TRIMming Type I Interferon-Mediated Innate Immune Response in Antiviral and Antitumor Defense
Abstract
1. Introduction
2. PRR Signaling Pathways to IFN-I Production
3. The TRIM Family
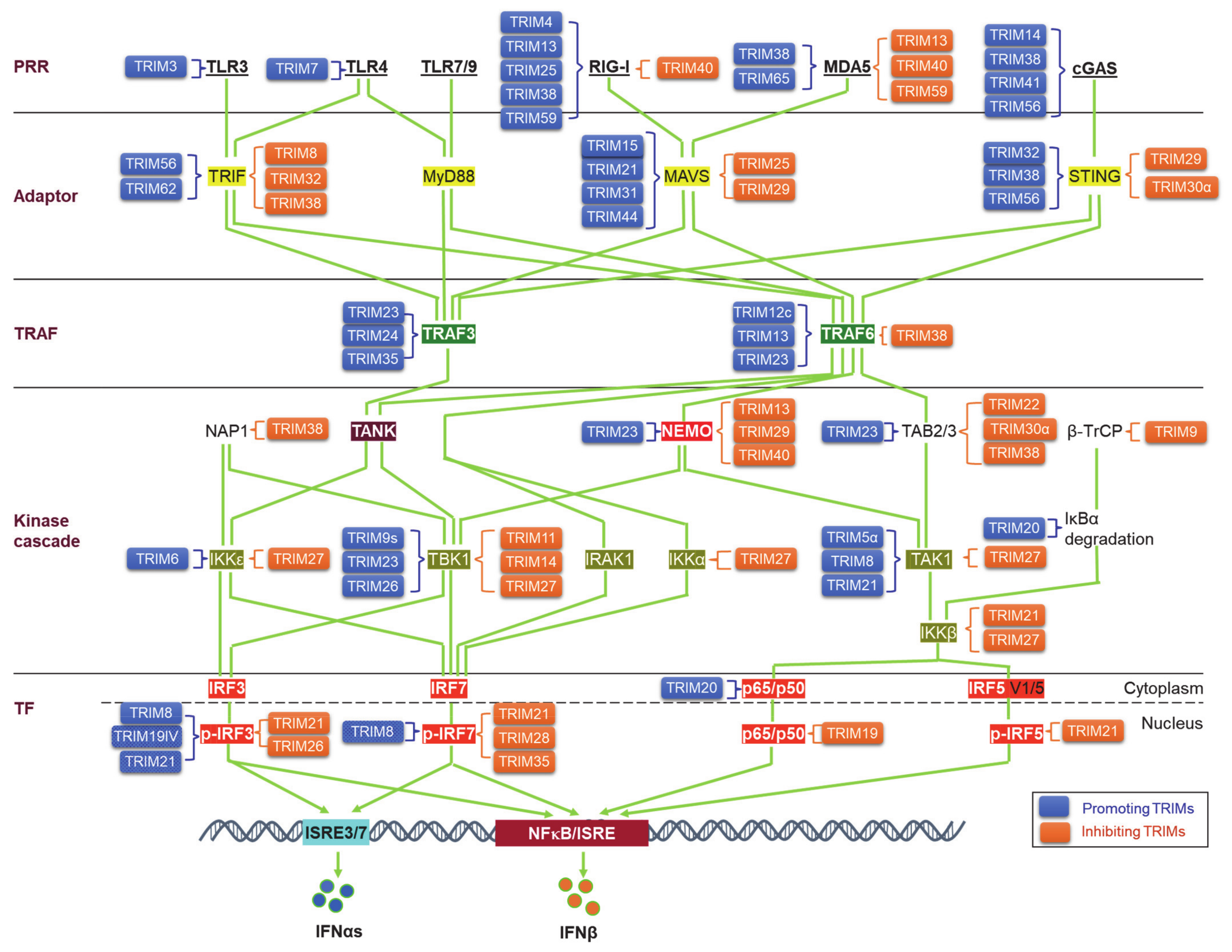
4. TRIMs in Regulating PRR Signaling Pathways to IFN-I Production
4.1. TRIMs in Regulating the PRRs
4.2. TRIMs in Regulating the Adaptors
4.3. TRIMs in Regulating TRAF3/6
4.4. TRIMs in Regulating the Kinase Cascades for Activation of IRFs and NFκB
4.5. TRIMs in Regulating the Transcription Factors IRFs and NF-κB for IFN-I Production
5. TRIMs in Regulating the Jak-STAT IFN-I Signaling
6. Viral Strategies to Subvert TRIM-Mediated IFN-I Regulatory Mechanisms
7. Perspectives
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, S.; Gong, M.; Zhao, F.; Shao, J.; Xie, Y.; Zhang, Y.; Chang, H. Type I Interferons: Distinct Biological Activities and Current Applications for Viral Infection. Cell. Physiol. Biochem. 2018, 51, 2377–2396. [Google Scholar] [CrossRef]
- Sandler, N.G.; Bosinger, S.E.; Estes, J.D.; Zhu, R.T.R.; Tharp, G.K.; Boritz, E.; Levin, D.; Wijeyesinghe, S.; Makamdop, K.N.; del Prete, G.Q.; et al. Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature 2014, 511, 601–605. [Google Scholar] [CrossRef]
- Mogensen, T.; Melchjorsen, J.; Larsen, C.; Paludan, S. Innate immune recognition and activation during HIV infection. Retrovirology 2010, 7, 54. [Google Scholar] [CrossRef]
- Cha, L.; Berry, C.M.; Nolan, D.; Castley, A.; Fernandez, S.; French, M.A. Interferon-alpha, immune activation and immune dysfunction in treated HIV infection. Clin. Trans. Immunol. 2014, 3, e10. [Google Scholar] [CrossRef]
- Crouse, J.; Kalinke, U.; Oxenius, A. Regulation of antiviral T cell responses by type I interferons. Nat. Rev. Immunol. 2015, 15, 231–242. [Google Scholar] [CrossRef] [PubMed]
- McNab, F.; Mayer-Barber, K.; Sher, A.; Wack, A.; O’Garra, A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015, 15, 87–103. [Google Scholar] [CrossRef]
- Catalfamo, M.; Wilhelm, C.; Tcheung, L.; Proschan, M.; Friesen, T.; Park, J.-H.; Adelsberger, J.; Baseler, M.; Maldarelli, F.; Davey, R.; et al. CD4 and CD8 T Cell Immune Activation during Chronic HIV Infection: Roles of Homeostasis, HIV, Type I IFN, and IL-7. J. Immunol. 2011, 186, 2106–2116. [Google Scholar] [CrossRef]
- Tough, D.F. Modulation of T-cell function by type I interferon. Immunol. Cell Biol. 2012, 90, 492–497. [Google Scholar] [CrossRef] [PubMed]
- Zitvogel, L.; Galluzzi, L.; Kepp, O.; Smyth, M.J.; Kroemer, G. Type I interferons in anticancer immunity. Nat. Rev. Immunol. 2015, 15, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Gajewski, T.F.; Corrales, L. New perspectives on type I IFNs in cancer. Cytokine Growth Factor Rev. 2015, 26, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Villar, M.; Gautron, A.S.; de Marcken, M.; Keller, M.J.; Hafler, D.A. TLR7 induces anergy in human CD4+ T cells. Nat. Immunol. 2015, 16, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Wilson, E.B.; Yamada, D.H.; Elsaesser, H.; Herskovitz, J.; Deng, J.; Cheng, G.; Aronow, B.J.; Karp, C.L.; Brooks, D.G. Blockade of chronic Type I interferon signaling to control persistent LCMV infection. Science 2013, 340, 202–207. [Google Scholar] [CrossRef] [PubMed]
- Teijaro, J.R.; Ng, C.; Lee, A.M.; Sullivan, B.M.; Sheehan, K.C.F.; Welch, M.; Schreiber, R.D.; Carlos de la Torre, J.; Oldstone, M.B.A. Persistent LCMV infection is controlled by blockade of Type I interferon signaling. Science 2013, 340, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Bosque, A.; Planelles, V. Induction of HIV-1 latency and reactivation in primary memory CD4+ T cells. Blood 2009, 113, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Navajas, J.M.; Lee, J.; David, M.; Raz, E. Immunomodulatory functions of type I interferons. Nat. Rev. Immunol. 2012, 12, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Bonjardim, C.A.; Ferreira, P.C.P.; Kroon, E.G. Interferons: Signaling, antiviral and viral evasion. Immunol. Lett. 2009, 122, 1–11. [Google Scholar] [CrossRef]
- Hervas-Stubbs, S.; Perez-Gracia, J.L.; Rouzaut, A.; Sanmamed, M.F.; Le Bon, A.; Melero, I. Direct Effects of Type I Interferons on Cells of the Immune System. Clin. Cancer Res. 2011, 17, 2619–2627. [Google Scholar] [CrossRef]
- Forster, S. Interferon signatures in immune disorders and disease. Immunol. Cell Biol. 2012, 90, 520–527. [Google Scholar] [CrossRef]
- Elkon, K.B.; Wiedeman, A. Type I IFN system in the development and manifestations of SLE. Curr.Opin. Rheumatol. 2012, 24, 499–505. [Google Scholar] [CrossRef]
- von Locquenghien, M.; Rozalén, C.; Celià-Terrassa, T. Interferons in cancer immunoediting: Sculpting metastasis and immunotherapy response. J. Clin. Investig. 2021, 131. [Google Scholar] [CrossRef]
- Arimoto, K.-I.; Miyauchi, S.; Stoner, S.A.; Fan, J.-B.; Zhang, D.-E. Negative regulation of type I IFN signaling. J. Leukoc. Biol. 2018, 103, 1099–1116. [Google Scholar] [CrossRef]
- Wang, L.; Ning, S. “Toll-free” pathways for production of type I interferons. Allergy Immunol. 2017, 1, 143–163. [Google Scholar] [CrossRef] [PubMed]
- McClellan, A.J.; Laugesen, S.H.; Ellgaard, L. Cellular functions and molecular mechanisms of non-lysine ubiquitination. Open Biol. 2019, 9, 190147. [Google Scholar] [CrossRef]
- Huang, Q.; Zhang, X. Emerging Roles and Research Tools of Atypical Ubiquitination. Proteomics 2020, 20, 1900100. [Google Scholar] [CrossRef]
- Hrdinka, M.; Gyrd-Hansen, M. The Met1-Linked Ubiquitin Machinery: Emerging Themes of (De)regulation. Mol. Cell 2017, 68, 265–280. [Google Scholar] [CrossRef] [PubMed]
- Iwai, K. Discovery of linear ubiquitination, a crucial regulator for immune signaling and cell death. FEBS J. 2020. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, D.; Sato, Y.; Ito, H.; Tokunaga, F. Linear Ubiquitin Code: Its Writer, Erasers, Decoders, Inhibitors, and Implications in Disorders. Int. J. Mol. Sci. 2020, 21, 3381. [Google Scholar] [CrossRef] [PubMed]
- Ferrarelli, L.K. Palmitoylation makes the switch for EGFR. Science 2020, 367, 1086–1088. [Google Scholar] [CrossRef]
- Wei, Y.; Xu, X. UFMylation: A Unique & Fashionable Modification for Life. Genom. Proteom. Bioinform. 2016, 14, 140–146. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y.; Zhu, C.; Robertson, E.S.; Cai, Q. Role of SUMOylation in Human Oncogenic Herpesvirus Infection. Virus Res. 2020. [Google Scholar] [CrossRef]
- Su, S.; Zhang, Y.; Liu, P. Roles of Ubiquitination and SUMOylation in DNA Damage Response. Curr. Issues Mol. Biol. 2019, 35, 59–84. [Google Scholar] [CrossRef]
- Yu, J.; Qin, B.; Lou, Z. Ubiquitin and ubiquitin-like molecules in DNA double strand break repair. Cell Biosci. 2020, 10, 13. [Google Scholar] [CrossRef]
- Chang, S.C.; Ding, J.L. Ubiquitination and SUMOylation in the chronic inflammatory tumor microenvironment. Biochim. Biophys. Acta BBA Rev. Cancer 2018, 1870, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Perng, Y.-C.; Lenschow, D.J. ISG15 in antiviral immunity and beyond. Nat. Rev. Microbiol. 2018, 16, 423–439. [Google Scholar] [CrossRef] [PubMed]
- Fiil, B.K.; Gyrd-Hansen, M. The Met1-linked ubiquitin machinery in inflammation and infection. Cell Death Differ. 2021, 28, 557–569. [Google Scholar] [CrossRef] [PubMed]
- Masucci, M.G. Epstein-Barr virus oncogenesis and the ubiquitin-proteasome system. Oncogene 2004, 23, 2107–2115. [Google Scholar] [CrossRef]
- Đukić, A.; Lulić, L.; Thomas, M.; Skelin, J.; Bennett Saidu, N.E.; Grce, M.; Banks, L.; Tomaić, V. HPV Oncoproteins and the Ubiquitin Proteasome System: A Signature of Malignancy? Pathogens 2020, 9, 133. [Google Scholar] [CrossRef]
- Shackelford, J.; Pagano, J. Role of the ubiquitin system and tumor viruses in AIDS-related cancer. BMC Biochem. 2007, 8, S8. [Google Scholar] [CrossRef]
- Zheng, Y.; Gao, C. Chapter Four—Fine-tuning of antiviral innate immunity by ubiquitination. Adv. Immunol. 2020, 145, 95–128. [Google Scholar]
