Encapsidation of Staufen-2 Enhances Infectivity of HIV-1
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Plasmids
2.3. Virus Production and Isolation
2.4. Generation of Stably over Expressing Staufen-2 and Vector HEK293T Cell Lines
2.5. HIV-1 Viral Core Isolation
2.6. Generation of Staufen-2 and Staufen-1 Knockout HEK293T Cell Lines
2.7. Luciferase-Based Infectivity Assay
2.8. TZM-bl Infectivity Assay
2.9. Immunoblot Analyses
2.10. Immunoprecipitation
2.11. GST Pull-Down Assay
2.12. Immunofluorescence
2.13. Sucrose Density Gradient Centrifugation
2.14. In Vitro DNA Cytidine Deamination Assay
2.15. A3G Degradation Assay
2.16. Statistical Analysis
3. Results
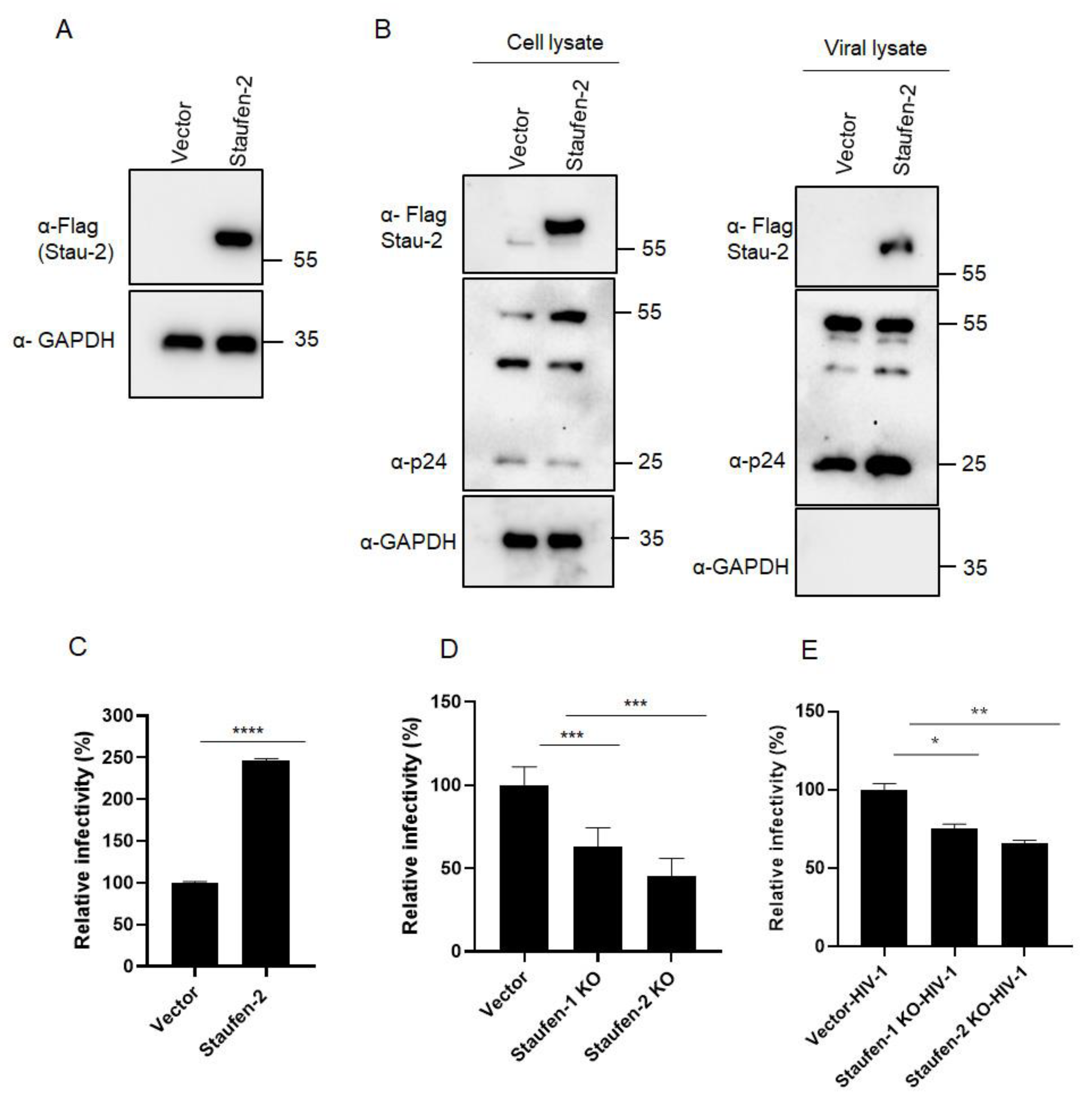
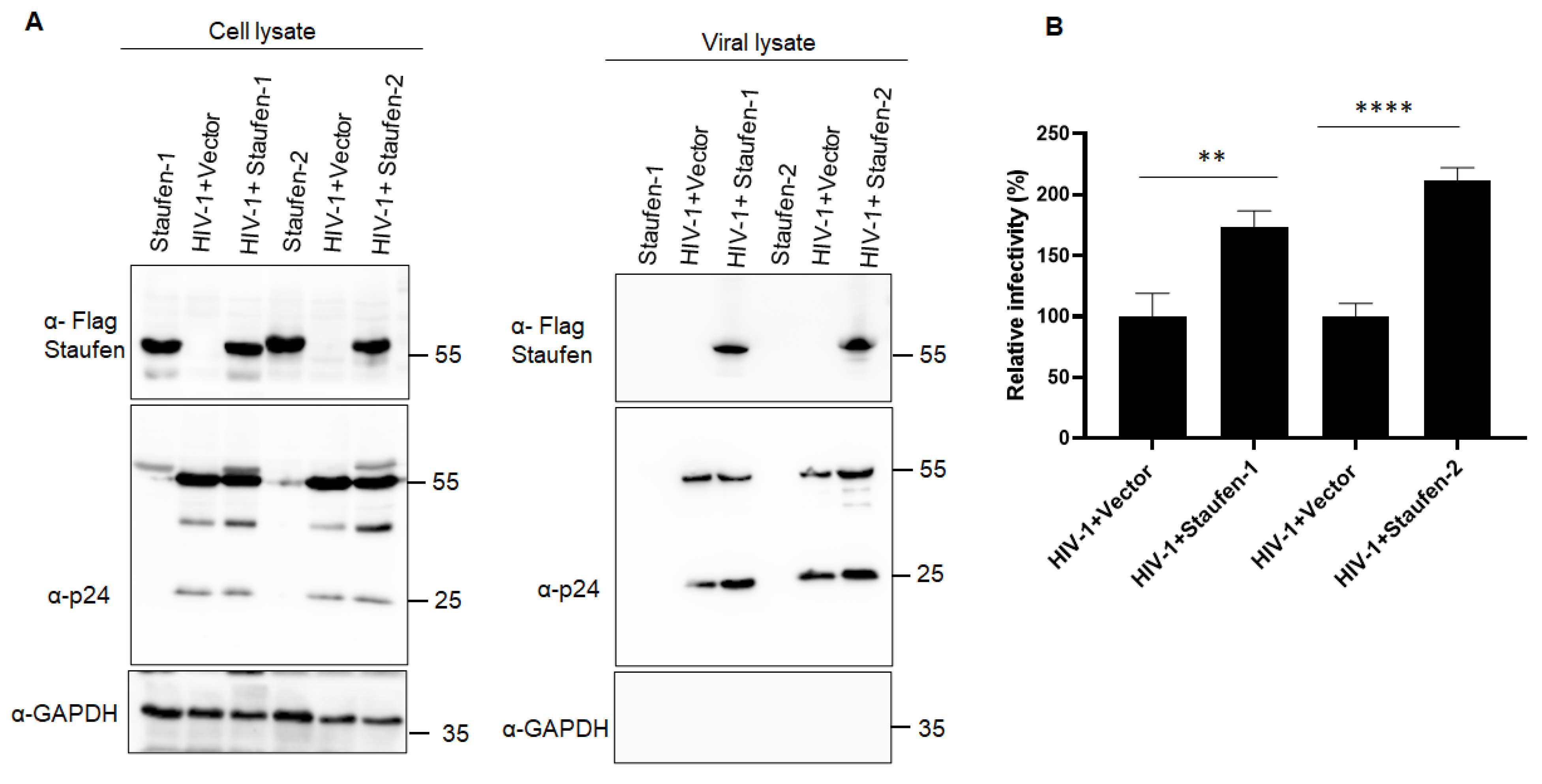
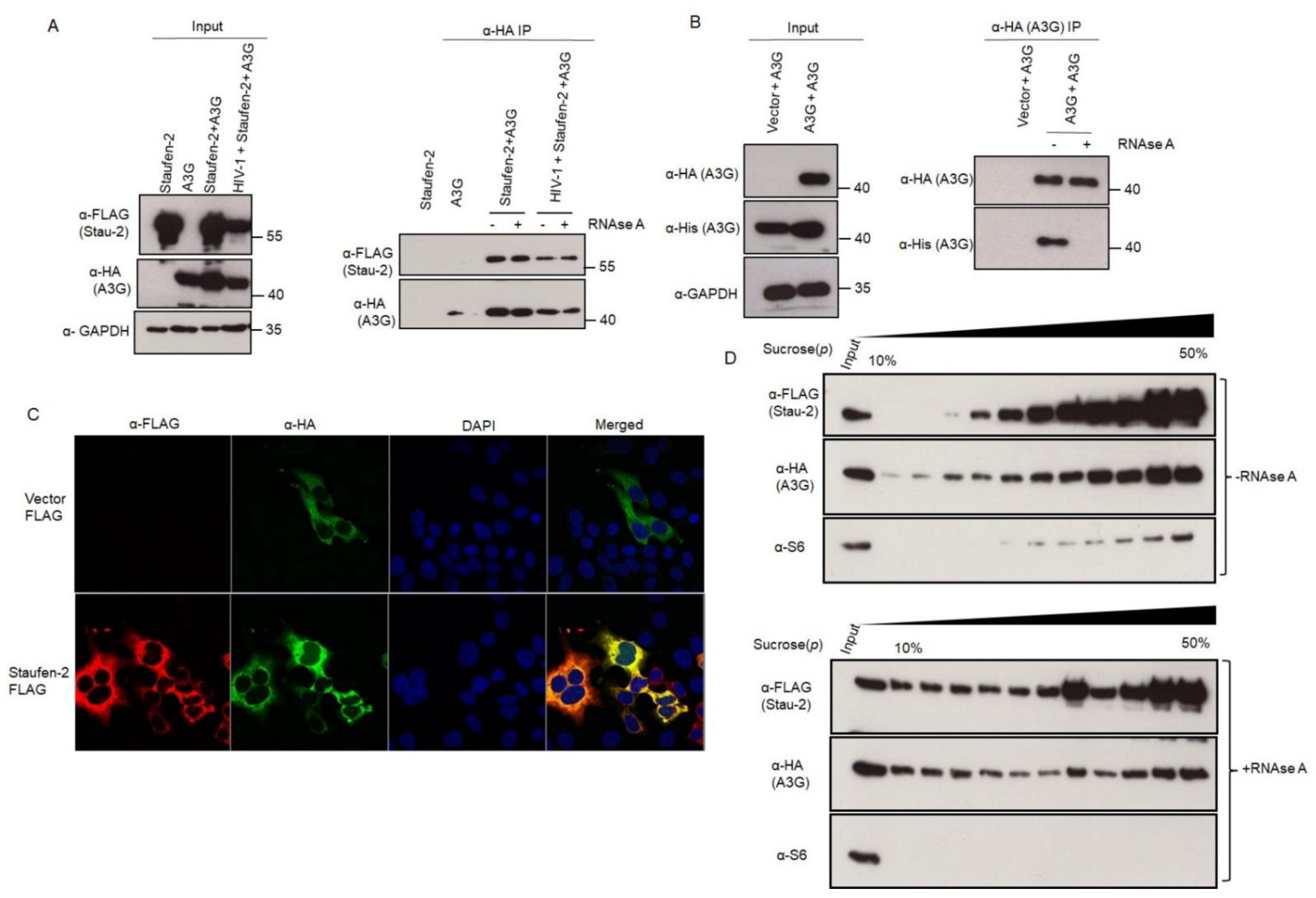
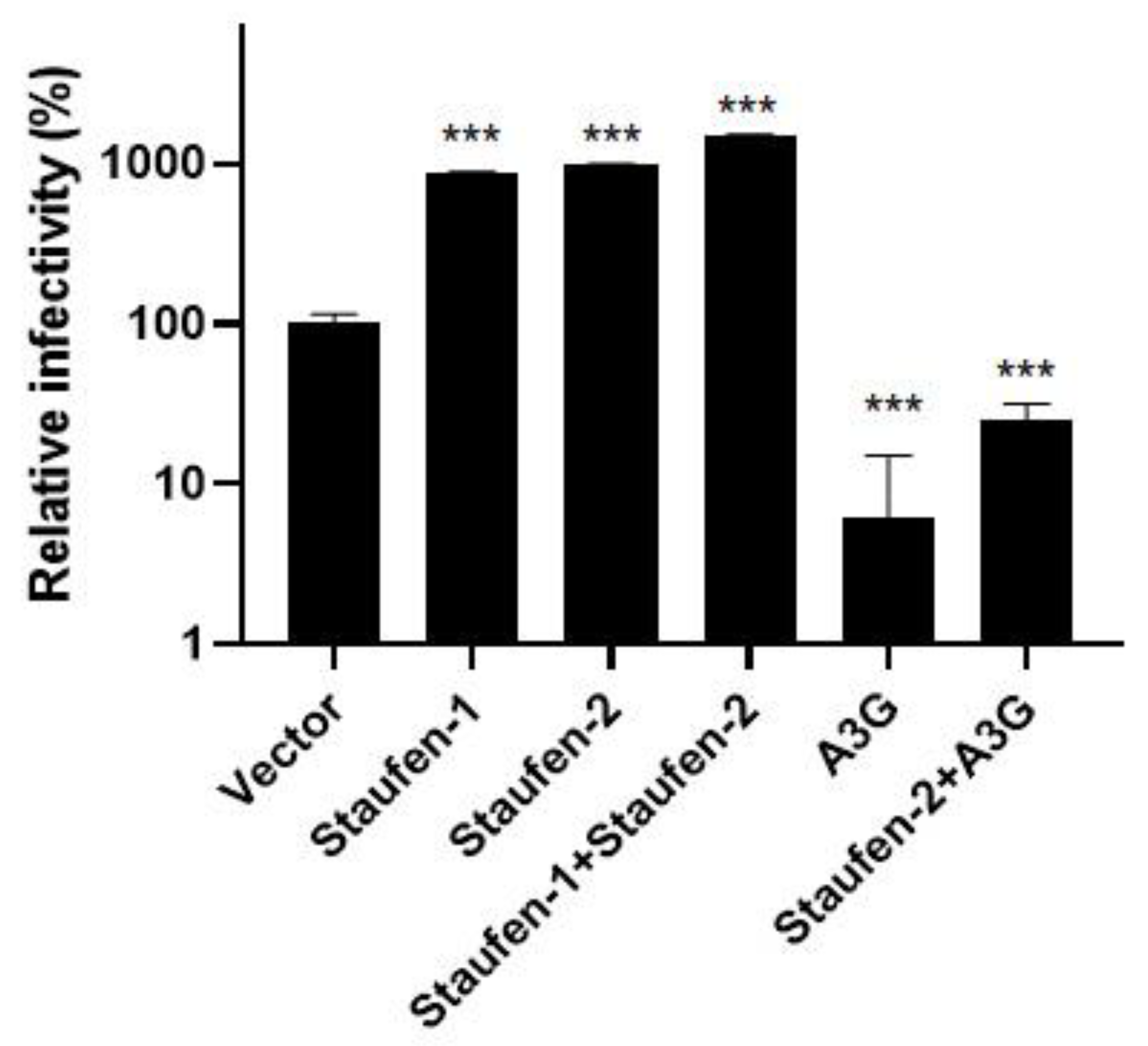
3.1. Staufen-2 Is Encapsidated into HIV-1 Virions, Enhancing Their Infectivity
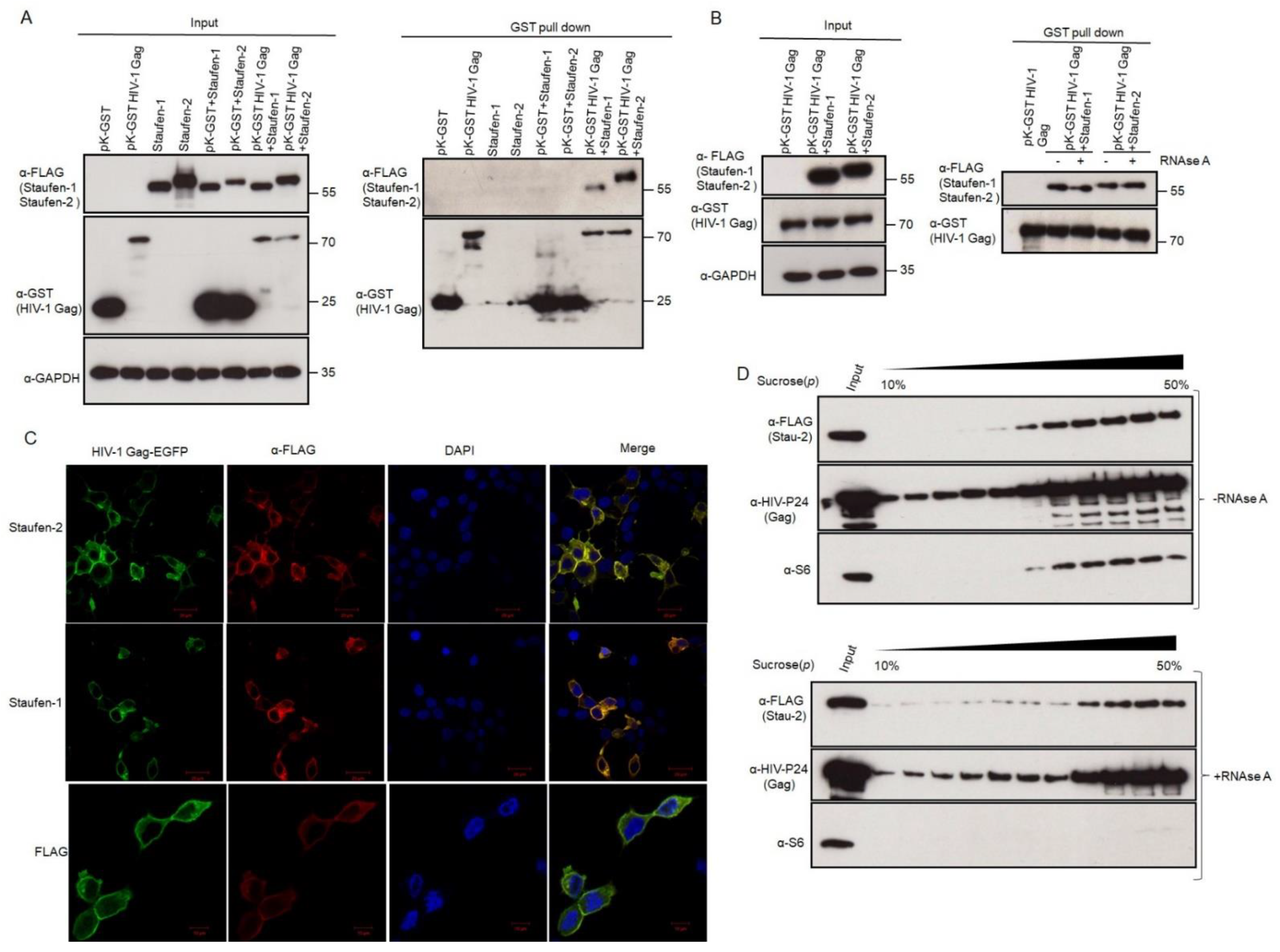
3.2. Staufen-2 Is Packaged into HIV-1 through Its Interaction with Gag
3.3. Multiple Pathways Mediate Staufen-2 Incorporation into Virions
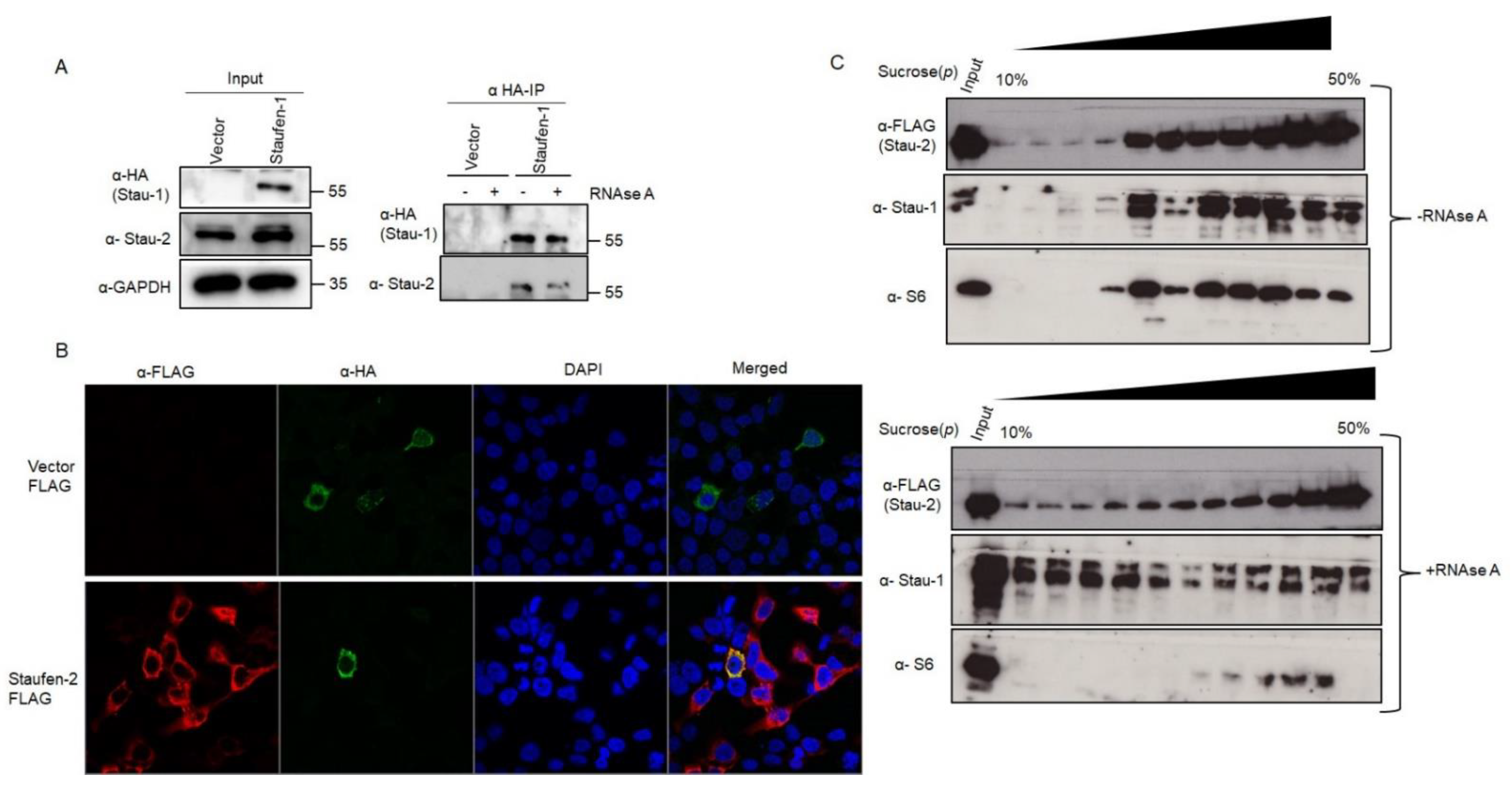
3.3.1. Staufen-2 Interacts with Staufen-1
3.3.2. Staufen-2 Interacts with APOBEC3G
3.4. Fate of the Viral Population with Encapsidated Staufen-2 Alone or with Its Partners Staufen-1 or A3G
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cochrane, A.W.; McNally, M.T.; Mouland, A.J. The retrovirus RNA trafficking granule: From birth to maturity. Retrovirology 2006, 3, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knoener, R.A.; Becker, J.T.; Scalf, M.; Sherer, N.M.; Smith, L.M. Elucidating the in vivo interactome of HIV-1 RNA by hybridization capture and mass spectrometry. Sci. Rep. 2017, 7, 16965. [Google Scholar] [CrossRef]
- Garcia-Moreno, M.; Järvelin, A.I.; Castello, A. Unconventional RNA-binding proteins step into the virus–host battlefront. Wiley Interdiscip. Rev. RNA 2018, 9, e1498. [Google Scholar] [CrossRef] [Green Version]
- Freed, E.O. HIV-1 Gag Proteins: Diverse Functions in the Virus Life Cycle. Virology 1998, 251, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Tuffy, K.M.; Maldonado, R.J.K.; Chang, J.; Rosenfeld, P.; Cochrane, A.; Parent, L.J. HIV-1 Gag Forms Ribonucleoprotein Complexes with Unspliced Viral RNA at Transcription Sites. Viruses 2020, 12, 1281. [Google Scholar] [CrossRef]
