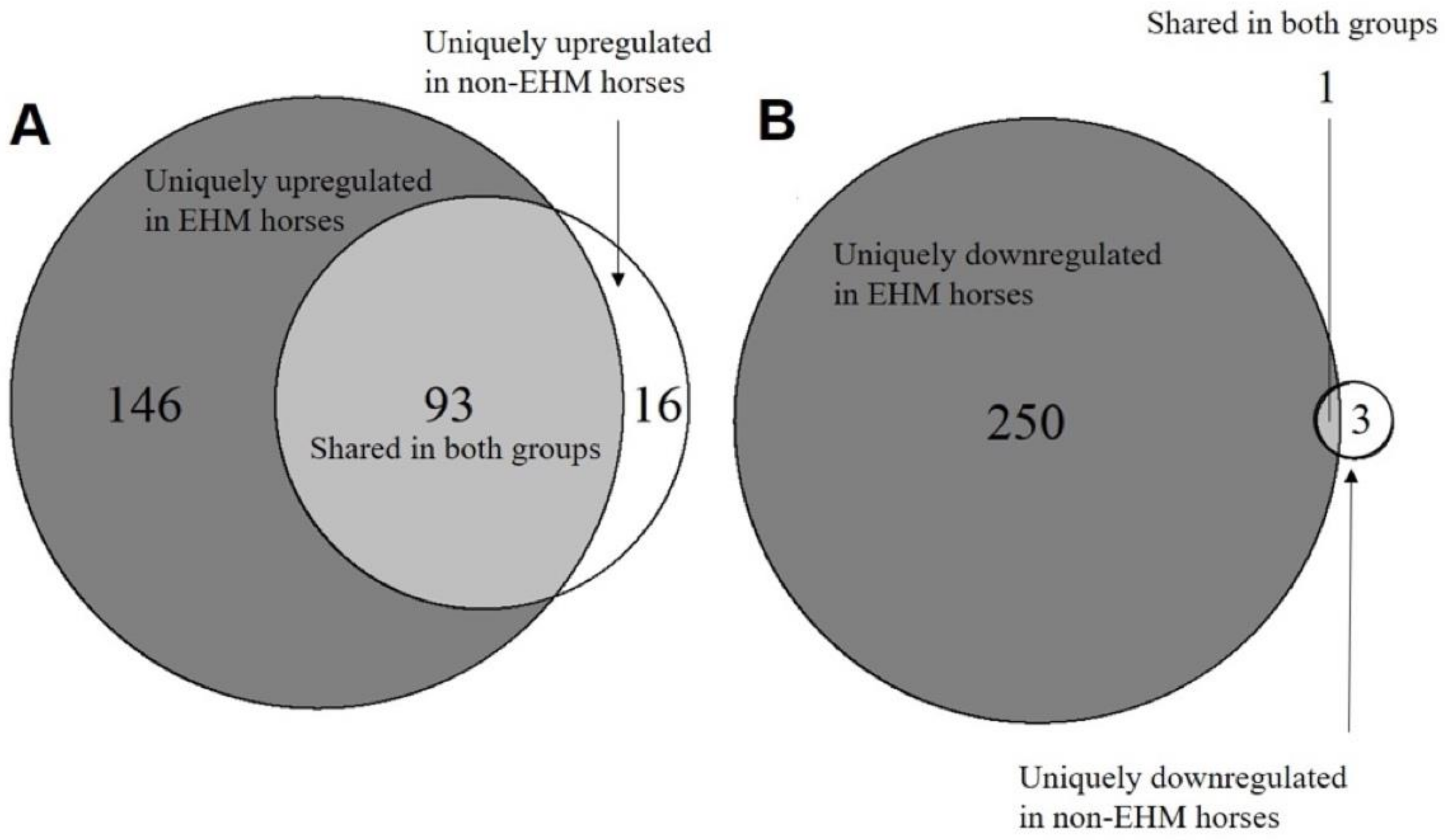
3.2. mRNA Sequencing Revealed Differential Host Gene Expression between EHM and Non-EHM Horses
3.3. Gene Ontology Overrepresentation Analysis Identified Enriched Biological Processes in EHM Horses
3.4. In Silico Cell Sorting Identified Differences in Cell Population Fractions between EHM and Non-EHM Horses in Response to EHV-1 Challenge
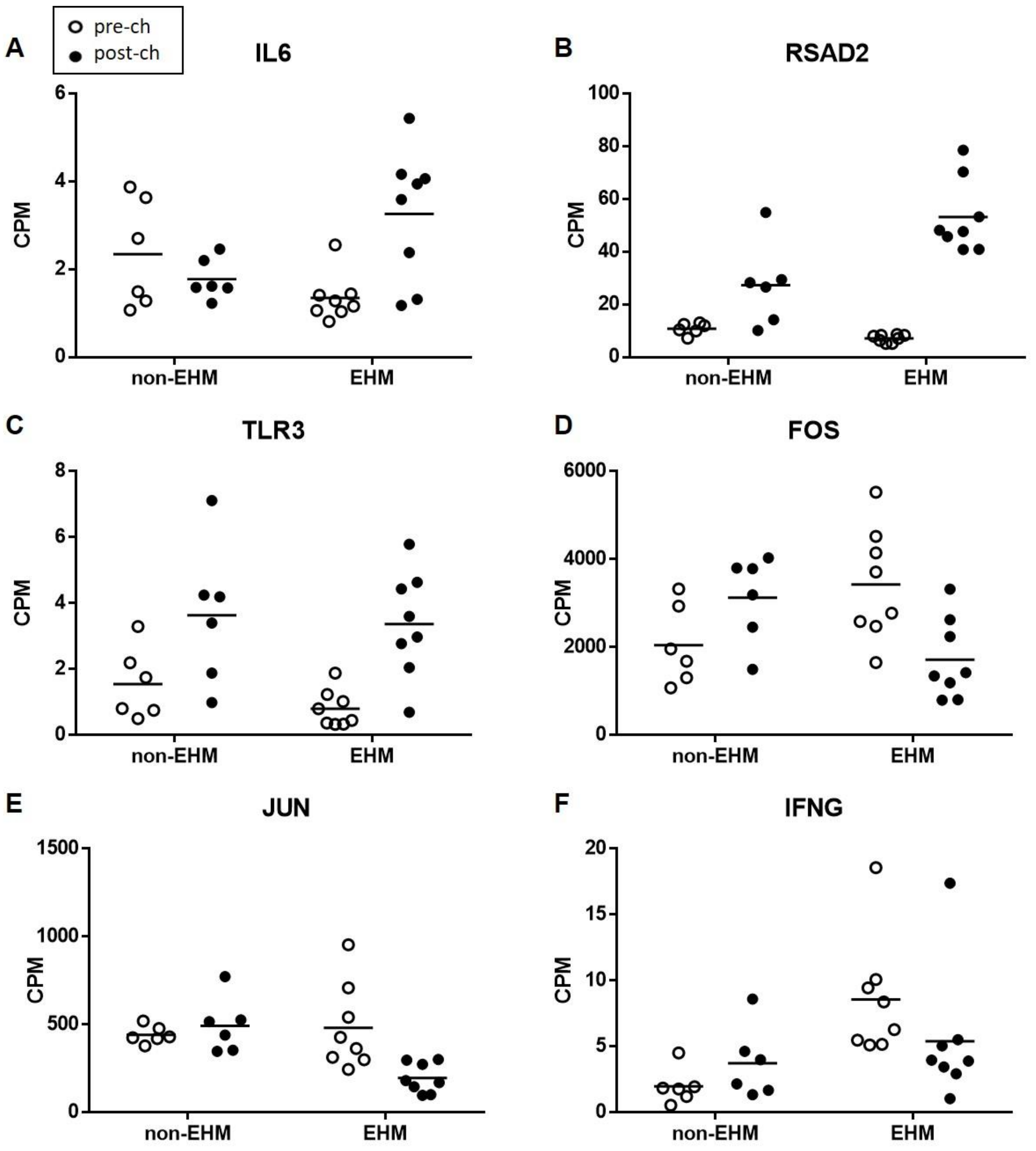
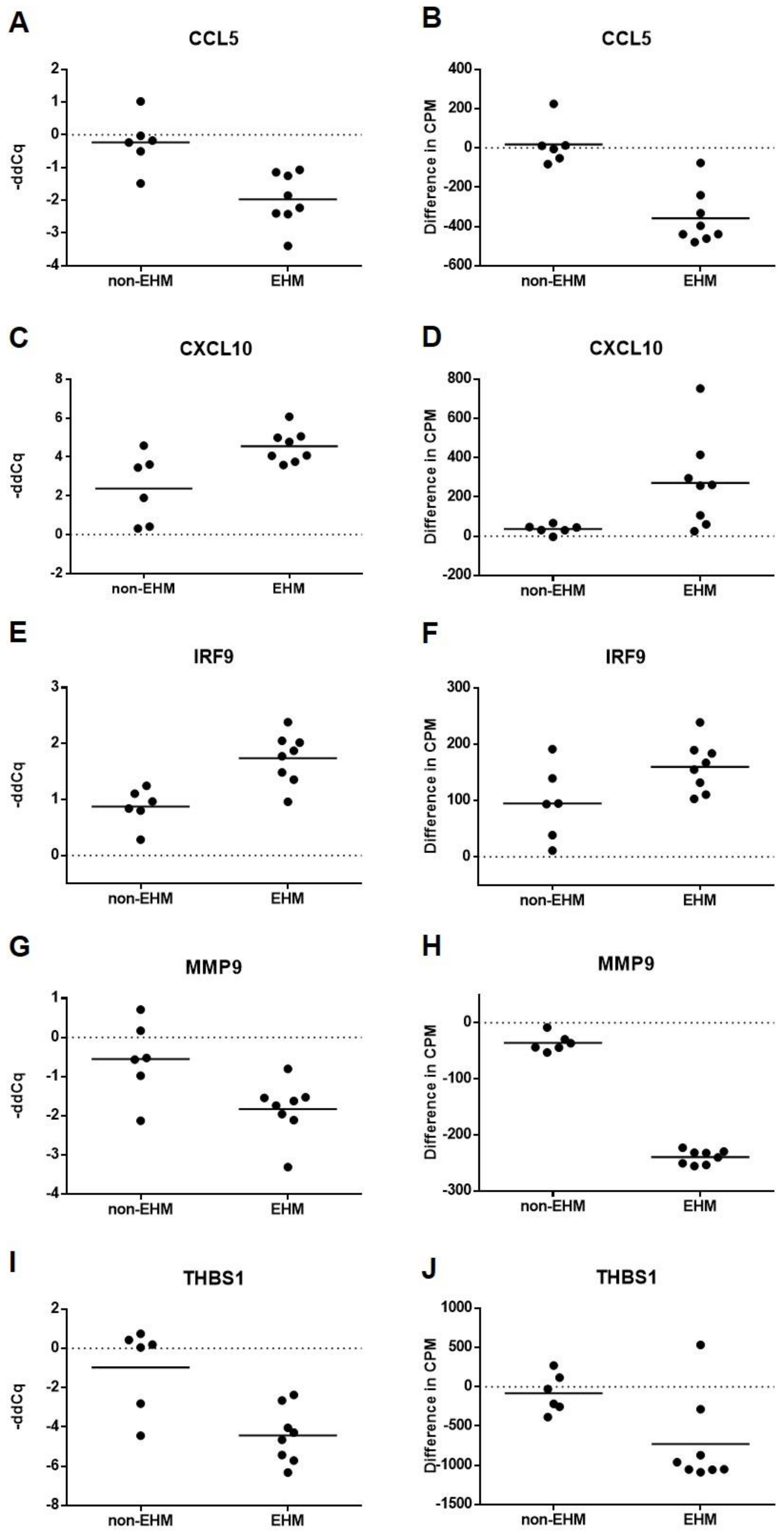
3.5. Whole Blood Cytokine RT-qPCR Confirmed Differential Expression of Select Genes between EHM and Non-EHM Horses
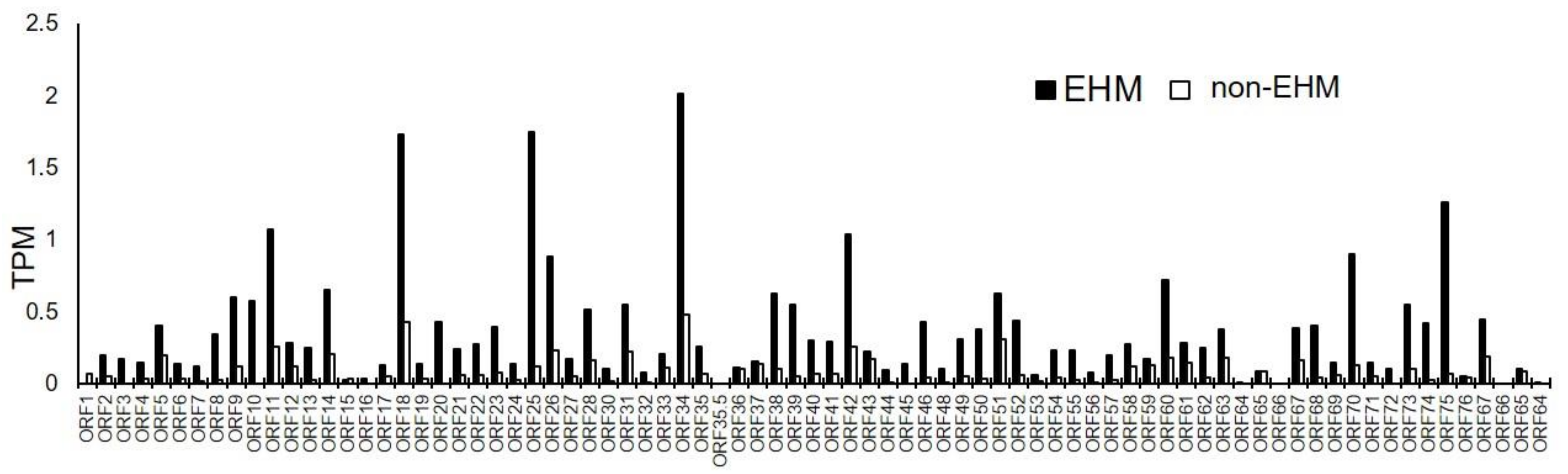
3.6. Viral mRNA Sequencing Identified EHV-1, EHV-2, and EHV-5 Transcripts in PBMCs Prior to and after EHV-1 Challenge Infection
3.7. Host, EHV-2, and EHV-5 miRNAs Were Identified in PBMCs of Horses