- Ashizawa, A.; Higashi, C.; Masuda, K.; Ohga, R.; Taira, T.; Fujimuro, M. The Ubiquitin System and Kaposi’s Sarcoma-Associated Herpesvirus. Front. Microbiol. 2012, 3, 66. [Google Scholar] [CrossRef]
- Lopez-Castejon, G. Control of the inflammasome by the ubiquitin system. FEBS J. 2020, 287, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Ning, S.; Pagano, J.; Barber, G. IRF7: Activation, regulation, modification, and function. Genes Immun. 2011, 12, 399–414. [Google Scholar] [CrossRef] [PubMed]
- Härtlova, A.; Erttmann, S.F.; Raffi, F.A.M.; Schmalz, A.M.; Resch, U.; Anugula, S.; Lienenklaus, S.; Nilsson, L.M.; Kröger, A.; Nilsson, J.A.; et al. DNA Damage Primes the Type I Interferon System via the Cytosolic DNA Sensor STING to Promote Anti-Microbial Innate Immunity. Immunity 2015, 42, 332–343. [Google Scholar] [CrossRef] [PubMed]
- Schlee, M.; Hartmann, G. Discriminating self from non-self in nucleic acid sensing. Nat. Rev. Immunol. 2016, 16, 566–580. [Google Scholar] [CrossRef] [PubMed]
- White, M.J.; McArthur, K.; Metcalf, D.; Lane, R.M.; Cambier, J.C.; Herold, M.J.; van Delft, M.F.; Bedoui, S.; Lessene, G.; Ritchie, M.E.; et al. Apoptotic Caspases Suppress mtDNA-Induced STING-Mediated Type I IFN Production. Cell 2014, 159, 1549–1562. [Google Scholar] [CrossRef]
- de Galarreta, M.R.; Lujambio, A. DNA sensing in senescence. Nat. Cell Biol. 2017, 19, 1008–1009. [Google Scholar] [CrossRef]
- Gluck, S.; Guey, B.; Gulen, M.F.; Wolter, K.; Kang, T.W.; Schmacke, N.A.; Bridgeman, A.; Rehwinkel, J.; Zender, L.; Ablasser, A. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat. Cell Biol. 2017, 19, 1061–1070. [Google Scholar] [CrossRef]
- Agalioti, T.; Lomvardas, S.; Parekh, B.; Yie, J.; Maniatis, T.; Thanos, D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-α promoter. Cell 2000, 103, 667–678. [Google Scholar] [CrossRef]
- Hatakeyama, S. TRIM Family Proteins: Roles in Autophagy, Immunity, and Carcinogenesis. Trends Biochem. Sci. 2017, 42, 297–311. [Google Scholar] [CrossRef] [PubMed]
- van Tol, S.; Hage, A.; Giraldo, M.; Bharaj, P.; Rajsbaum, R. The TRIMendous Role of TRIMs in Virus–Host Interactions. Vaccines 2017, 5, 23. [Google Scholar] [CrossRef] [PubMed]
- Sardiello, M.; Cairo, S.; Fontanella, B.; Ballabio, A.; Meroni, G. Genomic analysis of the TRIM family reveals two groups of genes with distinct evolutionary properties. BMC Evol. Biol. 2008, 8, 225. [Google Scholar] [CrossRef] [PubMed]
- Versteeg, G.A.; Benke, S.; García-Sastre, A.; Rajsbaum, R. InTRIMsic immunity: Positive and negative regulation of immune signaling by tripartite motif proteins. Cytokine Growth Factor Rev. 2014, 25, 563–576. [Google Scholar] [CrossRef]
- Yang, W.; Gu, Z.; Zhang, H.; Hu, H. To TRIM the Immunity: From Innate to Adaptive Immunity. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef]
- Giraldo, M.I.; Hage, A.; van Tol, S.; Rajsbaum, R. TRIM Proteins in Host Defense and Viral Pathogenesis. Curr. Clin. Microbiol. Rep. 2020, 7, 101–114. [Google Scholar] [CrossRef]
- Patil, G.; Li, S. Tripartite motif proteins: An emerging antiviral protein family. Future Virol. 2019, 14, 107–122. [Google Scholar] [CrossRef]
- Di Rienzo, M.; Romagnoli, A.; Antonioli, M.; Piacentini, M.; Fimia, G.M. TRIM proteins in autophagy: Selective sensors in cell damage and innate immune responses. Cell Death Differ. 2020, 27, 887–902. [Google Scholar] [CrossRef]
- Khan, R.; Khan, A.; Ali, A.; Idrees, M. The interplay between viruses and TRIM family proteins. Rev. Med. Virol. 2019, 29, e2028. [Google Scholar] [CrossRef]
- Hage, A.; Rajsbaum, R. To TRIM or not to TRIM: The balance of host-virus interactions mediated by the ubiquitin system. J. Gen. Virol. 2019, 100, 1641–1662. [Google Scholar] [CrossRef] [PubMed]
- Li, W.-W.; Nie, Y.; Yang, Y.; Ran, Y.; Luo, W.-W.; Xiong, M.-G.; Wang, S.-Y.; Xu, Z.-S.; Wang, Y.-Y. Ubiquitination of TLR3 by TRIM3 signals its ESCRT-mediated trafficking to the endolysosomes for innate antiviral response. Proc. Natl. Acad. Sci. USA 2020, 117, 23707–23716. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Li, Q.; Mao, A.P.; Hu, M.M.; Shu, H.B. TRIM4 modulates type I interferon induction and cellular antiviral response by targeting RIG-I for K63-linked ubiquitination. J. Mol. Cell Biol. 2014, 6, 154–163. [Google Scholar] [CrossRef]
- Ganser-Pornillos, B.K.; Pornillos, O. Restriction of HIV-1 and other retroviruses by TRIM5. Nat. Rev. Microbiol. 2019, 17, 546–556. [Google Scholar] [CrossRef]
- Chang, T.-H.; Yoshimi, R.; Ozato, K. TRIM12c, a Mouse Homolog of TRIM5, Is a Ubiquitin Ligase That Stimulates Type I IFN and NF-κB Pathways along with TNFR-Associated Factor 6. J. Immunol. 2015, 195, 5367–5379. [Google Scholar] [CrossRef]
- Pertel, T.; Hausmann, S.; Morger, D.; Zuger, S.; Guerra, J.; Lascano, J.; Reinhard, C.; Santoni, F.A.; Uchil, P.D.; Chatel, L.; et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 2011, 472, 361–365. [Google Scholar] [CrossRef]
- Bharaj, P.; Atkins, C.; Luthra, P.; Giraldo, M.I.; Dawes, B.E.; Miorin, L.; Johnson, J.R.; Krogan, N.J.; Basler, C.F.; Freiberg, A.N.; et al. The Host E3-Ubiquitin Ligase TRIM6 Ubiquitinates the Ebola Virus VP35 Protein and Promotes Virus Replication. J. Virol. 2017, 91, e00833-17. [Google Scholar] [CrossRef]
- Rajsbaum, R.; Versteeg, G.A.; Schmid, S.; Maestre, A.M.; Belicha-Villanueva, A.; Martínez-Romero, C.; Patel, J.R.; Morrison, J.; Pisanelli, G.; Miorin, L.; et al. Unanchored K48-linked polyubiquitin synthesized by the E3-ubiquitin ligase TRIM6 stimulates the interferon-IKKε kinase-mediated antiviral response. Immunity 2014, 40, 880–895. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, M.I.; Xia, H.; Aguilera-Aguirre, L.; Hage, A.; van Tol, S.; Shan, C.; Xie, X.; Sturdevant, G.L.; Robertson, S.J.; McNally, K.L.; et al. Envelope protein ubiquitination drives entry and pathogenesis of Zika virus. Nature 2020, 585, 414–419. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Zhu, X.; Yang, Z.; Zhang, W.; Sun, Z.; Ji, Q.; Chen, X.; Zhu, J.; Wang, C.; Nie, S. E3 ubiquitin ligase TRIM7 positively regulates the TLR4-mediated immune response via its E3 ligase domain in macrophages. Mol. Immunol. 2019, 109, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Hu, M.-M.; Lei, C.-Q.; Zhou, Q.; Lin, H.; Sun, M.-S.; Shu, H.-B. TRIM8 Negatively Regulates TLR3/4-Mediated Innate Immune Response by Blocking TRIF–TBK1 Interaction. J. Immunol. 2017, 199, 1856–1864. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Yan, J.; Mao, A.P.; Li, C.; Ran, Y.; Shu, H.B.; Wang, Y.Y. TRIM8 modulates TNFà- and IL-1á-triggered NF- k B activation by targeting TAK1 for K63-linked polyubiquitination. Proc. Natl. Acad. Sci. USA 2011, 108, 19341–19346. [Google Scholar] [CrossRef]
- Guo, L.; Dong, W.; Fu, X.; Lin, J.; Dong, Z.; Tan, X.; Zhang, T. Tripartite Motif 8 (TRIM8) Positively Regulates Pro-inflammatory Responses in Pseudomonas aeruginosa-Induced Keratitis Through Promoting K63-Linked Polyubiquitination of TAK1 Protein. Inflammation 2017, 40, 454–463. [Google Scholar] [CrossRef]
- Maarifi, G.; Smith, N.; Maillet, S.; Moncorgé, O.; Chamontin, C.; Edouard, J.; Sohm, F.; Blanchet, F.P.; Herbeuval, J.-P.; Lutfalla, G.; et al. TRIM8 is required for virus-induced IFN response in human plasmacytoid dendritic cells. Sci. Adv. 2019, 5, eaax3511. [Google Scholar] [CrossRef]
- Toniato, E.; Chen, X.P.; Losman, J.; Flati, V.; Donahue, L.; Rothman, P. TRIM8/GERP RING finger protein interacts with SOCS-1. J. Biol. Chem. 2002, 277, 37315–37322. [Google Scholar] [CrossRef] [PubMed]
- Okumura, F.; Matsunaga, Y.; Katayama, Y.; Nakayama, K.I.; Hatakeyama, S. TRIM8 modulates STAT3 activity through negative regulation of PIAS3. J. Cell Sci. 2010, 123, 2238–2245. [Google Scholar] [CrossRef] [PubMed]
- Tomar, D.; Sripada, L.; Prajapati, P.; Singh, R.; Singh, A.K.; Singh, R. Nucleo-Cytoplasmic Trafficking of TRIM8, a Novel Oncogene, Is Involved in Positive Regulation of TNF Induced NF-κB Pathway. PLoS ONE 2012, 7, e48662. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Liu, Q.; Tian, S.; Xie, W.; Cui, J.; Wang, R.F. TRIM9 short isoform preferentially promotes DNA and RNA virus-induced production of type I interferon by recruiting GSK3β to TBK1. Cell Res. 2016, 26, 613–628. [Google Scholar] [CrossRef]