- Xu, W.; Byun, H.; Dudley, J. The Role of APOBECs in Viral Replication. Microorganisms 2020, 8, 1899. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.S.; Dudley, J.P. APOBECs and virus restriction. Virology 2015, 479–480, 131–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Browne, E.P.; Allers, C.; Landau, N.R. Restriction of HIV-1 by APOBEC3G is cytidine deaminase-dependent. Virology 2009, 387, 313–321. [Google Scholar] [CrossRef] [Green Version]
- Soros, V.B.; Yonemoto, W.; Greene, W.C. Newly Synthesized APOBEC3G Is Incorporated into HIV Virions, Inhibited by HIV RNA, and Subsequently Activated by RNase H. PLoS Pathog. 2007, 3, e15. [Google Scholar] [CrossRef] [Green Version]
- Chiu, Y.-L.; Soros, V.B.; Kreisberg, J.F.; Stopak, K.; Yonemoto, W.; Greene, W.C. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature 2005, 435, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Chatel-Chaix, L.; Boulay, K.; Mouland, A.J.; DesGroseillers, L. The host protein Staufen1 interacts with the Pr55Gagzinc fingers and regulates HIV-1 assembly via its N-terminus. Retrovirology 2008, 5, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, S.; Hassine, S.; Monette, A.; Amorim, R.; DesGroseillers, L.; Mouland, A.J. HIV-1 requires Staufen1 to dissociate stress granules and to produce infectious viral particles. RNA 2019, 25, 727–736. [Google Scholar] [CrossRef]
- Abrahamyan, L.G.; Chatel-Chaix, L.; Ajamian, L.; Milev, M.P.; Monette, A.; Clément, J.-F.; Song, R.; Lehmann, M.; DesGroseillers, L.; Laughrea, M.; et al. Novel Staufen1 ribonucleoproteins prevent formation of stress granules but favour encapsidation of HIV-1 genomic RNA. J. Cell Sci. 2010, 123, 369–383. [Google Scholar] [CrossRef] [Green Version]
- Mouland, A.J.; Mercier, J.; Luo, M.; Bernier, L.; DesGroseillers, L.; Cohen, E.A. The Double-Stranded RNA-Binding Protein Staufen Is Incorporated in Human Immunodeficiency Virus Type 1: Evidence for a Role in Genomic RNA Encapsidation. J. Virol. 2000, 74, 5441–5451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heraud-Farlow, J.; Kiebler, M.A. The multifunctional Staufen proteins: Conserved roles from neurogenesis to synaptic plasticity. Trends Neurosci. 2014, 37, 470–479. [Google Scholar] [CrossRef] [Green Version]
