First Detection of SARS-CoV-2 Delta (B.1.617.2) Variant of Concern in a Dog with Clinical Signs in Spain
Abstract
:1. Introduction
2. Materials and Methods
2.1. Clinical Evaluation and Sample Collection
2.2. RNA Extraction and Detection by RT-qPCR
2.3. SARS-CoV-2 Genome Sequencing
2.4. Neutralizing Antibody Detection by SARS-CoV-2 Receptor-Binding Inhibition ELISA
2.5. SARS-CoV-2 Pseudoneutralization Assay
2.6. SARS-CoV-2 Neutralization Assay
3. Results
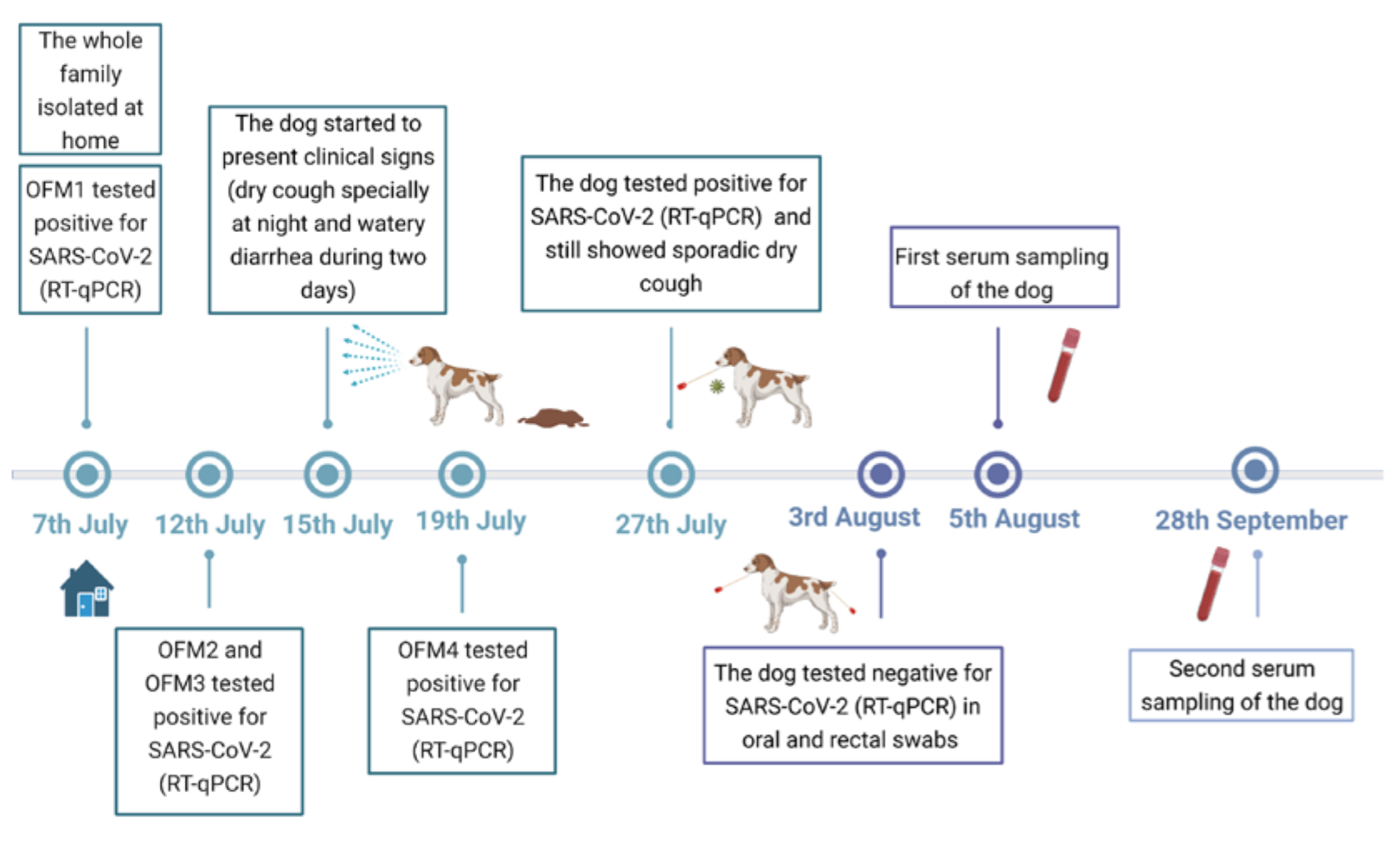
3.1. Clinical Follow-Up
3.2. RNA Detection and SARS-CoV-2 Sequencing
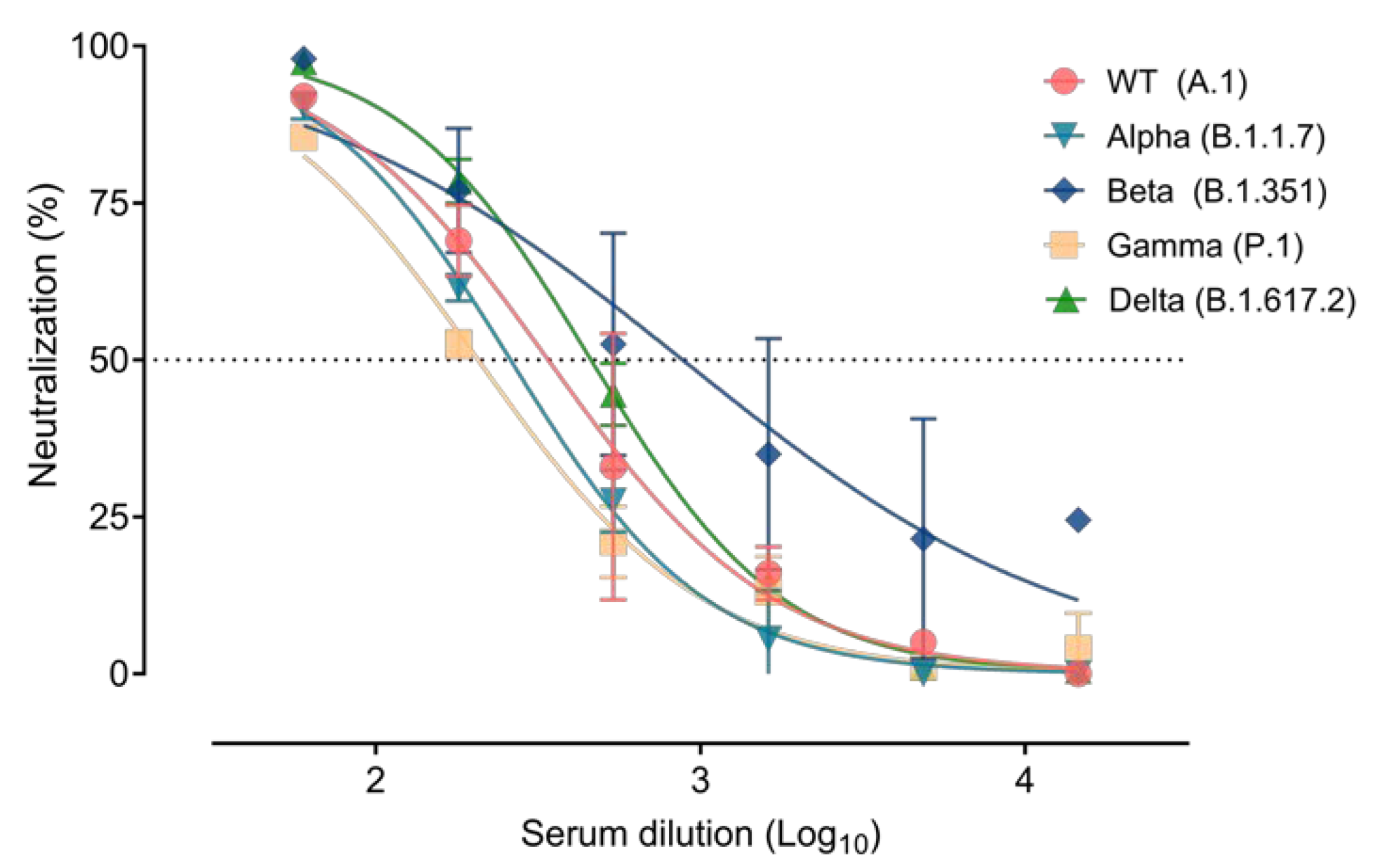
3.3. Immune Response Elicited after SARS-CoV-2 Infection
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization (WHO). Official COVID-19 Information. Available online: https://covid19.who.int/ (accessed on 15 December 2021).