- Shi, M.; Cho, H.; Inn, K.-S.; Yang, A.; Zhao, Z.; Liang, Q.; Versteeg, G.A.; Amini-Bavil-Olyaee, S.; Wong, L.-Y.; Zlokovic, B.V.; et al. Negative regulation of NF-κB activity by brain-specific TRIpartite Motif protein 9. Nat. Commun. 2014, 5, 4820. [Google Scholar] [CrossRef]
- Lee, Y.; Song, B.; Park, C.; Kwon, K.S. TRIM11 negatively regulates IFNβ production and antiviral activity by targeting TBK1. PLoS ONE 2013, 8, e63255. [Google Scholar] [CrossRef]
- Uchil, P.D.; Quinlan, B.D.; Chan, W.T.; Luna, J.M.; Mothes, W. TRIM E3 ligases interfere with early and late stages of the retroviral life cycle. PLoS Pathog. 2008, 4, e16. [Google Scholar] [CrossRef] [PubMed]
- Narayan, K.; Waggoner, L.; Pham, S.T.; Hendricks, G.L.; Waggoner, S.N.; Conlon, J.; Wang, J.P.; Fitzgerald, K.A.; Kang, J. TRIM13 Is a Negative Regulator of MDA5-Mediated Type I Interferon Production. J. Virol. 2014, 88, 10748–10757. [Google Scholar] [CrossRef]
- Huang, B.; Baek, S.H. Trim13 Potentiates Toll-Like Receptor 2-Mediated Nuclear Factor κB Activation via K29-Linked Polyubiquitination of Tumor Necrosis Factor Receptor-Associated Factor 6. Mol. Pharm. 2017, 91, 307–316. [Google Scholar] [CrossRef]
- Tomar, D.; Singh, R. TRIM13 regulates ubiquitination and turnover of NEMO to suppress TNF induced NF-κB activation. Cell. Signal. 2014, 26, 2606–2613. [Google Scholar] [CrossRef]
- Wang, S.; Chen, Y.; Li, C.; Wu, Y.; Guo, L.; Peng, C.; Huang, Y.; Cheng, G.; Qin, F. TRIM14 inhibits hepatitis C virus infection by SPRY domain-dependent targeted degradation of the viral NS5A protein. Sci. Rep. 2016, 6, 32336. [Google Scholar] [CrossRef]
- Hoffpauir, C.T.; Bell, S.L.; West, K.O.; Jing, T.; Wagner, A.R.; Torres-Odio, S.; Cox, J.S.; West, A.P.; Li, P.; Patrick, K.L.; et al. TRIM14 Is a Key Regulator of the Type I IFN Response during Mycobacterium tuberculosis Infection. J. Immunol. 2020, 205, 153–167. [Google Scholar] [CrossRef]
- Zhou, Z.; Jia, X.; Xue, Q.; Dou, Z.; Ma, Y.; Zhao, Z.; Jiang, Z.; He, B.; Jin, Q.; Wang, J. TRIM14 is a mitochondrial adaptor that facilitates RIG-I–like receptor-mediated innate immune response. Proc. Natl. Acad. Sci. USA 2014, 111, E245–E254. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Meng, Q.; Qin, Y.; Liang, P.; Tan, P.; He, L.; Zhou, Y.; Chen, Y.; Huang, J.; Wang, R.-F.; et al. TRIM14 Inhibits cGAS Degradation Mediated by Selective Autophagy Receptor p62 to Promote Innate Immune Responses. Mol. Cell 2016, 64, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Uchil, P.D.; Hinz, A.; Siegel, S.; Coenen-Stass, A.; Pertel, T.; Luban, J.; Mothes, W. TRIM Protein-Mediated Regulation of Inflammatory and Innate Immune Signaling and Its Association with Antiretroviral Activity. J. Virol. 2013, 87, 257–272. [Google Scholar] [CrossRef] [PubMed]
- Turelli, P.; Doucas, V.; Craig, E.; Mangeat, B.; Klages, N.; Evans, R.; Kalpana, G.; Trono, D. Cytoplasmic recruitment of INI1 and PML on incoming HIV preintegration complexes: Interference with early steps of viral replication. Mol. Cell 2001, 7, 1245–1254. [Google Scholar] [CrossRef]
- Regad, T.; Saib, A.; Lallemand-Breitenbach, V.; Pandolfi, P.P.; de Thé, H.; Chelbi-Alix, M.K. PML mediates the interferon-induced antiviral state against a complex retrovirus via its association with the viral transactivator. EMBO J. 2001, 20, 3495–3505. [Google Scholar] [CrossRef] [PubMed]
- Djavani, M.; Rodas, J.; Lukashevich, I.S.; Horejsh, D.; Pandolfi, P.P.; Borden, K.L.B.; Salvato, M.S. Role of the Promyelocytic Leukemia Protein PML in the Interferon Sensitivity of Lymphocytic Choriomeningitis Virus. J. Virol. 2001, 75, 6204–6208. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-E.; Ahn, J.-H. Positive Role of PML Protein in Type I Interferon Response and Its Regulation by Human Cytomegalovirus. PLoS Pathog. 2015, 11, e1004785. [Google Scholar] [CrossRef]
- El Asmi, F.; Maroui, M.A.; Dutrieux, J.; Blondel, D.; Nisole, S.; Chelbi-Alix, M.K. Implication of PMLIV in both intrinsic and innate immunity. PLoS Pathog. 2014, 10, e1003975. [Google Scholar] [CrossRef] [PubMed]
- Maroui, M.A.; Maarifi, G.; McManus, F.P.; Lamoliatte, F.; Thibault, P.; Chelbi-Alix, M.K. Promyelocytic Leukemia Protein (PML) Requirement for Interferon-induced Global Cellular SUMOylation. Mol. Cell. Proteom. 2018, 17, 1196–1208. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.-S.; Xu, Z.-X.; Hittelman, W.N.; Salomoni, P.; Pandolfi, P.P.; Chang, K.-S. Promyelocytic Leukemia Protein Sensitizes Tumor Necrosis Factor alpha-Induced Apoptosis by Inhibiting the NF-kB Survival Pathway. J. Biol. Chem. 2003, 278, 12294–12304. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.; Wan, X.; Mitxitorena, I.; Lindsay, A.J.; Paolo Pandolfi, P.; McCaffrey, M.W.; Keeshan, K.; Chen, Y.H.; Carmody, R.J. Regulation of NF-κB by PML and PML-RARα. Sci. Rep. 2017, 7, 44539. [Google Scholar] [CrossRef] [PubMed]
- Niwa-Kawakita, M.; Ferhi, O.; Soilihi, H.; Le Bras, M.; Lallemand-Breitenbach, V.; de Thé, H. PML is a ROS sensor activating p53 upon oxidative stress. J. Exp. Med. 2017, 214, 3197–3206. [Google Scholar] [CrossRef] [PubMed]
- Chae, J.J.; Wood, G.; Richard, K.; Jaffe, H.; Colburn, N.T.; Masters, S.L.; Gumucio, D.L.; Shoham, N.G.; Kastner, D.L. The familial Mediterranean fever protein, pyrin, is cleaved by caspase-1 and activates NF-kappaB through its N-terminal fragment. Blood 2008, 112, 1794–1803. [Google Scholar] [CrossRef]
- Zhang, Z.; Bao, M.; Lu, N.; Weng, L.; Yuan, B.; Liu, Y.J. The E3 ubiquitin ligase TRIM21 negatively regulates the innate immune response to intracellular double-stranded DNA. Nat. Immunol. 2013, 14, 172–178. [Google Scholar] [CrossRef]
- Xue, B.; Li, H.; Guo, M.; Wang, J.; Xu, Y.; Zou, X.; Deng, R.; Li, G.; Zhu, H. TRIM21 Promotes Innate Immune Response to RNA Viral Infection through Lys27-Linked Polyubiquitination of MAVS. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Young, J.A.; Sermwittayawong, D.; Kim, H.J.; Nandu, S.; An, N.; Erdjument-Bromage, H.; Tempst, P.; Coscoy, L.; Winoto, A. Fas-associated death domain (FADD) and the E3 ubiquitin-protein ligase TRIM21 interact to negatively regulate virus-induced interferon production. J. Biol. Chem. 2011, 286, 6521–6531. [Google Scholar] [CrossRef]
- McEwan, W.A.; Tam, J.C.H.; Watkinson, R.E.; Bidgood, S.R.; Mallery, D.L.; James, L.C. Intracellular antibody-bound pathogens stimulate immune signaling via the Fc receptor TRIM21. Nat. Immunol. 2013, 14, 327–336. [Google Scholar] [CrossRef]
- Geijtenbeek, T.B.; Gringhuis, S.I. An inside job for antibodies: Tagging pathogens for intracellular sensing. Nat. Immunol. 2013, 14, 309–311. [Google Scholar] [CrossRef]
- Wada, K.; Niida, M.; Tanaka, M.; Kamitani, T. Ro52-mediated monoubiquitination of IKKβ down-regulates NF-κB signalling. J Biochem. 2009, 146, 821–832. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Shi, H.X.; Liu, X.Y.; Shan, Y.F.; Wei, B.; Chen, S.; Wang, C. TRIM21 is essential to sustain IFN Regulatory Factor 3 activation during antiviral response. J. Immunol. 2009, 182, 3782–3792. [Google Scholar] [CrossRef] [PubMed]
- Stacey, K.B.; Breen, E.; Jefferies, C.A. Tyrosine phosphorylation of the E3 ubiquitin ligase TRIM21 positively regulates interaction with IRF3 and hence TRIM21 activity. PLoS ONE 2012, 7, e34041. [Google Scholar] [CrossRef] [PubMed]
- Higgs, R.; Gabhann, J.N.; Larbi, N.B.; Breen, E.P.; Fitzgerald, K.A.; Jefferies, C.A. The E3 ubiquitin ligase Ro52 negatively regulates IFN-á production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J. Immunol. 2008, 181, 1780–1786. [Google Scholar] [CrossRef]
- Kimura, T.; Jain, A.; Choi, S.W.; Mandell, M.A.; Johansen, T.; Deretic, V. TRIM-directed selective autophagy regulates immune activation. Autophagy 2017, 13, 989–990. [Google Scholar] [CrossRef] [PubMed]