- Park, E.; Maquat, L.E. Staufen-mediated mRNA decay. Wiley Interdiscip. Rev. RNA 2013, 4, 423–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miki, T.; Takano, K.; Yoneda, Y. The Role of Mammalian Staufen on mRNA Traffic: A View from Its Nucleocytoplasmic Shuttling Function. Cell Struct. Funct. 2005, 30, 51–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furic, L.; Maher-Laporte, M.; DesGroseillers, L. A genome-wide approach identifies distinct but overlapping subsets of cellular mRNAs associated with Staufen1- and Staufen2-containing ribonucleoprotein complexes. RNA 2007, 14, 324–335. [Google Scholar] [CrossRef] [Green Version]
- Heber, S.; Gáspár, I.; Tants, J.-N.; Günther, J.; Moya, S.M.F.; Janowski, R.; Ephrussi, A.; Sattler, M.; Niessing, D. Staufen2-mediated RNA recognition and localization requires combinatorial action of multiple domains. Nat. Commun. 2019, 10, 1659. [Google Scholar] [CrossRef] [Green Version]
- Lebeau, G.; Miller, L.C.; Tartas, M.; McAdam, R.; Laplante, I.; Badeaux, F.; DesGroseillers, L.; Sossin, W.S.; Lacaille, J.-C. Staufen 2 regulates mGluR long-term depression and Map1b mRNA distribution in hippocampal neurons. Learn. Mem. 2011, 18, 314–326. [Google Scholar] [CrossRef] [Green Version]
- Macchi, P.; Brownawell, A.M.; Grunewald, B.; DesGroseillers, L.; Macara, I.G.; Kiebler, M. The Brain-specific Double-stranded RNA-binding Protein Staufen2. J. Biol. Chem. 2004, 279, 31440–31444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, E.; Gleghorn, M.L.; Maquat, L.E. Staufen2 functions in Staufen1-mediated mRNA decay by binding to itself and its paralog and promoting UPF1 helicase but not ATPase activity. Proc. Natl. Acad. Sci. USA 2012, 110, 405–412. [Google Scholar] [CrossRef] [Green Version]
- Goetze, B.; Tuebing, F.; Xie, Y.; Dorostkar, M.; Thomas, S.; Pehl, U.; Boehm, S.; Macchi, P.; Kiebler, M. The brain-specific double-stranded RNA-binding protein Staufen2 is required for dendritic spine morphogenesis. J. Cell Biol. 2006, 172, 221–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, Y.; Hu, Z.; Wu, J.; Dai, F.; Lee, F.; Xu, Y. STAU1 selectively regulates the expression of inflammatory and immune response genes and alternative splicing of the nerve growth factor receptor signaling pathway. Oncol. Rep. 2020, 44, 1863–1874. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.-S.; Mogilicherla, K.; Gurusamy, D.; Chen, X.; Chereddy, S.; Palli, S.R. Double-stranded RNA binding protein, Staufen, is required for the initiation of RNAi in coleopteran insects. Proc. Natl. Acad. Sci. USA 2018, 115, 8334–8339. [Google Scholar] [CrossRef] [Green Version]