3.8. Host miRNAs Were Differentially Expressed between EHM and Non-EHM Horses in Response to EHV-1 Challenge Infection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lunn, D.P.; Davis-Poynter, N.; Flaminio, M.J.B.F.; Horohov, D.W.; Osterrieder, K.; Pusterla, N.; Townsend, H.G.G. Equine herpesvirus-1 consensus statement. J. Vet. Intern. Med. 2009, 23, 450–461. [Google Scholar] [CrossRef] [PubMed]
- USDA; APHIS. Equine Herpesvirus (EHV-1)—FINAL Situation Report. Available online: https://www.aphis.usda.gov/vs/nahss/equine/ehv/ehv_2011_final_sitrep_062311.pdf (accessed on 24 October 2018).
- USDA; APHIS; VS; CEAH. Equine Herpesvirus Myeloencephalopathy: A Potentially Emerging Disease. Available online: https://www.aphis.usda.gov/animal_health/emergingissues/downloads/ehv1final.pdf (accessed on 24 October 2018).
- USAHA. Equine Herpesvirus Myeloencephalopathy Incident Guidelines for State Animal Health Officials. Available online: http://www.usaha.org/upload/Publication/TopSpecific/EHM_Guidance_Document_Revised_Fe.pdf (accessed on 24 October 2018).