- Lazzari, E.; Korczeniewska, J.; Ni, G.J.; Smith, S.; Barnes, B.J.; Jefferies, C.A. TRIM21 Differentially Regulates the Stability of Interferon Regulatory Factor 5 (IRF5) Isoforms. PLoS ONE 2014, 9, e103609. [Google Scholar] [CrossRef]
- Higgs, R.; Lazzari, E.; Wynne, C.; NÃ Gabhann, J.; Espinosa, A.; Wahren-Herlenius, M.; Jefferies, C.A. Self protection from anti-Viral responses--Ro52 promotes degradation of the transcription factor IRF7 downstream of the viral toll-like receptors. PLoS ONE 2010, 5, e11776. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.J.; Anderson, D.E.; Lee, C.H.; Jang, M.K.; Tamura, T.; Tailor, P.; Cho, H.K.; Cheong, J.; Xiong, H.; Morse, H.C., III; et al. Cutting edge: Autoantigen Ro52 is an interferon inducible E3 ligase that ubiquitinates IRF8 and enhances cytokine expression in macrophages. J. Immunol. 2007, 179, 26–30. [Google Scholar] [CrossRef]
- Vicenzi, E.; Poli, G. The interferon-stimulated gene TRIM22: A double-edged sword in HIV-1 infection. Cytokine Growth Factor Rev. 2018, 40, 40–47. [Google Scholar] [CrossRef]
- Di Pietro, A.; Kajaste-Rudnitski, A.; Oteiza, A.; Nicora, L.; Towers, G.J.; Mechti, N.; Vicenzi, E. TRIM22 Inhibits Influenza A Virus Infection by Targeting the Viral Nucleoprotein for Degradation. J. Virol. 2013, 87, 4523–4533. [Google Scholar] [CrossRef]
- Yang, C.; Zhao, X.; Sun, D.; Yang, L.; Chong, C.; Pan, Y.; Chi, X.; Gao, Y.; Wang, M.; Shi, X.; et al. Interferon alpha (IFNα)-induced TRIM22 interrupts HCV replication by ubiquitinating NS5A. Cell. Mol. Immunol. 2016, 13, 94–102. [Google Scholar] [CrossRef]
- Qiu, H.; Huang, F.; Xiao, H.; Sun, B.; Yang, R. TRIM22 inhibits the TRAF6-stimulated NF-κB pathway by targeting TAB2 for degradation. Virol. Sin. 2013, 28, 209–215. [Google Scholar] [CrossRef]
- Arimoto, K.i.; Funami, K.; Saeki, Y.; Tanaka, K.; Okawa, K.; Takeuchi, O.; Akira, S.; Murakami, Y.; Shimotohno, K. Polyubiquitin conjugation to NEMO by triparite motif protein 23 (TRIM23) is critical in antiviral defense. Proc. Natl. Acad. Sci. USA 2010, 107, 15856–15861. [Google Scholar] [CrossRef]
- Poole, E.; Groves, I.; MacDonald, A.; Pang, Y.; Alcami, A.; Sinclair, J. Identification of TRIM23 as a cofactor involved in the regulation of NF-kappaB by human cytomegalovirus. J. Virol. 2009, 83, 3581–3590. [Google Scholar] [CrossRef]
- Sparrer, K.M.J.; Gableske, S.; Zurenski, M.A.; Parker, Z.M.; Full, F.; Baumgart, G.J.; Kato, J.; Pacheco-Rodriguez, G.; Liang, C.; Pornillos, O.; et al. TRIM23 mediates virus-induced autophagy via activation of TBK1. Nat. Microbiol. 2017, 2, 1543–1557. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Yu, T.; Gan, S.; Wang, Y.; Pei, Y.; Zhao, Q.; Pei, S.; Hao, S.; Yuan, J.; Xu, J.; et al. TRIM24 facilitates antiviral immunity through mediating K63-linked TRAF3 ubiquitination. J. Exp. Med. 2020, 217. [Google Scholar] [CrossRef] [PubMed]
- Tisserand, J.; Khetchoumian, K.; Thibault, C.; Dembélé, D.; Chambon, P.; Losson, R. TRIM24 (Trim24/Tif1α) Tumor Suppressor Protein Is a Novel Negative Regulator of Interferon (IFN)/Signal Transducers and Activators of Transcription (STAT) Signaling Pathway Acting through Retinoic Acid Receptor α (Rarα) Inhibition. J. Biol. Chem. 2011, 286, 33369–33379. [Google Scholar] [CrossRef] [PubMed]
- Allton, K.; Jain, A.K.; Herz, H.-M.; Tsai, W.-W.; Jung, S.Y.; Qin, J.; Bergmann, A.; Johnson, R.L.; Barton, M.C. Trim24 targets endogenous p53 for degradation. Proc. Natl. Acad. Sci. USA 2009, 106, 11612–11616. [Google Scholar] [CrossRef]
- Meyerson, N.R.; Zhou, L.; Guo, Y.R.; Zhao, C.; Tao, Y.J.; Krug, R.M.; Sawyer, S.L. Nuclear TRIM25 Specifically Targets Influenza Virus Ribonucleoproteins to Block the Onset of RNA Chain Elongation. Cell Host Microbe 2017, 22, 627–638.e627. [Google Scholar] [CrossRef]
- Gack, M.U.; Shin, Y.C.; Joo, C.H.; Urano, T.; Liang, C.; Sun, L.; Takeuchi, O.; Akira, S.; Chen, Z.; Inoue, S.; et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 2007, 446, 916–920. [Google Scholar] [CrossRef] [PubMed]
- Oshiumi, H.; Matsumoto, M.; Hatakeyama, S.; Seya, T. Riplet/RNF135, a RING-finger protein, ubiquitinates RIG-I to promote interferon-beta induction during the early phase of viral infection. J. Biol. Chem. 2009, 284, 807–817. [Google Scholar] [CrossRef] [PubMed]
- Castanier, C.; Zemirli, N.; Portier, A.; Garcin, D.; Bidère, N.; Vazquez, A.; Arnoult, D. MAVS ubiquitination by the E3 ligase TRIM25 and degradation by the proteasome is involved in type I interferon production after activation of the antiviral RIG-I-like receptors. BMC Biol. 2012, 10, 44. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.; Zhang, D.-E. The Interferon-inducible Ubiquitin-protein Isopeptide Ligase (E3) EFP Also Functions as an ISG15 E3 Ligase. J. Biol. Chem. 2006, 281, 3989–3994. [Google Scholar] [CrossRef]
- Zheng, X.; Wang, X.; Tu, F.; Wang, Q.; Fan, Z.; Gao, G. TRIM25 Is Required for the Antiviral Activity of Zinc Finger Antiviral Protein. J. Virol. 2017, 91, e00088-17. [Google Scholar] [CrossRef]
- Ran, Y.; Zhang, J.; Liu, L.L.; Pan, Z.Y.; Nie, Y.; Zhang, H.Y.; Wang, Y.Y. Autoubiquitination of TRIM26 links TBK1 to NEMO in RLR-mediated innate antiviral immune response. J. Mol. Cell Biol. 2016, 8, 31–43. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhao, W.; Zhao, K.; Zhang, L.; Gao, C. TRIM26 Negatively Regulates Interferon-β Production and Antiviral Response through Polyubiquitination and Degradation of Nuclear IRF3. PLoS Pathog. 2015, 11, e1004726. [Google Scholar] [CrossRef]
- Cai, J.; Chen, H.Y.; Peng, S.J.; Meng, J.L.; Wang, Y.; Zhou, Y.; Qian, X.P.; Sun, X.Y.; Pang, X.W.; Zhang, Y.; et al. USP7-TRIM27 axis negatively modulates antiviral type I IFN signaling. FASEB J. 2018, 32, 5238–5249. [Google Scholar] [CrossRef]
- Zheng, F.; Xu, N.; Zhang, Y. TRIM27 Promotes Hepatitis C Virus Replication by Suppressing Type I Interferon Response. Inflammation 2019, 42, 1317–1325. [Google Scholar] [CrossRef]
- Zheng, Q.; Hou, J.; Zhou, Y.; Yang, Y.; Xie, B.; Cao, X. Siglec1 suppresses antiviral innate immune response by inducing TBK1 degradation via the ubiquitin ligase TRIM27. Cell Res. 2015, 25, 1121–1136. [Google Scholar] [CrossRef]
- Zha, J.; Han, K.-J.; Xu, L.-G.; He, W.; Zhou, Q.; Chen, D.; Zhai, Z.; Shu, H.-B. The Ret Finger Protein Inhibits Signaling Mediated by the Noncanonical and Canonical IκB Kinase Family Members. J. Immunol. 2006, 176, 1072–1080. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; Deng, H.; Li, X.; Wu, X.; Tang, Q.; Chang, T.H.; Peng, H.; Rauscher, F.J.; Ozato, K.; Zhu, F. TRIM28 is a small ubiquitin-related modifier E3 ligase and negative regulator of IFN Regulatory Factor 7. J. Immunol. 2011, 187, 4754–4763. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Lin, L.; Tong, Y.; Liu, Y.; Mou, J.; Wang, X.; Wang, X.; Gong, Y.; Zhao, Y.; Liu, Y.; et al. TRIM29 negatively controls antiviral immune response through targeting STING for degradation. Cell Discov. 2018, 4, 13. [Google Scholar] [CrossRef]
- Xing, J.; Zhang, A.; Zhang, H.; Wang, J.; Li, X.C.; Zeng, M.-S.; Zhang, Z. TRIM29 promotes DNA virus infections by inhibiting innate immune response. Nat. Commun. 2017, 8, 945. [Google Scholar] [CrossRef]
- Xing, J.; Zhang, A.; Minze, L.J.; Li, X.C.; Zhang, Z. TRIM29 Negatively Regulates the Type I IFN Production in Response to RNA Virus. J. Immunol. 2018, 201, 183–192. [Google Scholar] [CrossRef]
- Xing, J.; Weng, L.; Yuan, B.; Wang, Z.; Jia, L.; Jin, R.; Lu, H.; Li, X.C.; Liu, Y.-J.; Zhang, Z. Identification of a role for TRIM29 in the control of innate immunity in the respiratory tract. Nat. Immunol. 2016, 17, 1373–1380. [Google Scholar] [CrossRef]
- Wang, Y.; Lian, Q.; Yang, B.; Yan, S.; Zhou, H.; He, L.; Lin, G.; Lian, Z.; Jiang, Z.; Sun, B. TRIM30α Is a Negative-Feedback Regulator of the Intracellular DNA and DNA Virus-Triggered Response by Targeting STING. PLoS Pathog. 2015, 11, e1005012. [Google Scholar] [CrossRef]