- Bilogan, C.K.; Horb, M.E. Xenopus staufen2 is required for anterior endodermal organ formation. Genesis 2011, 50, 251–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Trépanier, V.; Beaujois, R.; Viranaicken, W.; Drobetsky, E.; DesGroseillers, L. The downregulation of the RNA-binding protein Staufen2 in response to DNA damage promotes apoptosis. Nucleic Acids Res. 2016, 44, 3695–3712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Condé, L.; Quesada, Y.G.; Bonnet-Magnaval, F.; Beaujois, R.; DesGroseillers, L. STAU2 protein level is controlled by caspases and the CHK1 pathway and regulates cell cycle progression in the non-transformed hTERT-RPE1 cells. BMC Mol. Cell Biol. 2021, 22, 16. [Google Scholar] [CrossRef]
- Cockburn, D.M.; Charish, J.; Tassew, N.G.; Eubanks, J.; Bremner, R.; Macchi, P.; Monnier, P.P. The double-stranded RNA-binding protein Staufen 2 regulates eye size. Mol. Cell. Neurosci. 2012, 51, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-M.; Ou, B.-T.; Chen, C.-Y.; Chan, H.-H.; Chen, C.-J.; Wang, R.Y. Staufen1 Protein Participates Positively in the Viral RNA Replication of Enterovirus 71. Viruses 2019, 11, 142. [Google Scholar] [CrossRef] [Green Version]
- de Lucas, S.; Peredo, J.; Marión, R.M.; Sánchez, C.; Ortín, J. Human Staufen1 Protein Interacts with Influenza Virus Ribonucleoproteins and Is Required for Efficient Virus Multiplication. J. Virol. 2010, 84, 7603–7612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blackham, S.L.; McGarvey, M. A host cell RNA-binding protein, Staufen1, has a role in hepatitis C virus replication before virus assembly. J. Gen. Virol. 2013, 94, 2429–2436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dixit, U.; Pandey, A.K.; Mishra, P.; Sengupta, A.; Pandey, V.N. Staufen1 promotes HCV replication by inhibiting protein kinase R and transporting viral RNA to the site of translation and replication in the cells. Nucleic Acids Res. 2016, 44, 5271–5287. [Google Scholar] [CrossRef] [Green Version]
- Hanke, K.; Hohn, O.; Liedgens, L.; Fiddeke, K.; Wamara, J.; Kurth, R.; Bannert, N. Staufen-1 Interacts with the Human Endogenous Retrovirus Family HERV-K(HML-2) Rec and Gag Proteins and Increases Virion Production. J. Virol. 2013, 87, 11019–11030. [Google Scholar] [CrossRef] [Green Version]
- Chatel-Chaix, L.; Clément, J.-F.; Martel, C.; Bériault, V.; Gatignol, A.; DesGroseillers, L.; Mouland, A.J. Identification of Staufen in the Human Immunodeficiency Virus Type 1 Gag Ribonucleoprotein Complex and a Role in Generating Infectious Viral Particles. Mol. Cell. Biol. 2004, 24, 2637–2648. [Google Scholar] [CrossRef] [Green Version]