- Pusterla, N.; Mapes, S.; Wilson, W.D. Prevalence of equine herpesvirus type 1 in trigeminal ganglia and submandibular lymph nodes of equids examined postmortem. Vet. Rec. 2010, 167, 376–378. [Google Scholar] [CrossRef]
- Allen, G.P. Antemortem detection of latent infection with neuropathogenic strains of equine herpesvirus-1 in horses. Am. J. Vet. Res. 2006, 67, 1401–1405. [Google Scholar] [CrossRef] [PubMed]
- Allen, G.P.; Bolin, D.C.; Bryant, U.; Carter, C.N.; Giles, R.C.; Harrison, L.R.; Hong, C.B.; Jackson, C.B.; Poonacha, K.; Wharton, R.; et al. Prevalence of latent, neuropathogenic equine herpesvirus-1 in the Thoroughbred broodmare population of central Kentucky. Equine Vet. J. 2008, 40, 105–110. [Google Scholar] [CrossRef]
- Slater, J.D.; Borchers, K.; Thackray, A.M.; Field, H.J. The trigeminal ganglion is a location for equine herpesvirus 1 latency and reactivation in the horse. J. Gen. Virol. 1994, 75, 2007–2016. [Google Scholar] [CrossRef]
- Kydd, J.H.; Townsend, H.G.G.; Hannant, D. The equine immune response to equine herpesvirus-1: The virus and its vaccines. Vet. Immunol. Immunopathol. 2006, 111, 15–30. [Google Scholar] [CrossRef]
- Hussey, S.B.; Clark, R.; Lunn, K.F.; Breathnach, C.; Soboll, G.; Whalley, J.M.; Lunn, D.P. Detection and quantification of equine herpesvirus-1 viremia and nasal shedding by real-time polymerase chain reaction. J. Vet. Diagn. Investig. 2006, 18, 335–342. [Google Scholar] [CrossRef]
- Patel, J.R.; Edington, N.; Mumford, J.A. Variation in cellular tropism between isolates of Equine herpesvirus-1 in foals. Arch. Virol. 1982, 74, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Osterrieder, N.; Van de Walle, G.R. Pathogenic potential of equine alphaherpesviruses: The importance of the mononuclear cell compartment in disease outcome. Vet. Microbiol. 2010, 143, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Allen, G.P. Risk factors for development of neurologic disease after experimental exposure to equine herpesvirus-1 in horses. Am. J. Vet. Res. 2008, 69, 1595–1600. [Google Scholar] [CrossRef] [PubMed]
- Holz, C.L.; Nelli, R.K.; Eilidh Wilson, M.; Zarski, L.M.; Azab, W.; Baumgardner, R.; Osterrieder, N.; Pease, A.; Zhang, L.; Hession, S.; et al. Viral genes and cellular markers associated with neurological complications during herpesvirus infections. J. Gen. Virol. 2017, 98. [Google Scholar] [CrossRef]
- Goodman, L.B.; Loregian, A.; Perkins, G.A.; Nugent, J.; Buckles, E.L.; Mercorelli, B.; Kydd, J.H.; Palù, G.; Smith, K.C.; Osterrieder, N.; et al. A point mutation in a herpesvirus polymerase determines neuropathogenicity. PLoS Pathog. 2007, 3, e160. [Google Scholar] [CrossRef]
- Van de Walle, G.R.; Goupil, R.; Wishon, C.; Damiani, A.; Perkins, G.A.; Osterrieder, N. A Single-Nucleotide Polymorphism in a Herpesvirus DNA Polymerase is Sufficient to Cause Lethal Neurological Disease. J. Infect. Dis. 2009, 200, 20–25. [Google Scholar] [CrossRef]
- Nugent, J.; Birch-Machin, I.; Smith, K.C.; Mumford, J.A.; Swann, Z.; Newton, J.R.; Bowden, R.J.; Allen, G.P.; Davis-Poynter, N. Analysis of equid herpesvirus 1 strain variation reveals a point mutation of the DNA polymerase strongly associated with neuropathogenic versus nonneuropathogenic disease outbreaks. J. Virol. 2006, 80, 4047–4060. [Google Scholar] [CrossRef]
- Allen, G.P.; Breathnach, C.C. Quantification by real-time PCR of the magnitude and duration of leucocyte-associated viraemia in horses infected with neuropathogenic vs. non-neuropathogenic strains of EHV-1. Equine Vet. J. 2006, 38, 252–257. [Google Scholar] [CrossRef]
- Pusterla, N.; Hussey, G.S. Equine Herpesvirus 1 Myeloencephalopathy. Vet. Clin. N. Am. Equine Pract. 2014, 30, 489–506. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, B.T.; Davis, T.P. The blood-brain barrier/neurovascular unit in health and disease. Pharmacol. Rev. 2005, 57, 173–185. [Google Scholar] [CrossRef] [PubMed]
- Bazzoni, G.; Dejana, E. Endothelial Cell-to-Cell Junctions: Molecular Organization and Role in Vascular Homeostasis. Physiol. Rev. 2004, 84, 869–901. [Google Scholar] [CrossRef] [PubMed]
- Abbruscato, T.J.; Davis, T.P. Combination of Hypoxia/Aglycemia Compromises In Vitro Blood-Brain Barrier Integrity. J. Pharmacol. Exp. Ther. 1999, 289, 668–675. [Google Scholar]
- Fischer, S.; Clauss, M.; Wiesnet, M.; Renz, D.; Schaper, W.; Karliczek, G.F. Hypoxia induces permeability in brain microvessel endothelial cells via VEGF and NO. Am. J. Physiol. Physiol. 1999, 276, C812–C820. [Google Scholar] [CrossRef]
- Mark, K.S.; Davis, T.P. Cerebral microvascular changes in permeability and tight junctions induced by hypoxia-reoxygenation. Am. J. Physiol. Circ. Physiol. 2002, 282, H1485–H1494. [Google Scholar] [CrossRef] [PubMed]
- Yeo, W.M.; Osterrieder, N.; Stokol, T. Equine herpesvirus type 1 infection induces procoagulant activity in equine monocytes. Vet. Res. 2013, 44, 16. [Google Scholar] [CrossRef] [PubMed]
- Stokol, T.; Yeo, W.M.; Burnett, D.; DeAngelis, N.; Huang, T.; Osterrieder, N.; Catalfamo, J. Equid Herpesvirus Type 1 Activates Platelets. PLoS ONE 2015, 10, e0122640. [Google Scholar] [CrossRef] [PubMed]
- Goehring, L.S.; Hussey, G.S.; Diez, M.G.; Benedict, K.; Maxwell, L.K.; Morley, P.S.; van Oldruitenborgh-Oosterbaan, M.M.S.; Lunn, D.P. Plasma D-Dimer Concentrations during Experimental EHV-1 Infection of Horses. J. Vet. Intern. Med. 2013, 27, 1535–1542. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.E.; Holz, C.L.; Kopec, A.K.; Dau, J.J.; Luyendyk, J.P.; Soboll Hussey, G. Coagulation parameters following equine herpesvirus type 1 infection in horses. Equine Vet. J. 2019, 51, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Henninger, R.W.; Reed, S.M.; Saville, W.J.; Allen, G.P.; Hass, G.F.; Kohn, C.W.; Sofaly, C. Outbreak of Neurologic Disease Caused by Equine Herpesvirus-1 at a University Equestrian Center. J. Vet. Intern. Med. 2008, 21, 157–165. [Google Scholar] [CrossRef]
- Slater, J. Equine Herpesviruses, 2nd ed.; Sellon, D.C., Long, M.T., Saunders, W.B., Eds.; Elsevier: St. Louis, MO, USA, 2014; Chapter 14; pp. 151–168. ISBN 978-1-4557-0891-8. [Google Scholar]
- Maxwell, L.K.; Bentz, B.G.; Gilliam, L.L.; Ritchey, J.W.; Pusterla, N.; Eberle, R.; Holbrook, T.C.; McFarlane, D.; Rezabek, G.B.; Meinkoth, J.; et al. Efficacy of the early administration of valacyclovir hydrochloride for the treatment of neuropathogenic equine herpesvirus type-1 infection in horses. Am. J. Vet. Res. 2017, 78, 1126–1139. [Google Scholar] [CrossRef]
- Aiello, A.; Farzaneh, F.; Candore, G.; Caruso, C.; Davinelli, S.; Gambino, C.M.; Ligotti, M.E.; Zareian, N.; Accardi, G. Immunosenescence and its Hallmarks: How to Oppose Aging Strategically? A Review of Potential Options for Therapeutic Intervention. Front. Immunol. 2019, 10, 2247. [Google Scholar] [CrossRef]
- Hansen, S.; Baptiste, K.E.; Fjeldborg, J.; Horohov, D.W. A review of the equine age-related changes in the immune system: Comparisons between human and equine aging, with focus on lung-specific immune-aging. Ageing Res. Rev. 2015, 20, 11–23. [Google Scholar] [CrossRef]
- Hussey, G.S.; Hussey, S.B.; Wagner, B.; Horohov, D.W.; Van de Walle, G.R.; Osterrieder, N.; Goehring, L.S.; Rao, S.; Lunn, D.P. Evaluation of immune responses following infection of ponies with an EHV-1 ORF1/2 deletion mutant. Vet. Res. 2011, 42. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 10 June 2020).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Subgroup. 1000 Genome Project Data Processing S: The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2014, 31, 166–169. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. Next Gener. Seq. Data Anal. 2011, 17. [Google Scholar] [CrossRef]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2011, 40, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S.; Saini, H.K.; van Dongen, S.; Enright, A.J. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2007, 36, D154–D158. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef]
- Chen, Y.; McCarthy, D.; Ritchie, M.; Robinson, M.; Smyth, G. edgeR: Differential Analysis of Sequence Read Count Data, User’s Guide. Available online: https://www.bioconductor.org/packages/release/bioc/vignettes/edgeR/inst/doc/edgeRUsersGuide.pdf (accessed on 10 June 2020).