- Shi, M.; Deng, W.; Bi, E.; Mao, K.; Ji, Y.; Lin, G.; Wu, X.; Tao, Z.; Li, Z.; Cai, X.; et al. TRIM30à negatively regulates TLR-mediated NF- k B activation by targeting TAB2 and TAB3 for degradation. Nat. Immunol. 2008, 9, 369–377. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, M.; Chu, H.; Zhang, H.; Wu, H.; Song, G.; Wang, P.; Zhao, K.; Hou, J.; Wang, X.; et al. The ubiquitin E3 ligase TRIM31 promotes aggregation and activation of the signaling adaptor MAVS through Lys63-linked polyubiquitination. Nat. Immunol. 2017, 18, 214–224. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.; Wang, L.; Ding, H.; Schwamborn, J.C.; Li, S.; Dorf, M.E. TRIM32 Senses and Restricts Influenza A Virus by Ubiquitination of PB1 Polymerase. PLoS Pathog. 2015, 11, e1004960. [Google Scholar] [CrossRef]
- Zhang, J.; Hu, M.M.; Wang, Y.Y.; Shu, H.B. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J. Biol. Chem. 2012, 287, 28646–28655. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Liu, T.-T.; Lin, H.; Zhang, M.; Wei, J.; Luo, W.-W.; Hu, Y.-H.; Zhong, B.; Hu, M.-M.; Shu, H.-B. TRIM32-TAX1BP1-dependent selective autophagic degradation of TRIF negatively regulates TLR3/4-mediated innate immune responses. PLoS Pathog. 2017, 13, e1006600. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.; Mano, M.; Braga, L.; Naseem, A.; Marini, B.; Vu, D.M.; Collesi, C.; Meroni, G.; Lusic, M.; Giacca, M. Cellular TRIM33 restrains HIV-1 infection by targeting viral integrase for proteasomal degradation. Nat. Commun. 2019, 10, 926. [Google Scholar] [CrossRef]
- Sun, N.; Jiang, L.; Ye, M.; Wang, Y.; Wang, G.; Wan, X.; Zhao, Y.; Wen, X.; Liang, L.; Ma, S.; et al. TRIM35 mediates protection against influenza infection by activating TRAF3 and degrading viral PB2. Protein Cell 2020, 11, 894–914. [Google Scholar] [CrossRef]
- Wang, Y.; Yan, S.; Yang, B.; Wang, Y.; Zhou, H.; Lian, Q.; Sun, B. TRIM35 negatively regulates TLR7- and TLR9-mediated type I interferon production by targeting IRF7. FEBS Lett. 2015, 589, 1322–1330. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.M.; Liao, C.Y.; Yang, Q.; Xie, X.Q.; Shu, H.B. Innate immunity to RNA virus is regulated by temporal and reversible sumoylation of RIG-I and MDA5. J. Exp. Med. 2017, 214, 973–989. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.-M.; Yang, Q.; Xie, X.-Q.; Liao, C.-Y.; Lin, H.; Liu, T.-T.; Yin, L.; Shu, H.-B. Sumoylation Promotes the Stability of the DNA Sensor cGAS and the Adaptor STING to Regulate the Kinetics of Response to DNA Virus. Immunity 2016, 45, 555–569. [Google Scholar] [CrossRef]
- Zhao, W.; Wang, L.; Zhang, M.; Yuan, C.; Gao, C. E3 Ubiquitin Ligase TRIM38 Negatively Regulates TLR-Mediated Immune Responses by Proteasomal Degradation of TNF Receptor-Associated Factor 6 in Macrophages. J. Immunol. 2012, 188, 2567–2574. [Google Scholar] [CrossRef]
- Zhao, W.; Wang, L.; Zhang, M.; Wang, P.; Yuan, C.; Qi, J.; Meng, H.; Gao, C. TRIM38 negatively regulates TLR3/4- and RIG-I-mediated IFN-β production and antiviral response by targeting NAP1. J. Immunol. 2012, 188, 5311–5318. [Google Scholar] [CrossRef]
- Hu, M.M.; Xie, X.Q.; Yang, Q.; Liao, C.Y.; Ye, W.; Lin, H.; Shu, H.B. TRIM38 Negatively Regulates TLR3/4-Mediated Innate Immune and Inflammatory Responses by Two Sequential and Distinct Mechanisms. J. Immunol. 2015, 195, 4415–4425. [Google Scholar] [CrossRef]
- Xue, Q.; Zhou, Z.; Lei, X.; Liu, X.; He, B.; Wang, J.; Hung, T. TRIM38 Negatively Regulates TLR3-Mediated IFN-β Signaling by Targeting TRIF for Degradation. PLoS ONE 2012, 7, e46825. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Watanabe, M.; Nakamaru, Y.; Takagi, D.; Takahashi, H.; Fukuda, S.; Hatakeyama, S. TRIM39 negatively regulates the NFκB-mediated signaling pathway through stabilization of Cactin. Cell Mol. Life Sci. 2016, 73, 1085–1101. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Jia, M.; Song, H.; Yu, Z.; Wang, W.; Li, Q.; Zhang, L.; Zhao, W.; Cao, X. The E3 Ubiquitin Ligase TRIM40 Attenuates Antiviral Immune Responses by Targeting MDA5 and RIG-I. Cell Rep. 2017, 21, 1613–1623. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, K.; Okumura, F.; Takahashi, N.; Kataoka, A.; Kamiyama, T.; Todo, S.; Hatakeyama, S. TRIM40 promotes neddylation of IKKγ and is downregulated in gastrointestinal cancers. Carcinogenesis 2011, 32, 995–1004. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-S.; Zhang, Z.-Y.; Cai, H.; Zhao, M.; Mao, J.; Dai, J.; Xia, T.; Zhang, X.-M.; Li, T. RINCK (TRIM41)-mediated monoubiquitination of cGAS promotes antiviral innate immune responses. Cell Biosci. 2018, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Wang, J.; Wang, Y.; Zhou, H.; Wu, X.; Tian, Z.; Sun, B. Novel Function of Trim44 Promotes an Antiviral Response by Stabilizing VISA. J. Immunol. 2013, 190, 3613–3619. [Google Scholar] [CrossRef] [PubMed]
- Shibata, M.; Sato, T.; Nukiwa, R.; Ariga, T.; Hatakeyama, S. TRIM45 negatively regulates NF-κB-mediated transcription and suppresses cell proliferation. Biochem. Biophys. Res. Commun. 2012, 423, 104–109. [Google Scholar] [CrossRef]
- Liu, B.; Li, N.L.; Shen, Y.; Bao, X.; Fabrizio, T.; Elbahesh, H.; Webby, R.J.; Li, K. The C-Terminal Tail of TRIM56 Dictates Antiviral Restriction of Influenza A and B Viruses by Impeding Viral RNA Synthesis. J. Virol. 2016, 90, 4369–4382. [Google Scholar] [CrossRef]
- Seo, G.J.; Kim, C.; Shin, W.-J.; Sklan, E.H.; Eoh, H.; Jung, J.U. TRIM56-mediated monoubiquitination of cGAS for cytosolic DNA sensing. Nat. Commun. 2018, 9, 613. [Google Scholar] [CrossRef]
- Tsuchida, T.; Zou, J.; Saitoh, T.; Kumar, H.; Abe, T.; Matsuura, Y.; Kawai, T.; Akira, S. The Ubiquitin Ligase TRIM56 Regulates Innate Immune Responses to Intracellular Double-Stranded DNA. Immunity 2010, 33, 765–776. [Google Scholar] [CrossRef]
- Shen, Y.; Li, N.L.; Wang, J.; Liu, B.; Lester, S.; Li, K. TRIM56 is an essential component of the TLR3 antiviral signaling pathway. J. Biol. Chem. 2012, 287, 36404–36413. [Google Scholar] [CrossRef]
- Kondo, T.; Watanabe, M.; Hatakeyama, S. TRIM59 interacts with ECSIT and negatively regulates NF-κB and IRF-3/7-mediated signal pathways. Biochem. Biophys. Res. Commun. 2012, 422, 501–507. [Google Scholar] [CrossRef]
- Wynne, C.; Lazzari, E.; Smith, S.; McCarthy, E.M.; Joan, N.G.; Kallal, L.E.; Higgs, R.; Greco, A.; Cryan, S.A.; Biron, C.A.; et al. TRIM68 negatively regulates IFN-β production by degrading TRK fused gene, a novel driver of IFN-β downstream of anti-viral detection systems. PLoS ONE 2014, 9, e101503. [Google Scholar] [CrossRef]
- Lang, X.; Tang, T.; Jin, T.; Ding, C.; Zhou, R.; Jiang, W. TRIM65-catalized ubiquitination is essential for MDA5-mediated antiviral innate immunity. J. Exp. Med. 2017, 214, 459–473. [Google Scholar] [CrossRef] [PubMed]
- Hayman, T.J.; Hsu, A.C.; Kolesnik, T.B.; Dagley, L.F.; Willemsen, J.; Tate, M.D.; Baker, P.J.; Kershaw, N.J.; Kedzierski, L.; Webb, A.I.; et al. RIPLET, and not TRIM25, is required for endogenous RIG-I-dependent antiviral responses. Immunol. Cell Biol. 2019, 97, 840–852. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, S. Role of Post-Translational Modifications of cGAS in Innate Immunity. Int. J. Mol. Sci. 2020, 21, 7842. [Google Scholar] [CrossRef] [PubMed]
- Prabakaran, T.; Bodda, C.; Krapp, C.; Zhang, B.C.; Christensen, M.H.; Sun, C.; Reinert, L.; Cai, Y.; Jensen, S.; Skouboe, M.; et al. Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. EMBO J. 2018, 37, e97858. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P.; Strickson, S. The role of hybrid ubiquitin chains in the MyD88 and other innate immune signalling pathways. Cell Death Differ. 2017, 24, 1153–1159. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, X.; Cui, Y.; Tang, Y.; Chen, W.; Li, S.; Yu, H.; Pan, Y.; Wang, C. The E3 ubiquitin ligase AMFR and INSIG1 bridge the activation of TBK1 kinase by modifying the adaptor STING. Immunity 2014, 41, 919–933. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, L.; Jin, J.; Luan, Y.; Chen, C.; Li, Y.; Chu, H.; Wang, X.; Liao, G.; Yu, Y.; et al. p38 inhibition provides anti-DNA virus immunity by regulation of USP21 phosphorylation and STING activation. J. Exp. Med. 2017, 214, 991–1010. [Google Scholar] [CrossRef]