- Chatel-Chaix, L.; Abrahamyan, L.; Fréchina, C.; Mouland, A.J.; DesGroseillers, L. The Host Protein Staufen1 Participates in Human Immunodeficiency Virus Type 1 Assembly in Live Cells by Influencing pr55 Gag Multimerization. J. Virol. 2007, 81, 6216–6230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bélanger, G.; Stocksley, M.A.; Vandromme, M.; Schaeffer, L.; Furic, L.; DesGroseillers, L.; Jasmin, B.J. Localization of the RNA-binding proteins Staufen1 and Staufen2 at the mammalian neuromuscular junction. J. Neurochem. 2003, 86, 669–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mallardo, M.; Deitinghoff, A.; Müller, J.; Goetze, B.; Macchi, P.; Peters, C.; Kiebler, M.A. Isolation and characterization of Staufen-containing ribonucleoprotein particles from rat brain. Proc. Natl. Acad. Sci. USA 2003, 100, 2100–2105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, M.G.; Tosar, L.J.M.; Loschi, M.; Pasquini, J.M.; Correale, J.; Kindler, S.; Boccaccio, G.L. Staufen Recruitment into Stress Granules Does Not Affect Early mRNA Transport in Oligodendrocytes. Mol. Biol. Cell 2005, 16, 405–420. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, A.; Benjamin, R.; Balakrishnan, K.; Ghosh, P.; Banerjee, S. Human protein Staufen-2 promotes HIV-1 proliferation by positively regulating RNA export activity of viral protein Rev. Retrovirology 2014, 11, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marin, M.; Rose, K.M.; Kozak, S.L.; Kabat, D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 2003, 9, 1398–1403. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, Y.; Vigilante, A.; Darbo, E.; Zirra, A.; Militti, C.; D’Ambrogio, A.; Luscombe, N.M.; Ule, J. hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature 2015, 519, 491–494. [Google Scholar] [CrossRef] [Green Version]
- Russell, R.A.; Wiegand, H.L.; Moore, R.; Schäfer, A.; McClure, M.O.; Cullen, B.R. Foamy Virus Bet Proteins Function as Novel Inhibitors of the APOBEC3 Family of Innate Antiretroviral Defense Factors. J. Virol. 2005, 79, 8724–8731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanjana, N.E.; Shalem, O.; Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 2014, 11, 783–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shalem, O.; Sanjana, N.E.; Hartenian, E.; Shi, X.; Scott, D.A.; Mikkelsen, T.S.; Heckl, D.; Ebert, B.L.; Root, D.E.; Doench, J.G.; et al. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science 2014, 343, 84–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunseri, N.; O’Brien, M.; Bhardwaj, N.; Landau, N.R. Human Immunodeficiency Virus Type 1 Modified to Package Simian Immunodeficiency Virus Vpx Efficiently Infects Macrophages and Dendritic Cells. J. Virol. 2011, 85, 6263–6274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dull, T.; Zufferey, R.; Kelly, M.; Mandel, R.J.; Nguyen, M.; Trono, D.; Naldini, L. A Third-Generation Lentivirus Vector with a Conditional Packaging System. J. Virol. 1998, 72, 8463–8471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srivastava, K.K.; Fernandez-Larsson, R.; Zinkus, D.M.; Robinson, H.L. Human immunodeficiency virus type 1 NL4-3 replication in four T-cell lines: Rate and efficiency of entry, a major determinant of permissiveness. J. Virol. 1991, 65, 3900–3902. [Google Scholar] [CrossRef] [Green Version]
- Mariani, R.; Chen, D.; Schröfelbauer, B.; Navarro, F.; König, R.; Bollman, B.; Münk, C.; Nymark-McMahon, H.; Landau, N.R. Species-Specific Exclusion of APOBEC3G from HIV-1 Virions by Vif. Cell 2003, 114, 21–31. [Google Scholar] [CrossRef] [Green Version]
- Vasudevan, A.A.J.; Hofmann, H.; Willbold, D.; Häussinger, D.; Koenig, B.; Münk, C. Enhancing the Catalytic Deamination Activity of APOBEC3C Is Insufficient to Inhibit Vif-Deficient HIV-1. J. Mol. Biol. 2017, 429, 1171–1191. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Li, S.-X.; Yang, H.; Chen, X.S. Crystal structures of APOBEC3G N-domain alone and its complex with DNA. Nat. Commun. 2016, 7, 12193. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Dolan, P.T.; Dang, Y.; Zheng, Y.-H. Biochemical Differentiation of APOBEC3F and APOBEC3G Proteins Associated with HIV-1 Life Cycle. J. Biol. Chem. 2007, 282, 1585–1594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasudevan, A.A.J.; Balakrishnan, K.; Gertzen, C.G.W.; Borvető, F.; Zhang, Z.; Sangwiman, A.; Held, U.; Küstermann, C.; Banerjee, S.; Schumann, G.G.; et al. Loop 1 of APOBEC3C Regulates its Antiviral Activity against HIV-1. J. Mol. Biol. 2020, 432, 6200–6227. [Google Scholar] [CrossRef]
- Marino, D.; Perković, M.; Hain, A.; Vasudevan, A.A.J.; Hofmann, H.; Hanschmann, K.-M.; Mühlebach, M.D.; Schumann, G.G.; König, R.; Cichutek, K.; et al. APOBEC4 Enhances the Replication of HIV-1. PLoS ONE 2016, 11, e0155422. [Google Scholar] [CrossRef] [Green Version]