- Thomas, P.D.; Campbell, M.J.; Kejariwal, A.; Mi, H.; Karlak, B.; Daverman, R.; Diemer, K.; Muruganujan, A.; Narechania, A. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003, 13, 2129–2141. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2018, 47, D506–D515. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Carlson, M. org.Hs.eg.db: Genome Wide Annotation for Human; R Foundation: London, UK, 2019. [Google Scholar]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Steen, C.B.; Liu, C.L.; Gentles, A.J.; Chaudhuri, A.A.; Scherer, F.; Khodadoust, M.S.; Esfahani, M.S.; Luca, B.A.; Steiner, D.; et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019, 37, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Giessler, K.S.; Goehring, L.S.; Jacobs, S.; McCauley, A.; Esser, M.M.; Lee, Y.; Zarski, L.M.; Soboll Hussey, G. Use of the old horse model to identify host factors contributing to EHM pathogenesis. In Proceedings of the Conference of Research Workers in Animal Disease Conference Proceedings, Chicago, IL, USA, 4–8 December 2020; p. 367. [Google Scholar]
- Goehring, L.S.; van Maanen, C.; Berendsen, M.; Cullinane, A.; de Groot, R.J.; Rottier, P.J.M.; Wesselingh, J.J.C.M.; Sloet van Oldruitenborgh-Oosterbaan, M.M. Experimental infection with neuropathogenic equid herpesvirus type 1 (EHV-1) in adult horses. Vet. J. 2010, 186, 180–187. [Google Scholar] [CrossRef]
- Shaafi, S.; Sharifipour, E.; Rahmanifar, R.; Hejazi, S.; Andalib, S.; Nikanfar, M.; Baradarn, B.; Mehdizadeh, R. Interleukin-6, a reliable prognostic factor for ischemic stroke. Iran. J. Neurol. 2014, 13, 70–76. [Google Scholar]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. Immunotherapeutic implications of IL-6 blockade for cytokine storm. Immunotherapy 2016, 8, 959–970. [Google Scholar] [CrossRef]
- Angel, P.; Karin, M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta Rev. Cancer 1991, 1072, 129–157. [Google Scholar] [CrossRef]
- Owen, J.A.; Punt, J.; Stranford, S.A.; Jones, P.P.; Kuby, J. Receptors and signaling: B and T-cell receptors. In Kuby Immunology; W.H. Freeman and Company: New York, NY, USA, 2013; Chapter 3; pp. 65–104. [Google Scholar]
- Shakya, A.K.; O’Callaghan, D.J.; Kim, S.K. Comparative Genomic Sequencing and Pathogenic Properties of Equine Herpesvirus 1 KyA and RacL11. Front. Vet. Sci. 2017, 4, 211. [Google Scholar] [CrossRef]
- Matsumura, T.; Kondo, T.; Sugita, S.; Damiani, A.M.; O’Callaghan, D.J.; Imagawa, H. An Equine Herpesvirus Type 1 Recombinant with a Deletion in the gE and gI Genes Is Avirulent in Young Horses. Virology 1998, 242, 68–79. [Google Scholar] [CrossRef]
- Badr, Y.; Okada, A.; Abo-Sakaya, R.; Beshir, E.; Ohya, K.; Fukushi, H. Equine herpesvirus type 1 ORF51 encoding UL11 as an essential gene for replication in cultured cells. Arch. Virol. 2018, 163, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Schimmer, C.; Neubauer, A. The equine herpesvirus 1 UL11 gene product localizes to the trans-golgi network and is involved in cell-to-cell spread. Virology 2003, 308, 23–36. [Google Scholar] [CrossRef][Green Version]
- Meng, P.; Tang, X.; Jiang, X.; Tang, Q.; Bai, L.; Xia, Y.; Zou, Z.; Qin, X.; Cao, X.; Chen, C.; et al. Maternal exposure to traffic pollutant causes impairment of spermatogenesis and alterations of genome-wide mRNA and microRNA expression in F2 male mice. Environ. Toxicol. Pharmacol. 2018, 64, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Gui, Y.; Xu, Z.; Jin, T.; Zhang, L.; Chen, L.; Hong, B.; Xie, F.; Lv, W.; Hu, X. Using Extracellular Circulating microRNAs to Classify the Etiological Subtypes of Ischemic Stroke. Transl. Stroke Res. 2019, 10, 352–361. [Google Scholar] [CrossRef]
- Pak, J.H.; Kim, I.K.; Kim, S.M.; Maeng, S.; Song, K.J.; Na, B.-K.; Kim, T.-S. Induction of cancer-related microRNA expression profiling using excretory-secretory products of Clonorchis sinensis. Parasitol. Res. 2014, 113, 4447–4455. [Google Scholar] [CrossRef]
- Soliman, B.; Salem, A.; Ghazy, M.; Abu-Shahba, N.; El Hefnawi, M. Bioinformatics functional analysis of let-7a, miR-34a, and miR-199a/b reveals novel insights into immune system pathways and cancer hallmarks for hepatocellular carcinoma. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2018, 40. [Google Scholar] [CrossRef]
- Hermeking, H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010, 17, 193–199. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, J.-W.; Castella, M.; Huntsman, D.G.; Taniguchi, T. p53 is Positively Regulated by miR-542-3p. Cancer Res. 2014, 74, 3218–3227. [Google Scholar] [CrossRef]
- Bray, I.; Tivnan, A.; Bryan, K.; Foley, N.H.; Watters, K.M.; Tracey, L.; Davidoff, A.M.; Stallings, R.L. MicroRNA-542-5p as a novel tumor suppressor in neuroblastoma. Cancer Lett. 2011, 303, 56–64. [Google Scholar] [CrossRef]
- Zhi, Y.; Xie, X.; Wang, R.; Wang, B.; Gu, W.; Ling, Y.; Dong, W.; Zhi, F.; Liu, Y. Serum level of miR-10-5p as a prognostic biomarker for acute myeloid leukemia. Int. J. Hematol. 2015, 102, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Ulivi, P.; Foschi, G.; Mengozzi, M.; Scarpi, E.; Silvestrini, R.; Amadori, D.; Zoli, W. Peripheral blood miR-328 expression as a potential biomarker for the early diagnosis of NSCLC. Int. J. Mol. Sci. 2013, 14, 10332–10342. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.T.; Xu, Q.; Tong, J.L.; Zhu, M.M.; Nie, F.; Chen, X.; Xiao, S.D.; Ran, Z.H. MicroRNA expression profiling identifies miR-328 regulates cancer stem cell-like SP cells in colorectal cancer. Br. J. Cancer 2012, 106, 1320–1330. [Google Scholar] [CrossRef] [PubMed]
- Sha, H.-H.; Wang, D.-D.; Chen, D.; Liu, S.-W.; Wang, Z.; Yan, D.-L.; Dong, S.-C.; Feng, J.-F. MiR-138: A promising therapeutic target for cancer. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2017, 39. [Google Scholar] [CrossRef]
- Twenter, H.; Klohonatz, K.; Davis, K.; Bass, L.; Coleman, S.J.; Bouma, G.J.; Bruemmer, J.E. Transfer of MicroRNAs From Epididymal Epithelium to Equine Spermatozoa. J. Equine Vet. Sci. 2020, 87, 102841. [Google Scholar] [CrossRef]
- Bogedale, K.; Jagannathan, V.; Gerber, V.; Unger, L. Differentially expressed microRNAs, including a large microRNA cluster on chromosome 24, are associated with equine sarcoid and squamous cell carcinoma. Vet. Comp. Oncol. 2019, 17, 155–164. [Google Scholar] [CrossRef]