- Hacker, H.; Redecke, V.; Blagoev, B.; Kratchmarova, I.; Hsu, L.C.; Wang, G.G.; Kamps, M.P.; Raz, E.; Wagner, H.; Hacker, G.; et al. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature 2006, 439, 204–207. [Google Scholar] [CrossRef]
- Oganesyan, G.; Saha, S.K.; Guo, B.; He, J.Q.; Shahangian, A.; Zarnegar, B.; Perry, A.; Cheng, G. Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature 2006, 439, 208–211. [Google Scholar] [CrossRef] [PubMed]
- Tseng, P.-H.; Matsuzawa, A.; Zhang, W.; Mino, T.; Vignali, D.A.A.; Karin, M. Different modes of ubiquitination of the adaptor TRAF3 selectively activate the expression of type I interferons and proinflammatory cytokines. Nat. Immunol. 2010, 11, 70–75. [Google Scholar] [CrossRef]
- Li, F.; Li, Y.; Liang, H.; Xu, T.; Kong, Y.; Huang, M.; Xiao, J.; Chen, X.; Xia, H.; Wu, Y.; et al. HECTD3 mediates TRAF3 polyubiquitination and type I interferon induction during bacterial infection. J. Clin. Investig. 2018, 128, 4148–4162. [Google Scholar] [CrossRef] [PubMed]
- Lei, C.Q.; Zhong, B.; Zhang, Y.; Zhang, J.; Wang, S.; Shu, H.B. Glycogen synthase kinase 3β regulates IRF3 transcription factor-mediated antiviral response via activation of the kinase TBK1. Immunity 2010, 33, 878–889. [Google Scholar] [CrossRef]
- Khan, K.A.; Marineau, A.; Doyon, P.; Acevedo, M.; Durette, É.; Gingras, A.C.; Servant, M.J. TRK-Fused Gene (TFG), a protein involved in protein secretion pathways, is an essential component of the antiviral innate immune response. PLoS Pathog. 2021, 17, e1009111. [Google Scholar] [CrossRef]
- Sjöstrand, M.; Ambrosi, A.; Brauner, S.; Sullivan, J.; Malin, S.; Kuchroo, V.K.; Espinosa, A.; Wahren-Herlenius, M. Expression of the immune regulator tripartite-motif 21 is controlled by IFN regulatory factors. J. Immunol. 2013, 191, 3753–3763. [Google Scholar] [CrossRef] [PubMed]
- Tun-Kyi, A.; Finn, G.; Greenwood, A.; Nowak, M.; Lee, T.H.; Asara, J.M.; Tsokos, G.C.; Fitzgerald, K.; Israel, E.; Li, X.; et al. Essential role for the prolyl isomerase Pin1 in Toll-like receptor signaling and type I interferon-mediated immunity. Nat. Immunol. 2011, 12, 733–741. [Google Scholar] [CrossRef]
- Yu, C.-F.; Peng, W.-M.; Schlee, M.; Barchet, W.; Eis-Hübinger, A.M.; Kolanus, W.; Geyer, M.; Schmitt, S.; Steinhagen, F.; Oldenburg, J.; et al. SOCS1 and SOCS3 Target IRF7 Degradation To Suppress TLR7-Mediated Type I IFN Production of Human Plasmacytoid Dendritic Cells. J. Immunol. 2018, 200, 4024–4035. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, K.; Yokosawa, H. PIAS3 induces SUMO-1 modification and transcriptional repression of IRF1. FEBS Lett. 2002, 530, 204–208. [Google Scholar] [CrossRef]
- Ng, S.L.; Friedman, B.A.; Schmid, S.; Gertz, J.; Myers, R.M.; tenOever, B.R.; Maniatis, T. IKKe regulates the balance between type I and type II interferon responses. Proc. Natl. Acad. Sci. USA 2011, 108, 21170–21175. [Google Scholar] [CrossRef]
- Everett, R.D.; Boutell, C.; Hale, B.G. Interplay between viruses and host sumoylation pathways. Nat. Rev. Microbiol. 2013, 11, 400–411. [Google Scholar] [CrossRef] [PubMed]
- Hannoun, Z.; Maarifi, G.; Chelbi-Alix, M.K. The implication of SUMO in intrinsic and innate immunity. Cytokine Growth Factor Rev. 2016, 29, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Harty, R.N.; Pitha, P.M.; Okumura, A. Antiviral activity of innate immune protein ISG15. J. Innate Immun. 2009, 1, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Freitas, B.T.; Scholte, F.E.M.; Bergeron, É.; Pegan, S.D. How ISG15 combats viral infection. Virus Res. 2020, 286, 198036. [Google Scholar] [CrossRef]
- El-Asmi, F.; McManus, F.P.; Brantis-de-Carvalho, C.E.; Valle-Casuso, J.C.; Thibault, P.; Chelbi-Alix, M.K. Cross-talk between SUMOylation and ISGylation in response to interferon. Cytokine 2020, 129, 155025. [Google Scholar] [CrossRef] [PubMed]
- Takaoka, A.; Hayakawa, S.; Yanai, H.; Stoiber, D.; Negishi, H.; Kikuchi, H.; Sasaki, S.; Imai, K.; Shibue, T.; Honda, K.; et al. Integration of interferon-à/á signalling to p53 responses in tumour suppression and antiviral defence. Nature 2003, 424, 516–523. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.K.; Allton, K.; Duncan, A.D.; Barton, M.C. TRIM24 is a p53-induced E3-ubiquitin ligase that undergoes ATM-mediated phosphorylation and autodegradation during DNA damage. Mol. Cell. Biol. 2014, 34, 2695–2709. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, C.; Wang, X.; Hu, W.; Feng, Z. Tumor suppressor p53 cross-talks with TRIM family proteins. Genes Dis. 2020. [Google Scholar] [CrossRef]
- Gupta, S.; Ylä-Anttila, P.; Callegari, S.; Tsai, M.-H.; Delecluse, H.-J.; Masucci, M.G. Herpesvirus deconjugases inhibit the IFN response by promoting TRIM25 autoubiquitination and functional inactivation of the RIG-I signalosome. PLoS Pathog. 2018, 14, e1006852. [Google Scholar] [CrossRef]
- Gupta, S.; Ylä-Anttila, P.; Sandalova, T.; Achour, A.; Masucci, M.G. Interaction With 14-3-3 Correlates With Inactivation of the RIG-I Signalosome by Herpesvirus Ubiquitin Deconjugases. Front. Immunol. 2020, 11, 437. [Google Scholar] [CrossRef]
- Gupta, S.; Ylä-Anttila, P.; Sandalova, T.; Sun, R.; Achour, A.; Masucci, M.G. 14-3-3 scaffold proteins mediate the inactivation of trim25 and inhibition of the type I interferon response by herpesvirus deconjugases. PLoS Pathog. 2019, 15, e1008146. [Google Scholar] [CrossRef]
- Bodda, C.; Reinert, L.S.; Fruhwürth, S.; Richardo, T.; Sun, C.; Zhang, B.-c.; Kalamvoki, M.; Pohlmann, A.; Mogensen, T.H.; Bergström, P.; et al. HSV1 VP1-2 deubiquitinates STING to block type I interferon expression and promote brain infection. J. Exp. Med. 2020, 217. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, S.; Liu, L.; Wang, H.; Yang, B. HTLV-1 Tax impairs K63-linked ubiquitination of STING to evade host innate immunity. Virus Res. 2017, 232, 13–21. [Google Scholar] [CrossRef]
- Liu, Y.; Li, J.; Chen, J.; Li, Y.; Wang, W.; Du, X.; Song, W.; Zhang, W.; Lin, L.; Yuan, Z. Hepatitis B Virus Polymerase Disrupts K63-Linked Ubiquitination of STING To Block Innate Cytosolic DNA-Sensing Pathways. J. Virol. 2015, 89, 2287–2300. [Google Scholar] [CrossRef]
- Daubeuf, S.; Singh, D.; Tan, Y.; Liu, H.; Federoff, H.J.; Bowers, W.J.; Tolba, K. HSV ICP0 recruits USP7 to modulate TLR-mediated innate response. Blood 2009, 113, 3264–3275. [Google Scholar] [CrossRef] [PubMed]
- Conwell, S.E.; White, A.E.; Harper, J.W.; Knipe, D.M. Identification of TRIM27 as a Novel Degradation Target of Herpes Simplex Virus 1 ICP0. J. Virol. 2015, 89, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Shahnazaryan, D.; Khalil, R.; Wynne, C.; Jefferies, C.A.; Joan, N.G.-D.; Murphy, C.C. Herpes simplex virus 1 targets IRF7 via ICP0 to limit type I IFN induction. Sci. Rep. 2020, 10, 22216. [Google Scholar] [CrossRef]
- Orzalli, M.H.; DeLuca, N.A.; Knipe, D.M. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc. Natl. Acad. Sci. USA 2012, 109, E3008–E3017. [Google Scholar] [CrossRef]
- Lee, H.R.; Choi, W.C.; Lee, S.; Hwang, J.; Hwang, E.; Guchhait, K.; Haas, J.; Toth, Z.; Jeon, Y.H.; Oh, T.K.; et al. Bilateral inhibition of HAUSP deubiquitinase by a viral interferon regulatory factor protein. Nat. Struct. Mol. Biol. 2011, 18, 1336–1344. [Google Scholar] [CrossRef]
- Sivachandran, N.; Sarkari, F.; Frappier, L. Epstein-Barr Nuclear Antigen 1 contributes to nasopharyngeal carcinoma through disruption of PML nuclear bodies. PLoS Pathog. 2008, 4, e1000170. [Google Scholar] [CrossRef] [PubMed]
- Kubota, T.; Matsuoka, M.; Chang, T.H.; Tailor, P.; Sasaki, T.; Tashiro, M.; Kato, A.; Ozato, K. Virus infection triggers SUMOylation of IRF3 and IRF7, leading to the negative regulation of type I interferon gene expression. J. Biol. Chem. 2008, 283, 25660–25670. [Google Scholar] [CrossRef] [PubMed]
