Oral Selective TLR8 Agonist Selgantolimod Induces Multiple Immune Cell Responses in Humans
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Samples
2.2. Flow Cytometry (FACS) and Intracellular Cytokine Staining (ICS)
2.3. Cell sorting, Total RNA Isolation and Microarray Analysis
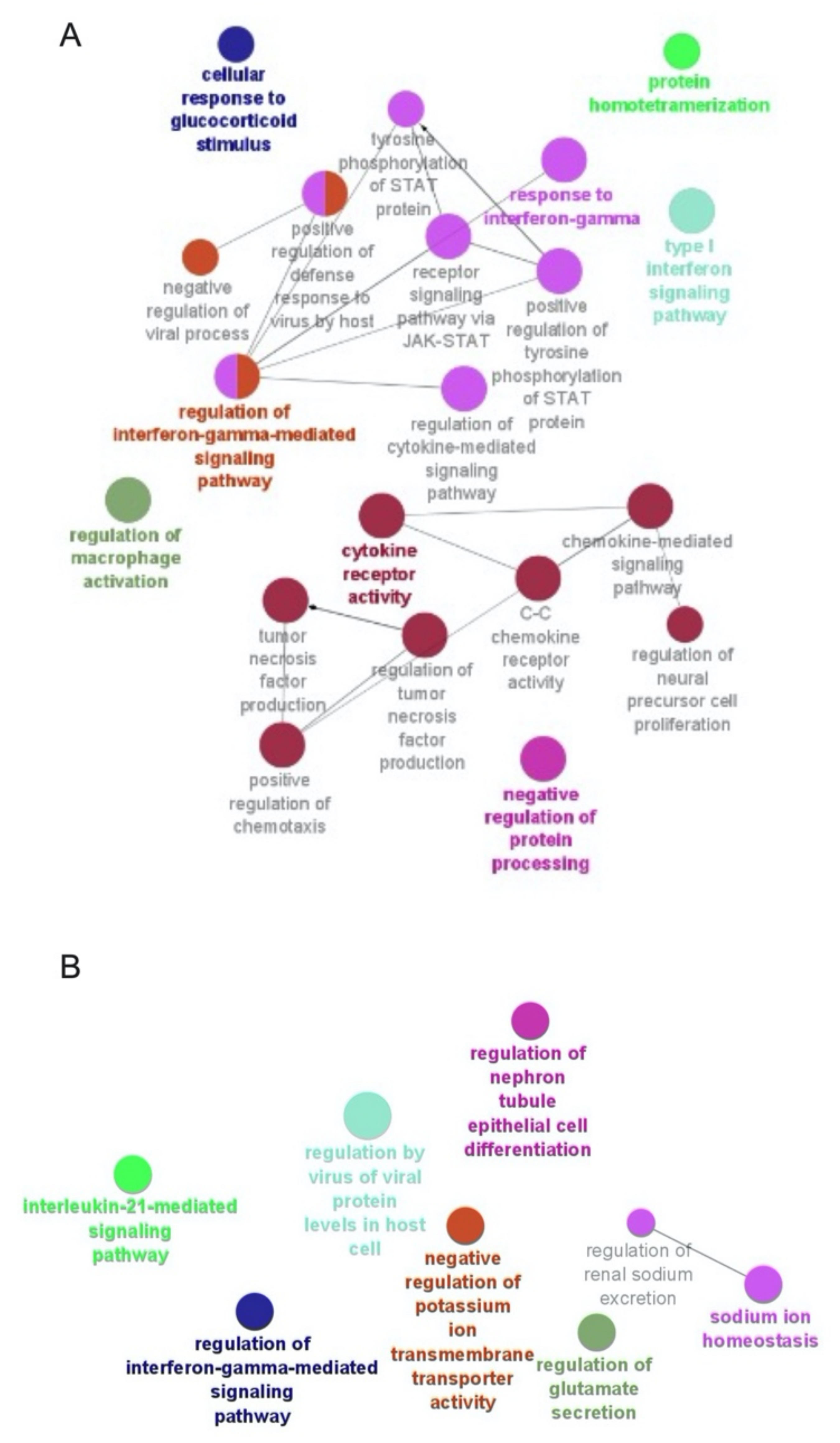
2.4. Functional Enrichment Analysis
2.5. Statistical Analysis
3. Results
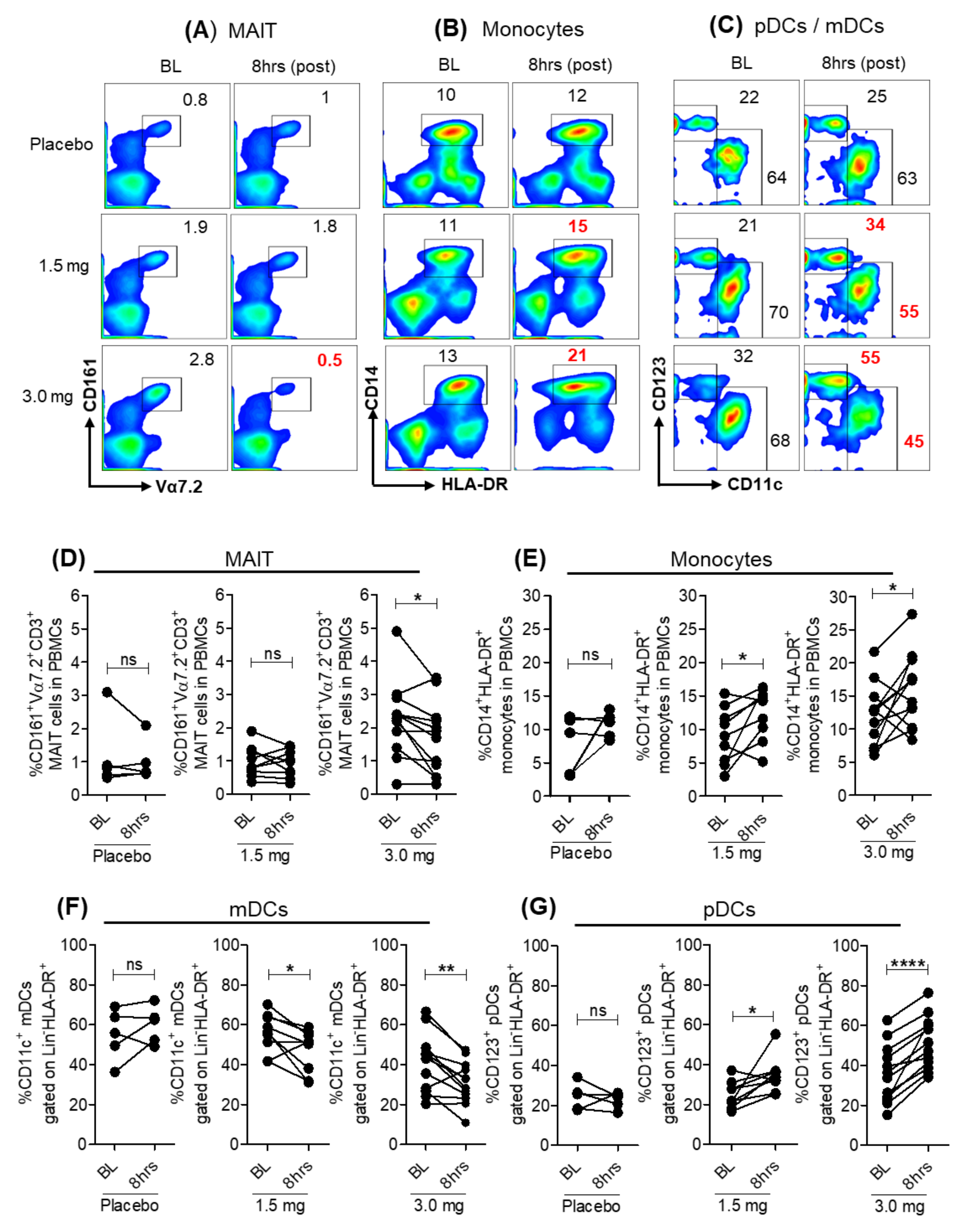
3.1. Modulation of Peripheral Blood Immune Cell Frequencies with SLGN Treatment in Healthy Subjects
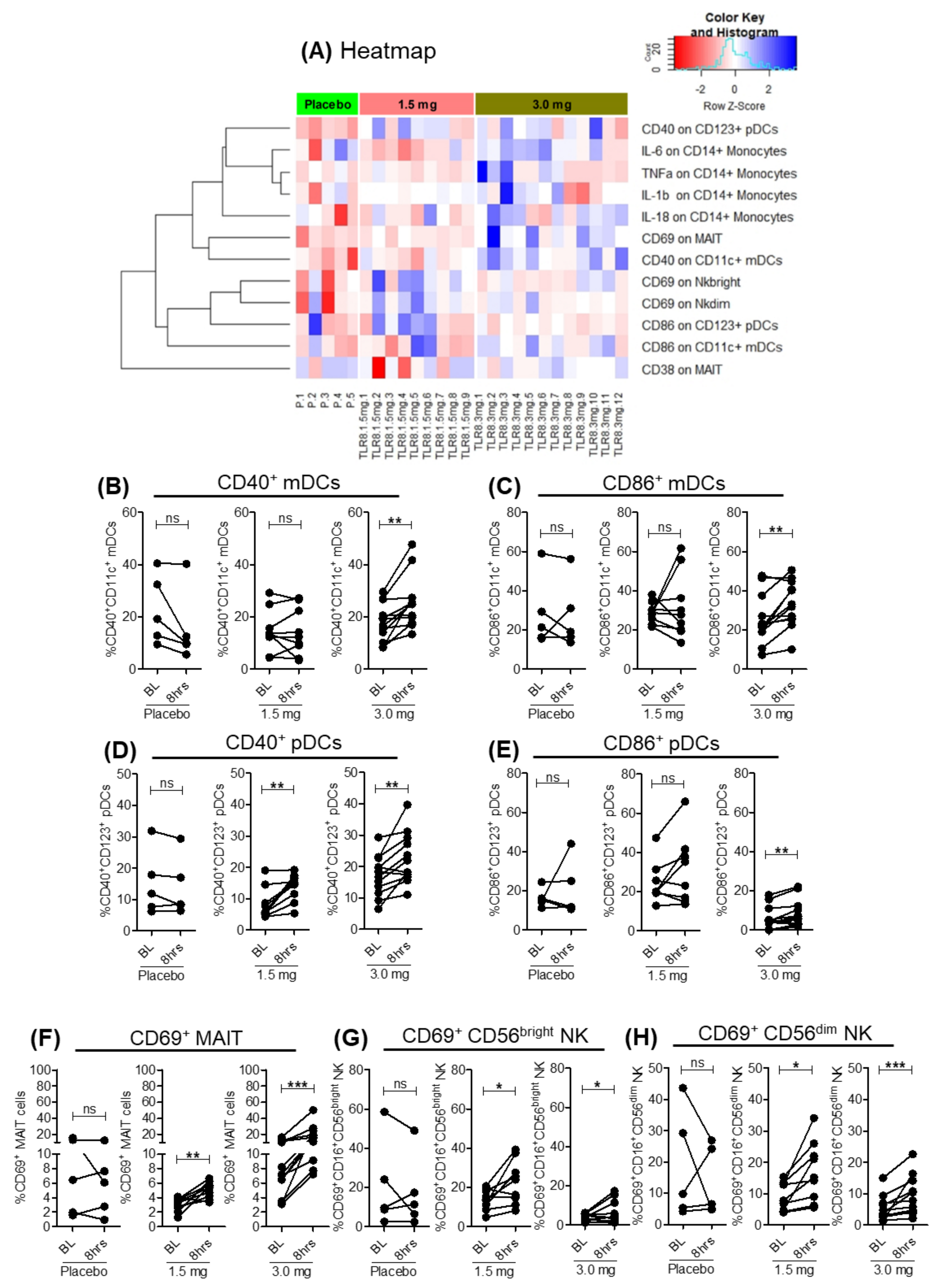
3.2. SLGN Activated Myeloid and Lymphoid Immune Cells in Peripheral Blood
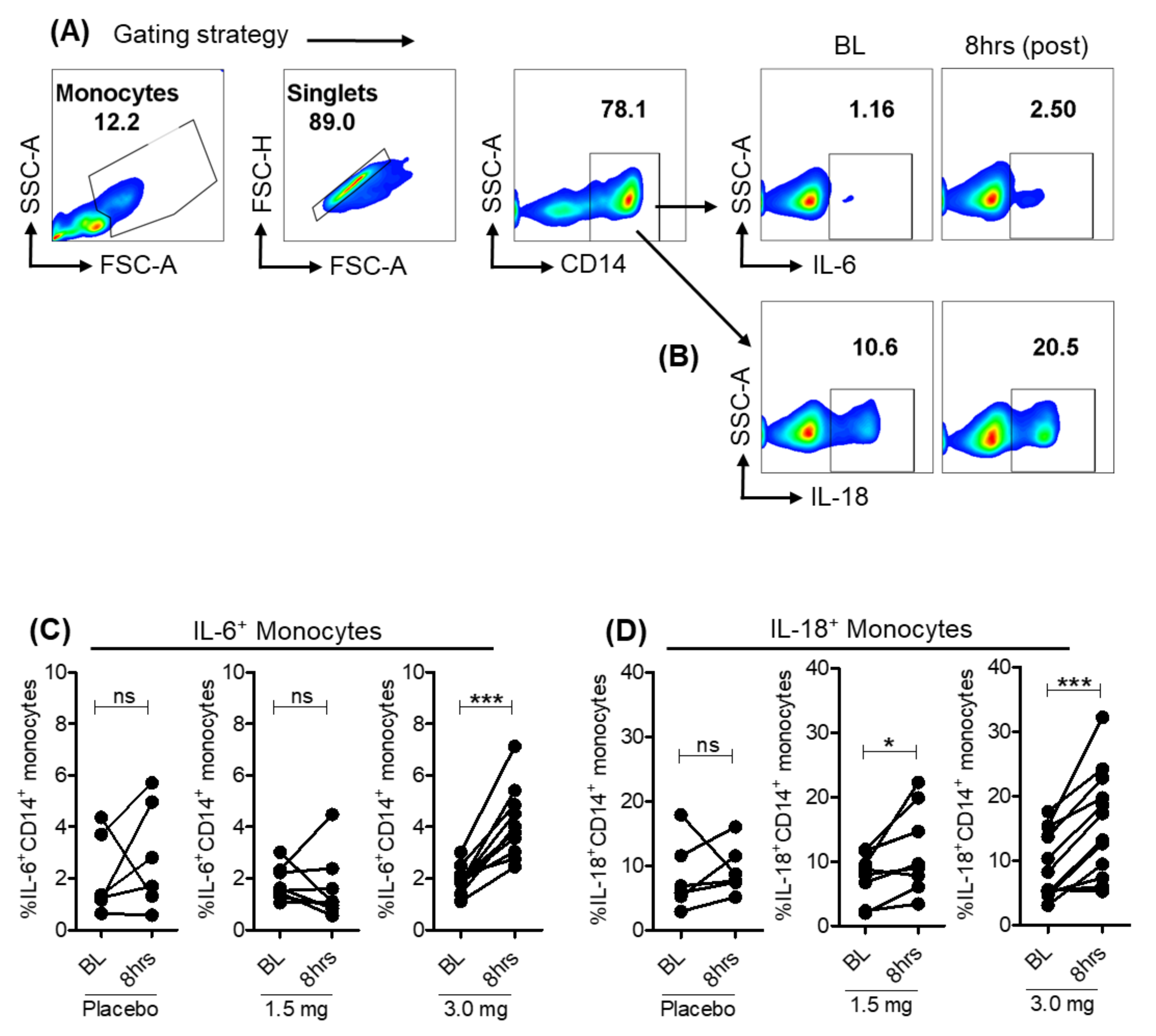
3.3. Pro-Inflammatory Cytokines Induced in Monocytes with Selgantolimod
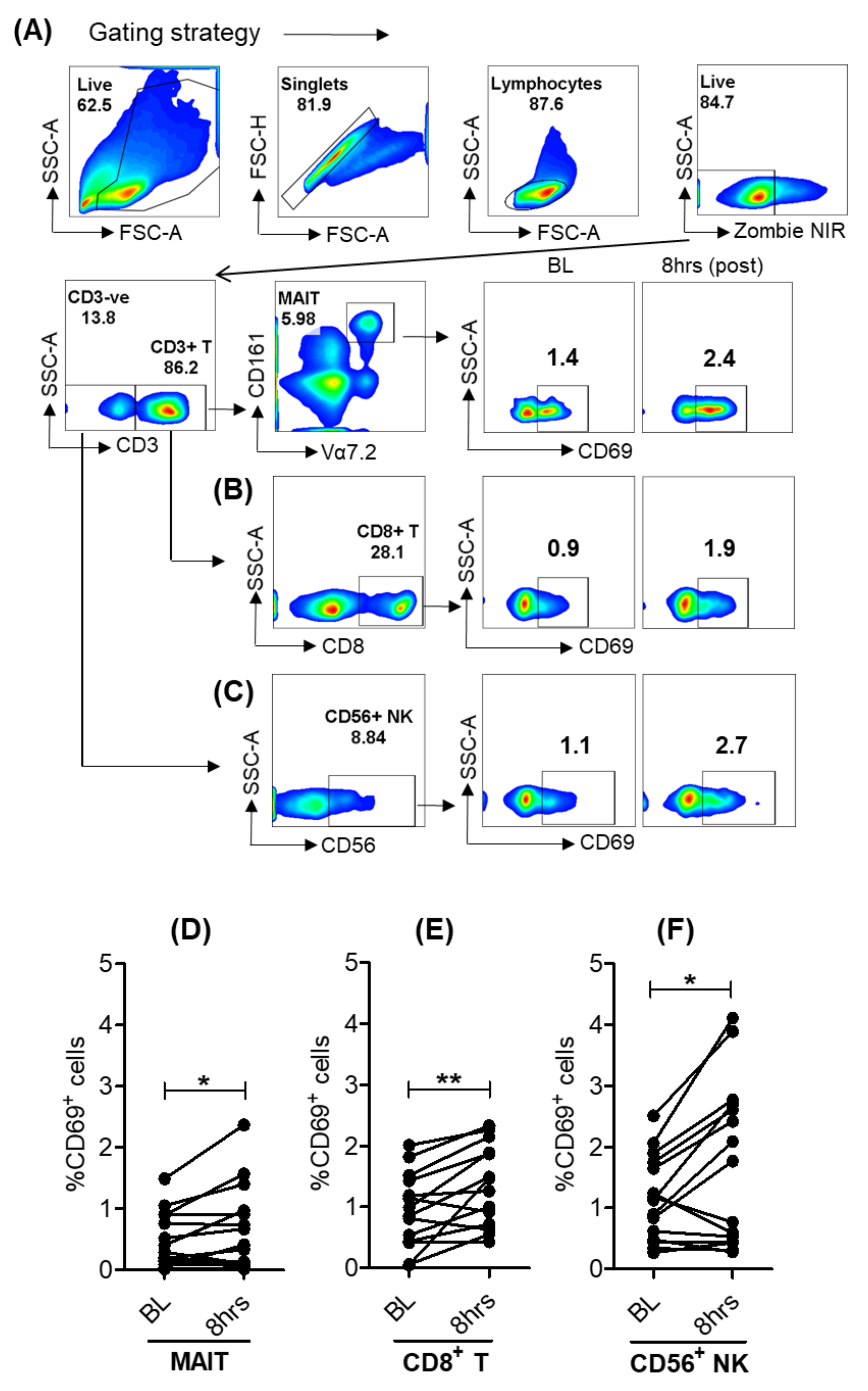
3.4. Selgantolimod Treatment Enhanced Production of Granzyme B and Perforin in MAIT and NK Cells