- Kuffour, E.O.; Schott, K.; Vasudevan, A.A.J.; Holler, J.; Schulz, W.A.; Lang, P.A.; Lang, K.S.; Kim, B.; Häussinger, D.; König, R.; et al. USP18 (UBP43) Abrogates p21-Mediated Inhibition of HIV-1. J. Virol. 2018, 92, e00592-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasudevan, A.A.J.; Perković, M.; Bulliard, Y.; Cichutek, K.; Trono, D.; Häussinger, D.; Münk, C. Prototype Foamy Virus Bet Impairs the Dimerization and Cytosolic Solubility of Human APOBEC3G. J. Virol. 2013, 87, 9030–9040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Draper, D.E.; Reynaldo, L.P. RNA binding strategies of ribosomal proteins. Nucleic Acids Res. 1999, 27, 381–388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horn, A.V.; Klawitter, S.; Held, U.; Berger, A.; Vasudevan, A.A.J.; Bock, A.; Hofmann, H.; Hanschmann, K.-M.O.; Trösemeier, J.-H.; Flory, E.; et al. Human LINE-1 restriction by APOBEC3C is deaminase independent and mediated by an ORF1p interaction that affects LINE reverse transcriptase activity. Nucleic Acids Res. 2013, 42, 396–416. [Google Scholar] [CrossRef] [Green Version]
- Huthoff, H.; Autore, F.; Gallois-Montbrun, S.; Fraternali, F.; Malim, M.H. RNA-Dependent Oligomerization of APOBEC3G Is Required for Restriction of HIV-1. PLoS Pathog. 2009, 5, e1000330. [Google Scholar] [CrossRef] [PubMed]
- Gallois-Montbrun, S.; Kramer, B.; Swanson, C.; Byers, H.; Lynham, S.; Ward, M.; Malim, M.H. Antiviral Protein APOBEC3G Localizes to Ribonucleoprotein Complexes Found in P Bodies and Stress Granules. J. Virol. 2007, 81, 2165–2178. [Google Scholar] [CrossRef] [Green Version]
- Milev, M.P.; Ravichandran, M.; Khan, M.F.; Schriemer, D.C.; Mouland, A.J. Characterization of Staufen1 Ribonucleoproteins by Mass Spectrometry and Biochemical Analyses Reveal the Presence of Diverse Host Proteins Associated with Human Immunodeficiency Virus Type 1. Front. Microbiol. 2012, 3, 367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sundquist, W.I.; Kräusslich, H.-G. HIV-1 Assembly, Budding, and Maturation. Cold Spring Harb. Perspect. Med. 2012, 2, a006924. [Google Scholar] [CrossRef] [PubMed]
- Freed, E.O. HIV-1 assembly, release and maturation. Nat. Rev. Genet. 2015, 13, 484–496. [Google Scholar] [CrossRef] [PubMed]
- Ramdas, P.; Sahu, A.K.; Mishra, T.; Bhardwaj, V.; Chande, A. From Entry to Egress: Strategic Exploitation of the Cellular Processes by HIV-1. Front. Microbiol. 2020, 11, 559792. [Google Scholar] [CrossRef] [PubMed]