- SARS-CoV-2 Variants of Concern as of 12 November 2021. Available online: https://www.ecdc.europa.eu/en/covid-19/variants-concern (accessed on 12 November 2021).
- Kirby, T. New variant of SARS-CoV-2 in UK causes surge of COVID-19. Lancet. Respir. Med. 2021, 9, e20–e21. [Google Scholar] [CrossRef]
- Happi, A.N.; Ugwu, C.A.; Happi, C.T. Tracking the emergence of new SARS-CoV-2 variants in South Africa. Nat. Med. 2021, 27, 372–373. [Google Scholar] [CrossRef]
- Faria, N.R.; Claro, I.M.; Candido, D.; Moyses Franco, L.A.; Andrade, P.S.; Coletti, T.M.; Silva, C.A.M.; Sales, F.C.; Manuli, E.R.; Aguiar, R.S.; et al. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: Preliminary findings—SARS-CoV-2 coronavirus/nCoV-2019 Genomic Epidemiology. Available online: https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586 (accessed on 18 October 2021).
- Weekly Epidemiological Update on COVID-19. 2021. Available online: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---10-august-2021 (accessed on 15 December 2021).
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 2021, 592, 438–443. [Google Scholar] [CrossRef]
- Tang, J.W.; Tambyah, P.A.; Hui, D.S. Emergence of a New SARS-CoV-2 Variant in the UK. J. Infect. 2021, 82(4), e27–e28. [Google Scholar] [CrossRef]
- Sabino, E.C.; Buss, L.F.; Carvalho, M.P.S.; Prete, C.A.; Crispim, M.A.E.; Fraiji, N.A.; Pereira, R.H.M.; Parag, K.V.; da Silva Peixoto, P.; Kraemer, M.U.G.; et al. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet 2021, 397, 452–455. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [Green Version]
- Deng, J.; Jin, Y.; Liu, Y.; Sun, J.; Hao, L.; Bai, J.; Huang, T.; Lin, D.; Jin, Y.; Tian, K. Serological survey of SARS-CoV-2 for experimental, domestic, companion and wild animals excludes intermediate hosts of 35 different species of animals. Transbound. Emerg. Dis. 2020, 67, 1745–1749. [Google Scholar] [CrossRef]
- Hobbs, E.C.; Reid, T.J. Animals and SARS-CoV-2: Species susceptibility and viral transmission in experimental and natural conditions, and the potential implications for community transmission. Transbound. Emerg. Dis. 2021, 68, 1850–1867. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Fontela, C.; Dowling, W.E.; Funnell, S.G.P.; Gsell, P.S.; Riveros-Balta, A.X.; Albrecht, R.A.; Andersen, H.; Baric, R.S.; Carroll, M.W.; Cavaleri, M.; et al. Animal models for COVID-19. Nature 2020, 586, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Segalés, J.; Puig, M.; Rodon, J.; Avila-Nieto, C.; Carrillo, J.; Cantero, G.; Terrón, M.T.; Cruz, S.; Parera, M.; Noguera-Julián, M.; et al. Detection of SARS-CoV-2 in a cat owned by a COVID-19-affected patient in Spain. Proc. Natl. Acad. Sci. USA 2020, 117, 24790–24793. [Google Scholar] [CrossRef]
- Fernández-Bellon, H.; Rodon, J.; Fernández-Bastit, L.; Almagro, V.; Padilla-Solé, P.; Lorca-Oró, C.; Valle, R.; Roca, N.; Grazioli, S.; Trogu, T.; et al. Monitoring natural SARS-CoV-2 infection in lions (Panthera leo) at the Barcelona zoo: Viral dynamics and host responses. Viruses 2021, 13, 1683. [Google Scholar] [CrossRef]
- Sit, T.H.C.; Brackman, C.J.; Ip, S.M.; Tam, K.W.S.; Law, P.Y.T.; To, E.M.W.; Yu, V.Y.T.; Sims, L.D.; Tsang, D.N.C.; Chu, D.K.W.; et al. Infection of dogs with SARS-CoV-2. Nature 2020, 586, 776–778. [Google Scholar] [CrossRef]