- Roos, J.; Enlund, E.; Funcke, J.-B.; Tews, D.; Holzmann, K.; Debatin, K.-M.; Wabitsch, M.; Fischer-Posovszky, P. miR-146a-mediated suppression of the inflammatory response in human adipocytes. Sci. Rep. 2016, 6, 38339. [Google Scholar] [CrossRef] [PubMed]
- Pacholewska, A.; Mach, N.; Mata, X.; Vaiman, A.; Schibler, L.; Barrey, E.; Gerber, V. Novel equine tissue miRNAs and breed-related miRNA expressed in serum. BMC Genom. 2016, 17, 831. [Google Scholar] [CrossRef]
- Fard, D.; Tamagnone, L. Semaphorins in health and disease. Cytokine Growth Factor Rev. 2020. [Google Scholar] [CrossRef]
- El-Mayet, F.S.; Sawant, L.; Thunuguntla, P.; Zhao, J.; Jones, C. Two Pioneer Transcription Factors, Krüppel-Like Transcription Factor 4 and Glucocorticoid Receptor, Cooperatively Transactivate the Bovine Herpesvirus 1 ICP0 Early Promoter and Stimulate Productive Infection. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Stanisic, D.I.; Cutts, J.; Eriksson, E.; Fowkes, F.J.I.; Rosanas-Urgell, A.; Siba, P.; Laman, M.; Davis, T.M.E.; Manning, L.; Mueller, I.; et al. γδ T cells and CD14+ monocytes are predominant cellular sources of cytokines and chemokines associated with severe malaria. J. Infect. Dis. 2014, 210, 295–305. [Google Scholar] [CrossRef]
- Smith, C.J.; Emsley, H.C.A.; Gavin, C.M.; Georgiou, R.F.; Vail, A.; Barberan, E.M.; del Zoppo, G.J.; Hallenbeck, J.M.; Rothwell, N.J.; Hopkins, S.J.; et al. Peak plasma interleukin-6 and other peripheral markers of inflammation in the first week of ischaemic stroke correlate with brain infarct volume, stroke severity and long-term outcome. BMC Neurol. 2004, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Nakase, T.; Yamazaki, T.; Ogura, N.; Suzuki, A.; Nagata, K. The impact of inflammation on the pathogenesis and prognosis of ischemic stroke. J. Neurol. Sci. 2008, 271, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Fassbender, K.; Rossol, S.; Kammer, T.; Daffertshofer, M.; Wirth, S.; Dollman, M.; Hennerici, M. Proinflammatory cytokines in serum of patients with acute cerebral ischemia: Kinetics of secretion and relation to the extent of brain damage and outcome of disease. J. Neurol. Sci. 1994, 122, 135–139. [Google Scholar] [CrossRef]
- Vila, N.; Castillo, J.; Dávalos, A.; Chamorro, Á. Proinflammatory cytokines and early neurological worsening in ischemic stroke. Stroke 2000, 31, 2325–2329. [Google Scholar] [CrossRef]
- Watson, C.; Whittaker, S.; Smith, N.; Vora, A.J.; Dumonde, D.C.; Brown, K.A. IL-6 acts on endothelial cells to preferentially increase their adherence for lymphocytes. Clin. Exp. Immunol. 1996, 105, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.H.; Vital, S.; Russell, J.; Seifert, H.; Granger, D.N. Interleukin-6 mediates enhanced thrombus development in cerebral arterioles following a brief period of focal brain ischemia. Exp. Neurol. 2015, 271, 351–357. [Google Scholar] [CrossRef]
- Basinska, K.; Marycz, K.; Śmieszek, A.; Nicpoń, J. The production and distribution of IL-6 and TNF-α in subcutaneous adipose tissue and their correlation with serum concentrations in Welsh ponies with equine metabolic syndrome. J. Vet. Sci. 2015, 16, 113–120. [Google Scholar] [CrossRef]
- Coppelman, E.B. The Use of Biomarkers to Determine the Severity of Osteoarthritis in the Tarsus of an Older Horse Population. Ph.D. Thesis, University of Minnesota, Minneapolis, MN, USA, December 2017. [Google Scholar]
- Adams, R.L.C.; Bird, R.J. Review article: Coagulation cascade and therapeutics update: Relevance to nephrology, Part 1: Overview of coagulation, thrombophilias and history of anticoagulants. Nephrology (Carlton) 2009, 14, 462–470. [Google Scholar] [CrossRef]
- Zarski, L.M.; Weber, P.S.D.; Lee, Y.; Hussey, G.S. Transcriptomic Profiling of Equine and Viral Genes in Peripheral Blood Mononuclear Cells in Horses during Equine Herpesvirus 1 Infection. Pathogens 2021, 10, 43. [Google Scholar] [CrossRef]
- Cevik, O.; Baykal, A.T.; Sener, A. Platelets Proteomic Profiles of Acute Ischemic Stroke Patients. PLoS ONE 2016, 11, e0158287. [Google Scholar] [CrossRef]
- Khaiboullina, S.F.; Morzunov, S.P.; Jeor, S.C.S.; Rizvanov, A.A.; Lombardi, V.C. Hantavirus Infection Suppresses Thrombospondin-1 Expression in Cultured Endothelial Cells in a Strain-Specific Manner. Front. Microbiol. 2016, 7, 1077. [Google Scholar] [CrossRef] [PubMed]
- Mar, C.; Tomás, S.; Mónica, M.; María, G.; Juan, A.; Florentino, N.; David, B.; de la Ossa, N.P.; Joaquín, S.; José, V.; et al. Serum Cellular Fibronectin and Matrix Metalloproteinase-9 as Screening Biomarkers for the Prediction of Parenchymal Hematoma After Thrombolytic Therapy in Acute Ischemic Stroke. Stroke 2007, 38, 1855–1859. [Google Scholar] [CrossRef]
- Channappanavar, R.; Fehr, A.R.; Zheng, J.; Wohlford-Lenane, C.; Abrahante, J.E.; Mack, M.; Sompallae, R.; McCray, P.B., Jr.; Meyerholz, D.K.; Perlman, S. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Investig. 2019, 129, 3625–3639. [Google Scholar] [CrossRef]
- Channappanavar, R.; Fehr, A.R.; Vijay, R.; Mack, M.; Zhao, J.; Meyerholz, D.K.; Perlman, S. Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice. Cell Host Microbe 2016, 19, 181–193. [Google Scholar] [CrossRef]
- Acharya, D.; Liu, G.; Gack, M.U. Dysregulation of type I interferon responses in COVID-19. Nat. Rev. Immunol. 2020, 20, 397–398. [Google Scholar] [CrossRef] [PubMed]
- Pavulraj, S.; Kamel, M.; Stephanowitz, H.; Liu, F.; Plendl, J.; Osterrieder, N.; Azab, W. Equine Herpesvirus Type 1 Modulates Cytokine and Chemokine Profiles of Mononuclear Cells for Efficient Dissemination to Target Organs. Viruses 2020, 12, 999. [Google Scholar] [CrossRef]
- Laval, K.; Favoreel, H.W.; Poelaert, K.C.K.; Van Cleemput, J.; Nauwynck, H.J. Equine herpesvirus type 1 enhances viral replication in CD172a+ monocytic cells upon adhesion to endothelial cells. J. Virol. 2015, 89, 10912–10923. [Google Scholar] [CrossRef] [PubMed]
- Linton, P.J.; Dorshkind, K. Age-related changes in lymphocyte development and function. Nat. Immunol. 2004, 5, 133–139. [Google Scholar] [CrossRef]
- Walford, R.L. The Immunologic Theory of Aging. Immunol. Rev. 1969, 2, 171. [Google Scholar] [CrossRef]
- Sikora, E.; Kamińska, B.; Radziszewska, E.; Kaczmarek, L. Loss of transcription factor AP-1 DNA binding activity during lymphocyte aging in vivo. FEBS Lett. 1992, 312, 179–182. [Google Scholar] [CrossRef]