3.5. Activation of Lymphoid Cells after Oral Selgantolimod Treatment in CHB Subjects
3.6. Selgantolimod (SLGN) Administration Modulated Global Gene Expression in Monocytes and Lymphocytes from PBMCs of Healthy Subjects
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eigenbrod, T.; Pelka, K.; Latz, E.; Kreikemeyer, B.; Dalpke, A.H. TLR8 Senses Bacterial RNA in Human Monocytes and Plays a Nonredundant Role for Recognition of Streptococcus pyogenes. J. Immunol. 2015, 195, 1092–1099. [Google Scholar] [CrossRef] [Green Version]
- Gorden, K.B.; Gorski, K.S.; Gibson, S.J.; Kedl, R.M.; Kieper, W.C.; Qiu, X.; Tomai, M.A.; Alkan, S.S.; Vasilakos, J.P. Synthetic TLR agonists reveal functional differences between human TLR7 and TLR8. J. Immunol. 2005, 174, 1259–1268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jurk, M.; Heil, F.; Vollmer, J.; Schetter, C.; Krieg, A.M.; Wagner, H.; Lipford, G.; Bauer, S. Human TLR7 or TLR8 independently confer responsiveness to the antiviral compound R-848. Nat. Immunol. 2002, 3, 499. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 2011, 34, 637–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, G.; Guo, Z.; Kiniwa, Y.; Voo, K.S.; Peng, W.; Fu, T.; Wang, D.Y.; Li, Y.; Wang, H.Y.; Wang, R.F. Toll-like receptor 8-mediated reversal of CD4+ regulatory T cell function. Science 2005, 309, 1380–1384. [Google Scholar] [CrossRef] [PubMed]
- Cervantes, J.L.; Weinerman, B.; Basole, C.; Salazar, J.C. TLR8: The forgotten relative revindicated. Cell Mol. Immunol. 2012, 9, 434–438. [Google Scholar] [CrossRef] [Green Version]
- Kieffer, M.E.; Patel, A.M.; Hollingsworth, S.A.; Seganish, W.M. Small molecule agonists of toll-like receptors 7 and 8: A patent review 2014–2020. Expert Opin. Ther. Pat. 2020, 30, 825–845. [Google Scholar] [CrossRef] [PubMed]