- Oreshkova, N.; Molenaar, R.-J.; Vreman, S.; Harders, F.; Munnink, B.B.O.; Hakze, R.; Gerhards, N.; Tolsma, P.; Bouwstra, R.; Sikkema, R.; et al. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Euro Surveill. 2020, 25, 2001005. [Google Scholar] [CrossRef]
- Montagutelli, X.; Prot, M.; Levillayer, L.; Salazar, E.B.; Jouvion, G.; Conquet, L.; Donati, F.; Albert, M.; Gambaro, F.; Behillil, S.; et al. The B1.351 and P.1 variants extend SARS-CoV-2 host range to mice. bioRxiv 2021. [Google Scholar] [CrossRef]
- Tarrés-Freixas, F.; Trinité, B.; Pons-Grífols, A.; Romero-Durana, M.; Riveira-Muñoz, E.; Ávila-Nieto, C.; Pérez, M.; Garcia-Vidal, E.; Pérez-Zsolt, D.; Muñoz-Basagoiti, J.; et al. SARS-CoV-2 B.1.351 (beta) variant shows enhanced infectivity in K18-hACE2 transgenic mice and expanded tropism to wildtype mice compared to B.1 variant. bioRxiv 2021. [Google Scholar] [CrossRef]
- Patterson, E.I.; Elia, G.; Grassi, A.; Giordano, A.; Desario, C.; Medardo, M.; Smith, S.L.; Anderson, E.R.; Prince, T.; Patterson, G.T.; et al. Evidence of exposure to SARS-CoV-2 in cats and dogs from households in Italy. Nat. Commun. 2020, 11, 6231. [Google Scholar] [CrossRef]
- Fritz, M.; Rosolen, B.; Krafft, E.; Becquart, P.; Elguero, E.; Vratskikh, O.; Denolly, S.; Boson, B.; Vanhomwegen, J.; Gouilh, M.A.; et al. High prevalence of SARS-CoV-2 antibodies in pets from COVID-19+ households. One Health 2021, 11, 100192. [Google Scholar] [CrossRef]
- Barroso-ar, S.; Dom, L.; Sánchez-Vizcaíno, J.M. First Detection of SARS-CoV-2 B.1.1.7 Variant of Concern in an Asymptomatic Dog in Spain. Viruses 2021, 13, 1379. [Google Scholar] [CrossRef] [PubMed]
- Miró, G.; Regidor-cerrillo, J.; Checa, R.; Diezma-díaz, C. SARS-CoV-2 Infection in One Cat and Three Dogs Living in COVID-19-Positive Households in Madrid, Spain. Front. Vet. Sci. 2021, 8, 341. [Google Scholar] [CrossRef]
- Doerksen, T.; Lu, A.; Noll, L.; Almes, K.; Bai, J.; Upchurch, D.; Palinski, R. Near-Complete Genome of SARS-CoV-2 Delta (AY.3) Variant Identified in a Dog in Kansas, USA. Viruses 2021, 13, 2104. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.W.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveil. 2020, 25, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Brustolin, M.; Rodon, J.; Rodríguez de la Concepción, M.L.; Ávila-Nieto, C.; Cantero, G.; Pérez, M.; Te, N.; Noguera-Julián, M.; Guallar, V.; Valencia, A.; et al. Protection against reinfection with D614- or G614-SARS-CoV-2 isolates in golden Syrian hamster. Emerg. Microbes Infect. 2021, 10, 797–809. [Google Scholar] [CrossRef]
- Rodon, J.; Muñoz-Basagoiti, J.; Perez-Zsolt, D.; Noguera-Julian, M.; Paredes, R.; Mateu, L.; Quiñones, C.; Perez, C.; Erkizia, I.; Blanco, I.; et al. Identification of Plitidepsin as Potent Inhibitor of SARS-CoV-2-Induced Cytopathic Effect After a Drug Repurposing Screen. Front. Pharmacol. 2021, 12, 676. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Trinité, B.; Tarrés-Freixas, F.; Rodon, J.; Pradenas, E.; Urrea, V.; Marfil, S.; Luisa, M.; De La Concepción, R.; Ávila-Nieto, C.; Aguilar-Gurrieri, C.; et al. SARS-CoV-2 infection elicits a rapid neutralizing antibody response that correlates with disease severity. Sci. Rep. 2021, 11, 2608. [Google Scholar] [CrossRef]