- Colomer-Lluch, M.; Ruiz, A.; Moris, A.; Prado, J.G. Restriction Factors: From Intrinsic Viral Restriction to Shaping Cellular Immunity Against HIV-1. Front. Immunol. 2018, 9, 2876. [Google Scholar] [CrossRef] [Green Version]
- Temple, J.; Tripler, T.N.; Shen, Q.; Xiong, Y. A snapshot of HIV-1 capsid–host interactions. Curr. Res. Struct. Biol. 2020, 2, 222–228. [Google Scholar] [CrossRef]
- Lama, J.; Planelles, V. Host factors influencing susceptibility to HIV infection and AIDS progression. Retrovirology 2007, 4, 52. [Google Scholar] [CrossRef] [Green Version]
- Ode, H.; Matsuda, M.; Matsuoka, K.; Hachiya, A.; Hattori, J.; Kito, Y.; Yokomaku, Y.; Iwatani, Y.; Sugiura, W. Quasispecies Analyses of the HIV-1 Near-full-length Genome with Illumina MiSeq. Front. Microbiol. 2015, 6, 1258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Azevedo, S.S.D.; Caetano, D.G.; Côrtes, F.H.; Teixeira, S.L.M.; Silva, K.D.S.; Hoagland, B.; Grinsztejn, B.; Veloso, V.G.; Morgado, M.G.; Bello, G. Highly divergent patterns of genetic diversity and evolution in proviral quasispecies from HIV controllers. Retrovirology 2017, 14, 29. [Google Scholar] [CrossRef] [PubMed]
- Sguanci, L.; Bagnoli, F.; Liò, P. Modeling HIV quasispecies evolutionary dynamics. BMC Evol. Biol. 2007, 7, S5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rebensburg, S.V.; Wei, G.; Larue, R.C.; Lindenberger, J.; Francis, A.C.; Annamalai, A.S.; Morrison, J.; Shkriabai, N.; Huang, S.-W.; KewalRamani, V.; et al. Sec24C is an HIV-1 host dependency factor crucial for virus replication. Nat. Microbiol. 2021, 6, 435–444. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balakrishnan, K.; Jaguva Vasudevan, A.A.; Mohareer, K.; Luedde, T.; Münk, C.; Banerjee, S. Encapsidation of Staufen-2 Enhances Infectivity of HIV-1. Viruses 2021, 13, 2459. https://doi.org/10.3390/v13122459
Balakrishnan K, Jaguva Vasudevan AA, Mohareer K, Luedde T, Münk C, Banerjee S. Encapsidation of Staufen-2 Enhances Infectivity of HIV-1. Viruses. 2021; 13(12):2459. https://doi.org/10.3390/v13122459
Chicago/Turabian StyleBalakrishnan, Kannan, Ananda Ayyappan Jaguva Vasudevan, Krishnaveni Mohareer, Tom Luedde, Carsten Münk, and Sharmistha Banerjee. 2021. "Encapsidation of Staufen-2 Enhances Infectivity of HIV-1" Viruses 13, no. 12: 2459. https://doi.org/10.3390/v13122459
APA StyleBalakrishnan, K., Jaguva Vasudevan, A. A., Mohareer, K., Luedde, T., Münk, C., & Banerjee, S. (2021). Encapsidation of Staufen-2 Enhances Infectivity of HIV-1. Viruses, 13(12), 2459. https://doi.org/10.3390/v13122459