- Whisler, R.L.; Newhouse, Y.G.; Bagenstose, S.E. Age-related reductions in the activation of mitogen-activated protein kinases p44mapk/ERK1 and p42mapk/ERK2 in human T cells stimulated via ligation of the T cell receptor complex. Cell. Immunol. 1996, 168, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Hutter, D.; Yo, Y.; Chen, W.; Liu, P.; Holbrook, N.J.; Roth, G.S.; Liu, Y. Age-related decline in Ras/ERK mitogen-activated protein kinase cascade is linked to a reduced association between Shc and EGF receptor. J. Gerontol. A Biol. Sci. Med. Sci. 2000, 55, B125–B134. [Google Scholar] [CrossRef]
- Gorgas, G.; Butch, E.R.; Guan, K.-L.; Miller, R.A. Diminished activation of the MAP kinase pathway in CD3-stimulated T lymphocytes from old mice. Mech. Ageing Dev. 1997, 94, 71–83. [Google Scholar] [CrossRef]
- Liu, B.; Carle, K.W.; Whisler, R.L. Reductions in the activation of ERK and JNK are associated with decreased IL-2 production in T cells from elderly humans stimulated by the TCR/CD3 complex and costimulatory signals. Cell. Immunol. 1997, 182, 79–88. [Google Scholar] [CrossRef]
- Douziech, N.; Seres, I.; Larbi, A.; Szikszay, E.; Roy, P.M.; Arcand, M.; Dupuis, G.; Fulop, T.J. Modulation of human lymphocyte proliferative response with aging. Exp. Gerontol. 2002, 37, 369–387. [Google Scholar] [CrossRef]
- Adams, A.A.; Breathnach, C.C.; Katepalli, M.P.; Kohler, K.; Horohov, D.W. Advanced age in horses affects divisional history of T cells and inflammatory cytokine production. Mech. Ageing Dev. 2008, 129, 656–664. [Google Scholar] [CrossRef]
- Horohov, D.W.; Kydd, J.H.; Hannant, D. The effect of aging on T cell responses in the horse. Dev. Comp. Immunol. 2002, 26, 121–128. [Google Scholar] [CrossRef]
- Katepalli, M.P.; Adams, A.A.; Lear, T.L.; Horohov, D.W. The effect of age and telomere length on immune function in the horse. Dev. Comp. Immunol. 2008, 32, 1409–1415. [Google Scholar] [CrossRef]
- Lunn, D.P.; Holmes, M.A.; Gibson, J.; Field, H.J.; Kydd, J.H.; Duffus, W.P.H. Haematological changes and equine lymphocyte subpopulation kinetics during primary infection and attempted re-infection of specific pathogen free foals with EHV-1. Equine Vet. J. 1991, 23, 35–40. [Google Scholar] [CrossRef]
- McCulloch, J.; Williamson, S.A.; Powis, S.J.; Edington, N. The effect of EHV-1 infection upon circulating leucocyte populations in the natural equine host. Vet. Microbiol. 1993, 37, 147–161. [Google Scholar] [CrossRef]
- Fulop, T.; Larbi, A.; Pawelec, G. Human T Cell Aging and the Impact of Persistent Viral Infections. Front. Immunol. 2013, 4, 271. [Google Scholar] [CrossRef] [PubMed]
- Lanna, A.; Henson, S.M.; Escors, D.; Akbar, A.N. The kinase p38 activated by the metabolic regulator AMPK and scaffold TAB1 drives the senescence of human T cells. Nat. Immunol. 2014, 15, 965–972. [Google Scholar] [CrossRef] [PubMed]
- Gilden, D.; Nagel, M.A.; Cohrs, R.J. Varicella-zoster. Handb. Clin. Neurol. 2014, 123, 265–283. [Google Scholar] [CrossRef]
- Goronzy, J.J.; Weyand, C.M. Understanding immunosenescence to improve responses to vaccines. Nat. Immunol. 2013, 14, 428–436. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, T.; Kydd, J.H.; Allen, G.P.; Wattrang, E.; Mumford, J.A.; Hannant, D. Determination of equid herpesvirus 1-specific, CD8+, cytotoxic T lymphocyte precursor frequencies in ponies. Vet. Immunol. Immunopathol. 1999, 70, 43–54. [Google Scholar] [CrossRef]
- Kydd, J.H.; Wattrang, E.; Hannant, D. Pre-infection frequencies of equine herpesvirus-1 specific, cytotoxic T lymphocytes correlate with protection against abortion following experimental infection of pregnant mares. Vet. Immunol. Immunopathol. 2003, 96, 207–217. [Google Scholar] [CrossRef]
- Perkins, G.A.; Wagner, B. The development of equine immunity: Current knowledge on immunology in the young horse. Equine Vet. J. 2015, 47, 267–274. [Google Scholar] [CrossRef]
- Groom, J.R.; Luster, A.D. CXCR3 ligands: Redundant, collaborative and antagonistic functions. Immunol. Cell Biol. 2011, 89, 207–215. [Google Scholar] [CrossRef]
- Wimer, C.L.; Damiani, A.; Osterrieder, N.; Wagner, B. Equine herpesvirus type-1 modulates CCL2, CCL3, CCL5, CXCL9, and CXCL10 chemokine expression. Vet. Immunol. Immunopathol. 2011, 140, 266–274. [Google Scholar] [CrossRef]
- Adams, D.H.; Rlloyd, A. Chemokines: Leucocyte recruitment and activation cytokines. Lancet 1997, 349, 490–495. [Google Scholar] [CrossRef]
- Mariani, E.; Pulsatelli, L.; Neri, S.; Dolzani, P.; Meneghetti, A.; Silvestri, T.; Ravaglia, G.; Forti, P.; Cattini, L.; Facchini, A. RANTES and MIP-1α production by T lymphocytes, monocytes and NK cells from nonagenarian subjects. Exp. Gerontol. 2002, 37, 219–226. [Google Scholar] [CrossRef]
- Gilkerson, J.R.; Whalley, J.M.; Drummer, H.E.; Studdert, M.J.; Love, D.N. Epidemiological studies of equine herpesvirus 1 (EHV-1) in Thoroughbred foals: A review of studies conducted in the Hunter Valley of New South Wales between 1995 and 1997. Vet. Microbiol. 1999, 68, 15–25. [Google Scholar] [CrossRef]
- Gilkerson, J.R.; Whalley, J.M.; Drummer, H.E.; Studdert, M.J.; Love, D.N. Epidemiology of EHV-1 and EHV-4 in the mare and foal populations on a Hunter Valley stud farm: Are mares the source of EHV-1 for unweaned foals. Vet. Microbiol. 1999, 68, 27–34. [Google Scholar] [CrossRef]
- Foote, C.E.; Love, D.N.; Gilkerson, J.R.; Whaley, J.M. Detection of EHV-1 and EHV-4 DNA in unweaned Thoroughbred foals from vaccinated mares on a large stud farm. Equine Vet. J. 2010, 36, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Mumford, J.A.; Rossdale, P.D.; Jessett, D.M.; Gann, S.J.; Ousey, J. Serological and virological investigations of an equid herpesvirus 1 (EHV-1) abortion storm on a stud farm in 1985. J. Reprod. Fertil. Suppl. 1987, 35, 509–518. [Google Scholar] [PubMed]
- Giessler, K.S.; Samoilowa, S.; Hussey, G.S.; Kiupel, M.; Matiasek, K.; Sledge, D.J.; Liesche, F.; Schlegel, J.; Fux, R.; Goehring, L.S. Viral load and cell tropism during early latent Equid Herpesvirus 1 infection differ over time in lymphoid and neural tissue samples from experimentally infected horses. Front. Vet. Sci. 2020, 7. [Google Scholar] [CrossRef]