- Ayithan, N.; Tang, L.; Tan, S.T.; Chen, D.; Wallin, J.J.; Fletcher, S.P.; Kottilil, S.; Poonia, B. Follicular helper T (TFH) cell targeting by TLR8 signaling for improving HBsAg-specific B cell response in chronic hepatitis B patients. Front. Immunol. 2021, 12, 735913. [Google Scholar] [CrossRef] [PubMed]
- Jo, J.; Tan, A.T.; Ussher, J.E.; Sandalova, E.; Tang, X.Z.; Tan-Garcia, A.; To, N.; Hong, M.; Chia, A.; Gill, U.S.; et al. Toll-like receptor 8 agonist and bacteria trigger potent activation of innate immune cells in human liver. PLoS Pathog. 2014, 10, e1004210. [Google Scholar] [CrossRef] [Green Version]
- Bennett, M.S.; Trivedi, S.; Iyer, A.S.; Hale, J.S.; Leung, D.T. Human mucosal-associated invariant T (MAIT) cells possess capacity for B cell help. J. Leukoc. Biol. 2017, 102, 1261–1269. [Google Scholar] [CrossRef]
- Ussher, J.E.; Willberg, C.B.; Klenerman, P. MAIT cells and viruses. Immunol. Cell Biol. 2018, 96, 630–641. [Google Scholar] [CrossRef]
- Alter, G.; Suscovich, T.J.; Teigen, N.; Meier, A.; Streeck, H.; Brander, C.; Altfeld, M. Single-stranded RNA derived from HIV-1 serves as a potent activator of NK cells. J. Immunol. 2007, 178, 7658–7666. [Google Scholar] [CrossRef]
- Schlaepfer, E.; Speck, R.F. Anti-HIV activity mediated by natural killer and CD8+ cells after toll-like receptor 7/8 triggering. PLoS ONE 2008, 3, e1999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giraldo, D.M.; Hernandez, J.C.; Urcuqui-Inchima, S. HIV-1-derived single-stranded RNA acts as activator of human neutrophils. Immunol. Res. 2016, 64, 1185–1194. [Google Scholar] [CrossRef] [PubMed]
- Mackman, R.L.; Mish, M.; Chin, G.; Perry, J.K.; Appleby, T.; Aktoudianakis, V.; Metobo, S.; Pyun, P.; Niu, C.; Daffis, S.; et al. Discovery of GS-9688 (Selgantolimod) as a Potent