- Ran, Y.; Liu, T.T.; Zhou, Q.; Li, S.; Mao, A.P.; Li, Y.; Liu, L.J.; Cheng, J.K.; Shu, H.B. SENP2 negatively regulates cellular antiviral response by deSUMOylating IRF3 and conditioning it for ubiquitination and degradation. J. Mol. Cell Biol. 2011, 3, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Selby, T.L.; Biel, N.; Varn, M.; Patel, S.; Patel, A.; Hilding, L.; Ray, A.; Ross, T.; Cramblet, W.T.; Moss, C.R.; et al. The Epstein-Barr Virus Oncoprotein, LMP1, Regulates the Function of SENP2, a SUMO-protease. Sci. Rep. 2019, 9, 9523. [Google Scholar] [CrossRef] [PubMed]
- Ning, S.; Pagano, J. The A20 deubiquitinase activity negatively regulates LMP1 activation of IRF7. J. Virol. 2010, 84, 6130–6138. [Google Scholar] [CrossRef] [PubMed]
- Ning, S.; Campos, A.D.; Darnay, B.; Bentz, G.; Pagano, J.S. TRAF6 and the three C-terminal lysine sites on IRF7 are required for its ubiquitination-mediated activation by the Tumor Necrosis Factor Receptor family member Latent Membrane Protein 1. Mol. Cell. Biol. 2008, 28, 6536–6546. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Y.; Zhao, J.; Ren, J.; Hall, K.H.; Moorman, J.P.; Yao, Z.Q.; Ning, S. LUBAC modulates LMP1 activation of NFκB and IRF7. J. Virol. 2017, 91. [Google Scholar]
- Everett, R.D.; Chelbi-Alix, M.K. PML and PML nuclear bodies: Implications in antiviral defence. Biochimie 2007, 89, 819–830. [Google Scholar] [CrossRef]
- Zakaryan, H.; Stamminger, T. Nuclear remodelling during viral infections. Cell. Microbiol. 2011, 13, 806–813. [Google Scholar] [CrossRef]
- Boutell, C.; Cuchet-Lourenço, D.; Vanni, E.; Orr, A.; Glass, M.; McFarlane, S.; Everett, R.D. A viral ubiquitin ligase has substrate preferential SUMO targeted ubiquitin ligase activity that counteracts intrinsic antiviral defence. PLoS Pathog. 2011, 7, e1002245. [Google Scholar] [CrossRef]
- Wang, S.; Long, J.; Zheng, C. The potential link between PML NBs and ICP0 in regulating lytic and latent infection of HSV-1. Protein Cell 2020, 3, 372–382. [Google Scholar] [CrossRef] [PubMed]
- Sivachandran, N.; Cao, J.Y.; Frappier, L. Epstein-Barr Nuclear Antigen 1 hijacks the host kinase CK2 to disrupt PML nuclear bodies. J. Virol. 2010, 84, 11113–11123. [Google Scholar] [CrossRef]
- Marcos-Villar, L.; Lopitz-Otsoa, F.; Gallego, P.; Muñoz-Fontela, C.; González-Santamaría, J.; Campagna, M.; Shou-Jiang, G.; Rodriguez, M.S.; Rivas, C. Kaposi’s Sarcoma-Associated Herpesvirus Protein LANA2 Disrupts PML Oncogenic Domains and Inhibits PML-Mediated Transcriptional Repression of the Survivin Gene. J. Virol. 2009, 83, 8849–8858. [Google Scholar] [CrossRef]
- Guion, L.G.; Sapp, M. The Role of Promyelocytic Leukemia Nuclear Bodies During HPV Infection. Front. Cell. Infect. Microbiol. 2020, 10, 35. [Google Scholar] [CrossRef]
- El McHichi, B.; Regad, T.; Maroui, M.-A.; Rodriguez, M.S.; Aminev, A.; Gerbaud, S.; Escriou, N.; Dianoux, L.; Chelbi-Alix, M.K. SUMOylation promotes PML degradation during encephalomyocarditis virus infection. J. Virol. 2010, 84, 11634–11645. [Google Scholar] [CrossRef] [PubMed]
- Blondel, D.; Regad, T.; Poisson, N.; Pavie, B.; Harper, F.; Pandolfi, P.P.; de Thé, H.; Chelbi-Alix, M.K. Rabies virus P and small P products interact directly with PML and reorganize PML nuclear bodies. Oncogene 2002, 21, 7957–7970. [Google Scholar] [CrossRef]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, Inflammation, and Cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef]
- Mantovani, A.; Allavena, P.; Sica, A.; Balkwill, F. Cancer-related inflammation. Nature 2008, 454, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A. Cancer: Inflaming metastasis. Nature 2009, 457, 36–37. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Taniguchi, K.; Karin, M. NF-κB, inflammation, immunity and cancer: Coming of age. Nat. Rev. Immunol. 2018, 18, 309–324. [Google Scholar] [CrossRef]
- Ng, K.W.; Marshall, E.A.; Bell, J.C.; Lam, W.L. cGAS–STING and Cancer: Dichotomous Roles in Tumor Immunity and Development. Trends Immunol. 2018, 39, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Harding, S.M.; Benci, J.L.; Irianto, J.; Discher, D.E.; Minn, A.J.; Greenberg, R.A. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 2017, 548, 466–470. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Chen, Z.J. The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J. Exp. Med. 2018, 215, 1287–1299. [Google Scholar] [CrossRef]
- Hong, S.; Li, Y.; Kaminski, P.J.; Andrade, J.; Laimins, L.A. Pathogenesis of Human Papillomaviruses Requires the ATR/p62 Autophagy-Related Pathway. mBio 2020, 11, e01628-20. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Howell, M.E.A.; Sparks-Wallace, A.; Hawkins, C.; Nicksic, C.; Kohne, C.; Moorman, J.P.; Yao, Z.Q.; Ning, S. p62-mediated Selective Autophagy Endows Virus-transformed Cells with Insusceptibility to DNA Damage under Oxidative Stress. PLoS Pathog. 2019, 15, e1007541. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.; Pauli, E.-K.; Biryukov, J.; Feister, K.F.; Meng, M.; White, E.A.; Münger, K.; Howley, P.M.; Meyers, C.; Gack, M.U. The Human Papillomavirus E6 Oncoprotein Targets USP15 and TRIM25 To Suppress RIG-I-Mediated Innate Immune Signaling. J. Virol. 2018, 92, e01737-17. [Google Scholar] [CrossRef]
- Poirson, J.; Biquand, E.; Straub, M.L.; Cassonnet, P.; Nominé, Y.; Jones, L.; van der Werf, S.; Travé, G.; Zanier, K.; Jacob, Y.; et al. Mapping the interactome of HPV E6 and E7 oncoproteins with the ubiquitin-proteasome system. FEBS J. 2017, 284, 3171–3201. [Google Scholar] [CrossRef] [PubMed]
- Inn, K.S.; Gack, M.U.; Tokunaga, F.; Shi, M.; Wong, L.Y.; Iwai, K.; Jung, J.U. Linear ubiquitin assembly complex negatively regulates RIG-I- and TRIM25-mediated type I interferon induction. Mol. Cell 2011, 41, 354–365. [Google Scholar] [CrossRef] [PubMed]
- Bharaj, P.; Wang, Y.E.; Dawes, B.E.; Yun, T.E.; Park, A.; Yen, B.; Basler, C.F.; Freiberg, A.N.; Lee, B.; Rajsbaum, R. The Matrix Protein of Nipah Virus Targets the E3-Ubiquitin Ligase TRIM6 to Inhibit the IKKε Kinase-Mediated Type-I IFN Antiviral Response. PLoS Pathog. 2016, 12, e1005880. [Google Scholar] [CrossRef]
- Röth, S.; Fulcher, L.J.; Sapkota, G.P. Advances in targeted degradation of endogenous proteins. Cell. Mol. Life Sci. 2019, 76, 2761–2777. [Google Scholar] [CrossRef]
- Sparrer, K.M.J.; Gack, M.U. TRIM proteins: New players in virus-induced autophagy. PLoS Pathog. 2018, 14, e1006787. [Google Scholar] [CrossRef] [PubMed]
- Shibutani, S.T.; Saitoh, T.; Nowag, H.; Munz, C.; Yoshimori, T. Autophagy and autophagy-related proteins in the immune system. Nat. Immunol. 2015, 16, 1014–1024. [Google Scholar] [CrossRef]
- Kuballa, P.; Nolte, W.M.; Castoreno, A.B.; Xavier, R.J. Autophagy and the Immune System. Annu. Rev. Immunol. 2012, 30, 611–646. [Google Scholar] [CrossRef] [PubMed]
- Germic, N.; Frangez, Z.; Yousefi, S.; Simon, H.-U. Regulation of the innate immune system by autophagy: Monocytes, macrophages, dendritic cells and antigen presentation. Cell Death Differ. 2019, 26, 715–727. [Google Scholar] [CrossRef]
- Jiang, G.-M.; Tan, Y.; Wang, H.; Peng, L.; Chen, H.-T.; Meng, X.-J.; Li, L.-L.; Liu, Y.; Li, W.-F.; Shan, H. The relationship between autophagy and the immune system and its applications for tumor immunotherapy. Mol. Cancer 2019, 18, 17. [Google Scholar] [CrossRef]
- Towers, C.G.; Wodetzki, D.; Thorburn, A. Autophagy and cancer: Modulation of cell death pathways and cancer cell adaptations. J. Cell Biol. 2019, 219. [Google Scholar] [CrossRef]
- Li, X.; He, S.; Ma, B. Autophagy and autophagy-related proteins in cancer. Mol. Cancer 2020, 19, 12. [Google Scholar] [CrossRef]
- Amaravadi, R.K.; Kimmelman, A.C.; Debnath, J. Targeting Autophagy in Cancer: Recent Advances and Future Directions. Cancer Discov. 2019, 9, 1167–1181. [Google Scholar] [CrossRef] [PubMed]
- Tao, S.; Drexler, I. Targeting Autophagy in Innate Immune Cells: Angel or Demon During Infection and Vaccination? Front. Immunol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]