- Kennedy, P.G.E.; Rovnak, J.; Badani, H.; Cohrs, R.J. A comparison of herpes simplex virus type 1 and varicella-zoster virus latency and reactivation. J. Gen. Virol. 2015, 96, 1581–1602. [Google Scholar] [CrossRef]
- Hoshino, Y.; Pesnicak, L.; Cohen, J.I.; Straus, S.E. Rates of reactivation of latent herpes simplex virus from mouse trigeminal ganglia ex vivo correlate directly with viral load and inversely with number of infiltrating CD8+ T cells. J. Virol. 2007, 81, 8157–8164. [Google Scholar] [CrossRef]
- Walker, J.; Leib, D.A. Protection from primary infection and establishment of latency by vaccination with a herpes simplex virus type 1 recombinant deficient in the virion host shutoff (vhs) function. Vaccine 1998, 16, 1–5. [Google Scholar] [CrossRef]
- Akkutay, A.Z.; Osterrieder, N.; Damiani, A.; Tischer, B.K.; Borchers, K.; Alkan, F. Prevalence of equine gammaherpesviruses on breeding farms in Turkey and development of a TaqMan MGB real-time PCR to detect equine herpesvirus 5 (EHV-5). Arch. Virol. 2014, 159, 2989–2995. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.A.; Balasuriya, U.B.R.; Gardner, I.A.; Barry, P.A.; Wilson, W.D.; Ferraro, G.L.; MacLachlan, N.J. Temporal detection of equine herpesvirus infections of a cohort of mares and their foals. Vet. Microbiol. 2006, 116, 249–257. [Google Scholar] [CrossRef]
- Smith, K.C.; Mumford, J.A.; Lakhani, K. A comparison of equid herpesvirus-1 (EHV-1) vascular lesions in the early versus late pregnant equine uterus. J. Comp. Pathol. 1996, 114, 231–247. [Google Scholar] [CrossRef]





| ID | Description | Gene Ratio | Bg Ratio | p Value | p.adjust | q Value | geneID | Count |
|---|---|---|---|---|---|---|---|---|
| Upregulated | ||||||||
| GO:0051607 | defense response to virus | 13/92 | 175/13,991 | 1.1 × 10−10 | 2.1 × 10−07 | 1.8 × 10−07 | EIF2AK2/TRIM56/ADAR/CXCL10/IRF9/RTP4/ISG15/AIM2/IRF7/TLR3/IL6/ISG20/IL15 | 13 |
| GO:0032479 | regulation of type I interferon production | 8/92 | 104/13,991 | 3.9 × 10−07 | 1.3 × 10−04 | 1.1 × 10−04 | TRIM56/UBA7/NMI/ISG15/IRF7/ACOD1/ TLR3/ZBP1 | 8 |
| GO:0042107 | cytokine metabolic process | 7/92 | 101/13,991 | 4.4 × 10−06 | 6.8 × 10−04 | 5.9 × 10−04 | LAG3/NMI/IGF2BP3/IRF7/TLR3/IL6/CYBB | 7 |
| GO:0043900 | regulation of multi-organism process | 11/92 | 313/13,991 | 6.3 × 10−06 | 9.0 × 10−04 | 7.9 × 10−04 | EIF2AK2/ADAR/PPID/ISG15/CD180/AIM2/ ACOD1/ISG20/IL15/SLPI/TIMP1 | 11 |
| GO:0048771 | tissue remodeling | 7/92 | 150/13,991 | 5.8 × 10−05 | 4.5 × 10−03 | 3.9 × 10−03 | TGM2/CD38/SNX10/IL6/IL15/VDR/TIMP1 | 7 |
| GO:0032570 | response to progesterone | 4/92 | 38/13,991 | 1.1 × 10−04 | 6.7 × 10−03 | 5.9 × 10−03 | CD38/ACOD1/NR1H3/RAMP2 | 4 |
| GO:0010883 | regulation of lipid storage | 4/92 | 41/13,991 | 1.5 × 10−04 | 7.9 × 10−03 | 6.9 × 10−03 | IL6/LPL/NR1H3/MSR1 | 4 |
| GO:0015012 | heparan sulfate proteoglycan biosynthetic process | 3/92 | 23/13,991 | 4.4 × 10−04 | 1.8 × 10−02 | 1.6 × 10−02 | EXT1/TCF7L2/EXTL2 | 3 |
| GO:0010743 | regulation of macrophage derived foam cell differentiation | 3/92 | 25/13,991 | 5.7 × 10−04 | 2.1 × 10−02 | 1.9 × 10−02 | LPL/NR1H3/MSR1 | 3 |
| GO:0070661 | leukocyte proliferation | 7/92 | 231/13,991 | 8.1 × 10−04 | 2.6 × 10−02 | 2.2 × 10−02 | TNFSF13B/CD38/CD180/IL6/GAPT/IL15/CCL8 | 7 |
| GO:0090077 | foam cell differentiation | 3/92 | 30/13,991 | 9.8 × 10−04 | 2.8 × 10−02 | 2.4 × 10−02 | LPL/NR1H3/MSR1 | 3 |
| GO:0097050 | type B pancreatic cell apoptotic process | 2/92 | 10/13,991 | 1.9 × 10−03 | 4.7 × 10−02 | 4.1 × 10−02 | IL6/TCF7L2 | 2 |
| Downregulated | ||||||||
| GO:0030335 | positive regulation of cell migration | 20/181 | 441/13,991 | 1.1 × 10−06 | 8.4 × 10−04 | 7.6 × 10−04 | DAPK2/MMP9/DOCK5/FGFR1/CASS4/CCL5/FN1/HSPA5/THBS1/JUN/INSR/ADAM8/PDGFB/PDGFD/SEMA6C/SEMA4C/TNFSF14/GATA2/RHOB/GPNMB | 20 |
| GO:0031952 | regulation of protein autophosphorylation | 6/181 | 43/13,991 | 1.8 × 10−05 | 8.9 × 10−03 | 8.0 × 10−03 | NLRP12/JUN/PDGFB/PDGFD/ERRFI1/GPNMB | 6 |
| GO:0046777 | protein autophosphorylation | 12/181 | 220/13,991 | 2.9 × 10−05 | 1.3 × 10−02 | 1.1 × 10−02 | DAPK2/PTK6/FGFR1/NLRP12/JUN/INSR/ PDGFB/PDGFD/ERRFI1/INSRR/BMX/GPNMB | 12 |
| GO:0070371 | ERK1 and ERK2 cascade | 13/181 | 292/13,991 | 1.1 × 10−04 | 3.0 × 10−02 | 2.7 × 10−02 | FGFR1/CCL5/FN1/NLRP12/FBLN1/JUN/INSR/PDGFB/PDGFD/ERRFI1/INSRR/ZFP36L2/ GPNMB | 13 |
| GO:0032103 | positive regulation of response to external stimulus | 12/181 | 260/13,991 | 1.5 × 10−04 | 3.6 × 10−02 | 3.3 × 10−02 | DAPK2/FGFR1/CCL5/FAM19A3/NLRP12/ THBS1/MAPK13/ADAM8/PDGFB/PDGFD/ NPY/TNFSF14 | 12 |
| GO:0010035 | response to inorganic substance | 17/181 | 491/13,991 | 2.2 × 10−04 | 4.6 × 10−02 | 4.2 × 10−02 | EEF1A2/MMP9/HSPA5/THBS1/MAPK13/JUN/SELENOP/PDGFD/PTCH1/GSN/FOSB/FOS/ SLC40A1/CHP2/JUND/RHOB/IL1A | 17 |
| GO:0071248 | cellular response to metal ion | 9/181 | 161/13,991 | 2.5 × 10−04 | 4.6 × 10−02 | 4.2 × 10−02 | MMP9/HSPA5/JUN/GSN/FOSB/FOS/SLC40A1/CHP2/JUND | 9 |
| EHM Pre Challenge (% of Total Cell Population) | EHM Post Challenge (% of Total Cell Population) | Non-EHM Pre Challenge (% of Total Cell Population) | Non-EHM Post Challenge (% of Total Cell Population) | |
|---|---|---|---|---|
| B cells naïve | 23.03 ± 1.84 | 24.75 ± 3.23 | 37.03 ± 1.53 | 32.55 ± 1.14 ** |
| B cells memory | 0.60 ± 0.60 | 0.41 ± 0.41 | 0.03 ± 0.03 | 0.00 ± 0.00 |
| Plasma cells | 0.27 ± 0.12 | 0.46 ± 0.29 | 0.28 ± 0.15 | 0.74 ± 0.06 * |
| T cells CD8 | 7.92 ± 1.56 | 2.36 ± 1.59 * | 2.18 ± 0.79 | 2.21 ± 1.03 |
| T cells CD4 naive | 1.64 ± 0.66 | 4.60 ± 2.05 | 6.21 ± 2.25 | 10.26 ± 1.62 |
| T cells CD4 memory resting | 0.77 ± 0.55 | 1.34 ± 0.77 | 0.02 ± 0.02 | 0.00 ± 0.00 |
| T cells CD4 memory activated | 7.88 ± 0.86 | 9.55 ± 1.62 | 0.55 ± 0.26 | 1.71 ± 0.52 * |
| T cells follicular helper | 16.62 ± 1.58 | 14.86 ± 1.84 | 23.27 ± 0.80 | 19.43 ± 1.44 * |
| T cells regulatory (Tregs) | 1.82 ± 0.62 | 0.16 ± 0.16 ** | 1.48 ± 0.49 | 0.41 ± 0.41 |
| T cells gamma delta | 0.88 ± 0.33 | 2.84 ± 0.66 * | 0.79 ± 0.32 | 2.67 ± 0.82 * |
| NK cells resting | 6.43 ± 1.10 | 2.75 ± 1.08 ** | 2.24 ± 0.82 | 0.86 ± 0.35 |
| NK cells activated | 0.13 ± 0.13 | 0.34 ± 0.22 | 0.63 ± 0.42 | 0.53 ± 0.26 |
| Monocytes | 12.71 ± 2.75 | 13.85 ± 3.72 | 7.27 ± 0.57 | 11.85 ± 2.22 |
| Macrophages M0 | 2.99 ± 0.91 | 0.00 ± 0.00 ** | 1.77 ± 1.01 | 0.15 ± 0.15 |
| Macrophages M1 | 0.00 ± 0.00 | 0.98 ± 0.30 ** | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Macrophages M2 | 6.34 ± 0.57 | 8.54 ± 0.80 * | 4.22 ± 0.84 | 5.54 ± 0.93 |