- Díez, J.M.; Romero, C.; Cruz, M.; Vandeberg, P.; Merritt, W.K.; Pradenas, E.; Trinité, B.; Blanco, J.; Clotet, B.; Willis, T.; et al. Anti-SARS-CoV-2 hyperimmune globulin demonstrates potent neutralization and antibody-dependent cellular cytotoxicity and phagocytosis through N and S proteins. J. Infect. Dis. 2021. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.; Smith, D.; Ghai, R.R.; Wallace, R.M.; Torchetti, M.K.; Loiacono, C.; Murrell, L.S.; Carpenter, A.; Moroff, S.; Rooney, J.A.; et al. First Reported Cases of SARS-CoV-2 Infection in Companion Animals—New York, March–April 2020. MMWR. Morb. Mortal. Wkly. Rep. 2020, 69, 710–713. [Google Scholar] [CrossRef]
- Shi, J.; Wen, Z.; Zhong, G.; Yang, H.; Wang, C.; Huang, B.; Liu, R.; He, X.; Shuai, L.; Sun, Z.; et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science 2020, 368, 1016–1020. [Google Scholar] [CrossRef] [Green Version]
- Mishra, A.; Kumar, N.; Bhatia, S.; Aasdev, A.; Kanniappan, S.; Sekhar, A.T.; Gopinadhan, A.; Silambarasan, R.; Sreekumar, C.; Dubey, C.K.; et al. Sars-cov-2 delta variant among asiatic lions, india. Emerg. Infect. Dis. 2021, 27, 2723–2725. [Google Scholar] [CrossRef] [PubMed]
- Hamer, S.A.; Pauvolid-Corrêa, A.; Zecca, I.B.; Davila, E.; Auckland, L.D.; Roundy, C.M.; Tang, W.; Torchetti, M.K.; Killian, M.L.; Jenkins-Moore, M.; et al. SARS-CoV-2 infections and viral isolations among serially tested cats and dogs in households with infected owners in texas, usa. Viruses 2021, 13, 938. [Google Scholar] [CrossRef] [PubMed]
- SARS-COV-2 Delta Variant Now Dominant in much of the European Region and Efforts must be Reinforced to Prevent TRANSMISSION, Warn WHO/Europe and ECDC. Available online: https://www.ecdc.europa.eu/en/news-events/sars-cov-2-delta-variant-now-dominant-european-region (accessed on 3 November 2021).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández-Bastit, L.; Rodon, J.; Pradenas, E.; Marfil, S.; Trinité, B.; Parera, M.; Roca, N.; Pou, A.; Cantero, G.; Lorca-Oró, C.; et al. First Detection of SARS-CoV-2 Delta (B.1.617.2) Variant of Concern in a Dog with Clinical Signs in Spain. Viruses 2021, 13, 2526. https://doi.org/10.3390/v13122526
Fernández-Bastit L, Rodon J, Pradenas E, Marfil S, Trinité B, Parera M, Roca N, Pou A, Cantero G, Lorca-Oró C, et al. First Detection of SARS-CoV-2 Delta (B.1.617.2) Variant of Concern in a Dog with Clinical Signs in Spain. Viruses. 2021; 13(12):2526. https://doi.org/10.3390/v13122526
Chicago/Turabian StyleFernández-Bastit, Leira, Jordi Rodon, Edwards Pradenas, Silvia Marfil, Benjamin Trinité, Mariona Parera, Núria Roca, Anna Pou, Guillermo Cantero, Cristina Lorca-Oró, and et al. 2021. "First Detection of SARS-CoV-2 Delta (B.1.617.2) Variant of Concern in a Dog with Clinical Signs in Spain" Viruses 13, no. 12: 2526. https://doi.org/10.3390/v13122526
APA StyleFernández-Bastit, L., Rodon, J., Pradenas, E., Marfil, S., Trinité, B., Parera, M., Roca, N., Pou, A., Cantero, G., Lorca-Oró, C., Carrillo, J., Izquierdo-Useros, N., Clotet, B., Noguera-Julián, M., Blanco, J., Vergara-Alert, J., & Segalés, J. (2021). First Detection of SARS-CoV-2 Delta (B.1.617.2) Variant of Concern in a Dog with Clinical Signs in Spain. Viruses, 13(12), 2526. https://doi.org/10.3390/v13122526