| TRIM | Synonym | Targets in the IFN-I Network | Ub Conjugation Type | Outcome of Conjugation | Selected References |
|---|---|---|---|---|---|
| TRIM3 | RNF97 | TLR3 | K63 | Promotes ESCRT-mediated TLR3 sorting to endosomes | [59] |
| TRIM4 | RNF87 | RIG-I | K63 | Activation | [60] |
| TRIM5α | RNF88 | HIV Gag | K48 | Degradation | [61,62] |
| TAK1 | K63 | Activation | [63] | ||
| TRIM12c in mice | TRAF6 | K63 (?) | Activation | [62] | |
| TRIM6 | RNF89 | Ebola VP35 | Poly | Promotes VP35 IFN-I inhibitory activity | [64] |
| IKKε | Free K48 | Activation of IKKε, leading to STAT1 activation | [65] | ||
| TRIM7 | RNF90 GNIP | Zika virus envelope (E) | K63 | Enhances virus attachment and entry into the cell | [66] |
| TLR4 | NA* | Promotes TLR4 activation | [67] | ||
| TRIM8 | RNF27 GERP | TRIF | K6, K33 | Disrupts the TRIF-TBK1 complex | [68] |
| TAK1 | K63 | Activation | [69,70] | ||
| IRF7 | Protects p-IRF7 from Pin1-mediated proteasomal degradation in the nucleus | [71] | |||
| SOCS1 | K48 (?) | Degradation | [72] | ||
| PIAS3 | K48 | Degradation | [73] | ||
| Interaction (?) | Promotes PIAS3 nucleus-to-cytoplasm translocation | [74] | |||
| TRIM9s | RNF91 SPRING | TBK1 | Interaction | Recruits GSK3β and TBK1, leading to TBK1 activation | [75] |
| TRIM9 | β-TrCP | Interaction | Stabilizes IκBα | [75,76] | |
| TRIM11 | RNF92 BIA1 | TBK1 | Interaction | Inhibits TBK1 activation | [77] |
| TRIM5 | NA* | Degradation | [78] | ||
| TRIM13 | RNF77 RFP2 CAR LEU5 DLEU5 | RIG-I | Interaction | Potentiates RIG-I activity | [79] |
| MDA5 | Interaction | Inhibition | [79] | ||
| TRAF6 | K29 | Activation | [80] | ||
| NEMO | K48 | Degradation | [81] | ||
| TRIM14 | KIAA0129 | HCV NS5A | K48 (?) | Degradation | [82] |
| cGAS, TBK1 | Interaction | Inhibition of autophagic degradation of cGAS | [83,84,85] | ||
| MAVS | Interaction | Recruitment of NEMO to MAVS signalosome | [84] | ||
| TRIM15 | RNF93 ZNF178 ZNFB7 | MAVS | NA* | Promotes RIG-I-mediated IFN production | [86] |
| TRIM19 | RNF71 PML MYL | HIV genome | Sequestrates HIV genome in the cytoplasm, blocking HIV transduction | [87] | |
| HFV Tas | Represses HFV transcription by preventing Tas binding to viral DNA | [88] | |||
| LCMV Z | Inhibits LCMV replication | [89] | |||
| hCMV IE1 | Interaction | IE1 forms a complex with TRIM19-STAT1/2 to impede IFN-I signaling | [90] | ||
| STAT1/2 | Induction and stabilization, promoting IFN-I signaling | [90] | |||
| Pin1 (by TRIM19IV) | Regulates the cellular distribution of Pin1 | [91] | |||
| Ubc9 (The only SUMO E2) | Required for IFN-induced global sumoylation | [92] | |||
| NFκB | Inhibits NFκB-mediated transcription and survival | [93] | |||
| Promotes IKKε-mediated p65 phosphorylation and NFκB activity | [94] | ||||
| ROS | Functions as an ROS sensor promoting p53 activation | [95] | |||
| TRIM20 | Pyrin MEFV | p65 | Interaction | Promotes p65 nuclear translocation | [96] |
| IκBα | Promotes IκBα degradation | [96] | |||
| TRIM21 | RNF81 Ro52 SSA1 | DDX41 | K48 | Degradation | [97] |
| MAVS | K27 | Activation | [98] | ||
| FADD | Interaction | Promotes IRF7 ubiquitination-mediated degradation | [99] | ||
| TAK1 | Free K63 | Activates TAK1, leading to the activation of NFκB, AP1, and IRFs | [100,101] | ||
| IKKβ | Mono-Ub | Autophagic degradation | [102] | ||
| IRF3 | Interaction | Protects p-IRF3 from Pin1-mediated proteasomal degradation | [103] | ||
| K48 | Targets IRF3 for proteosomal degradation | [104,105] | |||
| Interacts with ULK1, Beclin1, and p62 | Targets IRF3 for autophagic degradation | [106] | |||
| IRF5 | Various | Degradation of isoforms V1 and V5, but not V2 or V3 | [107] | ||
| IRF7 | K48 | Degradation | [108] | ||
| IRF8 | NA* | Activation | [109] | ||
| TRIM22 | RNF94 STAF50 | HIV Gag, LTR | Degradation | [110] | |
| Influenza A Virus NP | Degradation | [111] | |||
| HCV NS5A | K48 (?) | Degradation | [112] | ||
| TAB2 | K48 (?) | Degradation | [113] | ||
| TRIM23 | RNF46 ARD1 ARFD1 | TRAF3 | Interaction | Function not clear, likely promoting TRAF3-mediated antiviral activity | [114] |
| TRAF6 | Interaction | Activation of NFκB mediated by HCMV UL144 | [115] | ||
| NEMO | K27 | Activation | [114] | ||
| TBK1 | K27 of TRIM23 (self) | Recruits and activates TBK1, inducing TBK1-mediated autophagy | [116] | ||
| TRIM24 | RNF82 TIF1A | TRAF3 | K63 | Activation | [117] |
| RARα | Interaction | Inhibits RARα activity and retinoic acid-induced STAT1 expression | [118] | ||
| p53 | K48 (?) | Promotes p53 ubiquitination and degradation | [119] | ||
| TRIM25 | RNF147 ZNF147 | Influenza virus vRNP | Blocks vRNA chain elongation | [120] | |
| RIG-I | K63 | Activation | [121,122] | ||
| MAVS | K48 | Degradation | [123] | ||
| ISG15 | Functions as an ISG15 E3 ligase | [124] | |||
| ZAP | K48, K63 | Critical for ZAP inhibition of viral genome translation | [125] | ||
| TRIM26 | RNF95 ZNF173 AFP | TBK1 | K27 of TRIM26 (self) | Bridges TBK1-NEMO interaction, leading to TBK1 activation | [126] |
| IRF3 | K48 | Degradation | [127] | ||
| TRIM27 | RNF76 RFP | TBK1 | K48 | Degradation | [128,129,130] |
| IKKα, IKKβ, IKKε | Interaction | Inhibition | [131] | ||
| TRIM28 | RNF96 KAP1 | IRF7 | Sumoylation | Inhibition | [132] |
| TRIM29 | ATDC | STING | K48 | Degradation | [133,134] |
| MAVS | K11 | Degradation | [135] | ||
| NEMO | K48 | Degradation | [136] | ||
| TRIM30α | RPT1 | STING | K48 | Degradation | [137] |
| TAB2/3 | Lysosomal degradation | [138] | |||
| TRIM31 | RNF HCG1 | MAVS | K63 | Promotes MAVS signalosome assembly | [139] |
| TRIM32 | TATIP BBS11 HT2A | Influenza PB1 | K48 | Degradation | [140] |
| STING | K63 | Activation | [141] | ||
| TRIF | NA* | Targets TRIF for TAX1BP1-mediated autophagic degradation | [142] | ||
| TRIM33 | TIF1γ | HIV integrase | K48 | Degradation | [143] |
| TRIM35 | HLS5 MAIR | TRAF3 | K63 | Activation | [144] |
| IRF7 | K48 | Degradation | [145] | ||
| TRIM38 | RNF15 RORET | RIG-I, MDA5 | Sumoylation | Stabilization | [146] |
| cGAS, STING | Sumoylation | Stabilization | [147] | ||
| TRAF6 | K48 | Degradation | [148] | ||
| NAP1 | K48 | Degradation | [149] | ||
| TAB2 | K48? | Degradation | [150] | ||
| TRIF | K48 | Degradation | [150,151] | ||
| TRIM39 | RNF23 TFP | Cactin | NA* | Stabilizes Cactin, inhibiting NFκB and IRFs | [152] |
| TRIM40 | RNF35 | RIG-I, MDA5 | K27, K48 | Degradation | [153] |
| NEMO | Neddylation | Inhibition | [154] | ||
| TRIM41 | RINCK MGC1127 | cGAS | Mono-Ub | Activation | [155] |
| TRIM44 | DIPB AN3 | MAVS | Interaction | Stabilization of MAVS by preventing its ubiquitination | [156] |
| TRIM45 | RNF99 | NFκB | E3 ligase activity not required | Inhibition of TNFα-mediated NFκB activation | [157] |
| TRIM56 | RNF109 | Influenza virus RNA | Inhibits vRNA synthesis | [158] | |
| cGAS | Mono-Ub | Activation | [159] | ||
| STING | K63 | Activation | [160] | ||
| TRIF | Interaction | Activation | [161] | ||
| TRIM59 | RNF104 TSBF1 MRF1 IFT80L | ECSIT | Interaction | Inhibition of TLR singling pathways to activate NFκB and IRFs | [162] |
| TRIM62 | DEAR1 | TRIF | NA* | Activation | [86] |
| TRIM65 | MDA5 | K63 | Activation | ||
| TRIM68 | RNF137 SS56 | TFG | various | Induces TFG lysosomal degradation | [163] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Ning, S. TRIMming Type I Interferon-Mediated Innate Immune Response in Antiviral and Antitumor Defense. Viruses 2021, 13, 279. https://doi.org/10.3390/v13020279
Wang L, Ning S. TRIMming Type I Interferon-Mediated Innate Immune Response in Antiviral and Antitumor Defense. Viruses. 2021; 13(2):279. https://doi.org/10.3390/v13020279
Chicago/Turabian StyleWang, Ling, and Shunbin Ning. 2021. "TRIMming Type I Interferon-Mediated Innate Immune Response in Antiviral and Antitumor Defense" Viruses 13, no. 2: 279. https://doi.org/10.3390/v13020279
APA StyleWang, L., & Ning, S. (2021). TRIMming Type I Interferon-Mediated Innate Immune Response in Antiviral and Antitumor Defense. Viruses, 13(2), 279. https://doi.org/10.3390/v13020279