| Dendritic cells resting | 0.04 ± 0.04 | 0.86 ± 0.37 * | 0.37 ± 0.29 | 0.35 ± 0.30 |
| Dendritic cells activated | 2.92 ± 0.48 | 4.88 ± 0.89 * | 4.24 ± 0.32 | 5.99 ± 0.42 * |
| Mast cells resting | 0.33 ± 0.33 | 0.23 ± 0.22 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Mast cells activated | 4.51 ± 0.78 | 1.80 ± 0.99 * | 5.43 ± 2.04 | 2.32 ± 1.80 |
| Eosinophils | 1.63 ± 0.77 | 3.83 ± 0.58 ** | 1.74 ± 0.60 | 1.56 ± 0.50 |
| Neutrophils | 0.54 ± 0.23 | 0.60 ± 0.24 | 0.25 ± 0.25 | 0.86 ± 0.56 |
| mirBase ID | Mouse or Human Orthologue | Mature Sequence | logFC | FDR | Predicted Differentially Expressed Target Genes in Contrast Comparison | Predicted Differentially Expressed Target Genes Unique in EHM Horses |
|---|---|---|---|---|---|---|
| Upregulated in EHM vs non-EHM horses: contrast comparison (between groups) | ||||||
| Novel (id: 764) | unknown | CCCGCCCGGCCCGGCCGCC | 2.63 | 0.04 | ND | ND |
| Novel | mmu-miR-7059-5p | GCCGGGGAGCCCGGCGGGC | 2.01 | 0.03 | ADAM22, CREB5, FADS2, INSR, KLF6, OAS2, ORAI2 | ADAM22, ARL4D, CREB5, F11R, FADS2, HIC1, IER5, INSR, JAM2, JUND, KLF6, LAYN, ORAI2, PARD6G, PSD3, PSMB9, SOBP, SRGAP1, TBXA2R |
| Downregulated in EHM vs non-EHM horses: contrast comparison (between groups) | ||||||
| Novel | mmu-miR-669k-5p | TGTGCATGTGTGCATGTAGGCAG | −1.3 | 0.05 | ADAM22, CMPK2, CREB5, DUSP1, EPHA4, INSR, KCNC4, KCNQ4, LONRF3, MEGF9, NHSL2, NUDT4, OAS2, PAQR8, PLCB1, SCD, SLC16A14, SLC18B1, SLC7A11, STARD13, TET1 | ADAM22, AGAP1, AKAP7, BAIAP3, CORO2A, CREB5, ELOVL7, EMP1, FOSB, HSPA2, HSPA5, HTRA1, INSR, INSRR, JAM2, KCNQ4, LONRF3, MEFV, MEGF9, NFASC, NHSL2, NUDT4, PLCB1, PARD6G, PLEKHA6, PSD3, PTPRF, QPCT, SDC3, SLC18B1, SLC7A11, ADAM22, CMPK2, CREB5, XCR1, DUSP1, SRGAP1, STARD13,TAP1,TOR1B, TMEM26, |
| eca-miR-199a/b-5p | hsa-miR-199a-3p | ACAGTAGTCTGCACATTGGTT | −1.33 | 0.008 | ACER2, FOS, FRY, SCD SLC7A11, LONRF3, PLCB1, | ACER2, TLE2, FRY, FOS, SCD, PTPRF, CXXC5, LONRF3, PLCB1, SLC7A11, FN1 |
| eca-miR-34c | hsa-miR-34a-5p | AGGCAGTGTAGTTAGCTGATTGC | −1.3 | 0.008 | ADAM22, CREB5, DIXDC1, FRMD4A, KLF4, LONRF3, MAP1A | ADAM22, CERS4, CREB5, DIXDC1, DLL1, FOSB, LONRF3, PSD3, RGMB, SEMA4C, TRANK1, VAT1, VWA5B2 |
| Novel | hsa-miR-542-5p | TCGGGGATTCAGGTGGCTGTTC | −1.24 | 0.008 | BCORL1, GPR137C, INSR, KLF6, ORAI2, OSM, PTCH1, TEX35, ZNF862 | ADAM33, ADAMTS2, FOSB, GFI1, IER5, INSR, JUND, KLF6, KLRB1, LTBP4, NECAB3, NPDC1, ORAI2, PARP12, PLEKHG5, PTCH1, RGS16, RHOB, SDC3, SRGAP1, TCF7L2, TMEM151B, TNFSF14, ZBTB16, ZFP36L2, ZNF827 |
| eca-miR-10b | hsa-miR-10a-5p | TACCCTGTAGAACCGAATTTGT | −1.20 | 0.035 | STARD13, EPHA4 | NFASC, SOBP, STARD13, ZBTB16, ZNF827 |
| eca-miR-328 | hsa-miR-328-3p | CTGGCCCTCTCTGCCCTTCCGT | −1.13 | 0.01 | NHSL2 | CHP2, HIC1, LHFPL2, MFSD2A, NHSL2, PLEKHA6, PTPRF, TCF7L2 |
| eca-miR-146a | hsa-miR-146a-5p | TGAGAACTGAATTCCATGGGTT | −1.05 | 0.001 | BCORL1, SGIP1 | CDS1, CNTFR, LAYN, MYBL1,SCN3B, VAT1 |
| Uniquely upregulated in EHM horses (within group) | ||||||
| Novel (id: 764) | unknown | CCCGCCCGGCCCGGCCGCC | 2.31 | 0.001 | ND | ND |
| Novel (id: 983) | unknown | CCGCCCGCCGCCGCCGCC | 1.74 | 0.01 | ND | ND |
| Novel (id: 187) | unknown | CCCGCCCGCCGCCGCCGCC | 1.73 | 0.01 | ND | ND |
| Novel | hsa-miR-7108-3p | CCCCGCCCGCCGCCGCCG | 1.7 | 0.01 | ATF3, CD7, DBP, INSR, LIPE, MAPK13, NEO1, NLRP12, ORAI2, OSCAR, OSM, PDGFB, PTCH1, SEMA6C, ZNF862 | ADAR, ARL4D, AXIN2, CD180, CD7, EMP1, FOSB, GATA2, GPNMB, HIC1, HSH2D, IER3, INSR, JUND, LAMTOR2, LIPE, MAPK13, MICAL2, MSR1, NEO1, NFASC, NLRP12, NPTX1, NTNG2, ORAI2, PDGFB, PLEKHG5, PLXND1, PSMB9, PTCH1, PTPRF, SDC3, SEMA4C, SEMA6C, SOBP, TNFSF14, VAT1, ZBTB16, ZDHHC1 |
| Novel | mmu-miR-7059-5p | GCCGGGGAGCCCGGCGGGC | 1.6 | 0.002 | ADAM22, CREB5, FADS2, INSR, KLF6, OAS2, ORAI2 | ADAM22, ARL4D, CREB5, F11R, FADS2, HIC1, IER5, INSR, JAM2, JUND, KLF6, LAYN, ORAI2, PARD6G, PSD3, PSMB9, SOBP, SRGAP1, TBXA2R |
| Uniquely downregulated in EHM horses (within group) | ||||||
| eca-miR-483 | unknown | CACTCCTCTCCTCCCGTCTTCT | −1.8 | <0.001 | ND | ND |
| eca-miR-146a | hsa-miR-146a-5p | TGAGAACTGAATTCCATGGGTT | −1.49 | <0.001 | BCORL1, SGIP1 | CDS1, CNTFR, LAYN, MYBL1,SCN3B, VAT1 |
| eca-miR-34c | hsa-miR-34a-5p | AGGCAGTGTAGTTAGCTGATTGC | −1.34 | <0.001 | ADAM22, CREB5, DIXDC1, FRMD4A, KLF4, LONRF3, MAP1A | ADAM22, CERS4, CREB5, DIXDC1, DLL1, FOSB, LONRF3, PSD3, RGMB, SEMA4C, TRANK1, VAT1, VWA5B2 |
| eca-miR-138 | hsa-miR-138-5p | AGCTGGTGTTGTGAATCAGGCCG | −1.31 | <0.001 | LONRF3, DAPK2, FRMD4A, FRMPD3 | AHDC1, DAPK2, EXT1, FOSB, LONRF3, NPTX1, SCN3B, SDC3, SEMA4C, SOBP, ZFP36L2 |
| eca-miR-199b-5p | hsa-miR-199a-3p | ACAGTAGTCTGCACATTGGTT | −1.03 | <0.001 | ACER2, FOS, FRY, SCD LONRF3, PLCB1, SLC7A11 | ACER2, TLE2, FRY, FOS, SCD, PTPRF, CXXC5, LONRF3, PLCB1, SLC7A11, FN1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zarski, L.M.; Giessler, K.S.; Jacob, S.I.; Weber, P.S.D.; McCauley, A.G.; Lee, Y.; Soboll Hussey, G. Identification of Host Factors Associated with the Development of Equine Herpesvirus Myeloencephalopathy by Transcriptomic Analysis of Peripheral Blood Mononuclear Cells from Horses. Viruses 2021, 13, 356. https://doi.org/10.3390/v13030356
Zarski LM, Giessler KS, Jacob SI, Weber PSD, McCauley AG, Lee Y, Soboll Hussey G. Identification of Host Factors Associated with the Development of Equine Herpesvirus Myeloencephalopathy by Transcriptomic Analysis of Peripheral Blood Mononuclear Cells from Horses. Viruses. 2021; 13(3):356. https://doi.org/10.3390/v13030356
Chicago/Turabian StyleZarski, Lila M., Kim S. Giessler, Sarah I. Jacob, Patty Sue D. Weber, Allison G. McCauley, Yao Lee, and Gisela Soboll Hussey. 2021. "Identification of Host Factors Associated with the Development of Equine Herpesvirus Myeloencephalopathy by Transcriptomic Analysis of Peripheral Blood Mononuclear Cells from Horses" Viruses 13, no. 3: 356. https://doi.org/10.3390/v13030356
APA StyleZarski, L. M., Giessler, K. S., Jacob, S. I., Weber, P. S. D., McCauley, A. G., Lee, Y., & Soboll Hussey, G. (2021). Identification of Host Factors Associated with the Development of Equine Herpesvirus Myeloencephalopathy by Transcriptomic Analysis of Peripheral Blood Mononuclear Cells from Horses. Viruses, 13(3), 356. https://doi.org/10.3390/v13030356