and Selective Oral Toll-Like Receptor 8 Agonist for the Treatment of Chronic Hepatitis, B. J. Med. Chem. 2020, 63, 10188–10203. [Google Scholar] [CrossRef]
- Daffis, S.; Balsitis, S.; Chamberlain, J.; Zheng, J.; Santos, R.; Rowe, W.; Ramakrishnan, D.; Pattabiraman, D.; Spurlock, S.; Chu, R.; et al. Toll-Like Receptor 8 Agonist GS-9688 Induces Sustained Efficacy in the Woodchuck Model of Chronic Hepatitis, B. Hepatology 2020, 73, 53–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reyes, M.; Lutz, J.D.; Lau, A.H.; Gaggar, A.; Grant, E.P.; Joshi, A.; Mackman, R.L.; Ling, J.; Tan, S.K.; Ayithan, A.; et al. Safety, pharmacokinetics and pharmacodynamics of selgantolimod, an oral Toll-like receptor 8 agonist: A Phase Ia study in healthy subjects. Antivir. Ther. 2020, 25, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Gane, E.J.; Kim, H.J.; Visvanathan, K.; Kim, Y.J.; Nguyen, A.H.; Wallin, J.J.; Chen, D.Y.; McDonald, C.; ARORA, p.; Tan, S.K.; et al. Safety, pharmacokinetics, and pharmacodynamics of the oral TLR8 agonist selgantolimod in chronic hepatitis B. Hepatology 2021, 74, 1737–1749. [Google Scholar] [CrossRef] [PubMed]
- Amin, O.E.; Colbeck, E.J.; Daffis, S.; Khan, S.; Ramakrishnan, D.; Pattabiraman, D.; Chu, R.; Steuer, H.M.; Lehar, S.; Peiser, L.; et al. Therapeutic potential of TLR8 agonist GS-9688 (selgantolimod) in chronic hepatitis B: Re-Modelling of antiviral and regulatory mediators. Hepatology 2021, 74, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jensen, L.J.; Kuhn, M.; Stark, M.; Chaffron, S.; Creevey, C.; Muller, J.; Doerks, T.; Julien, P.; Roth, A.; Simonovic, M.; et al. STRING 8—A global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009, 37 (Suppl. 1), D412–D416. [Google Scholar] [CrossRef] [PubMed]
- Ussher, J.E.; Bilton, M.; Attwod, E.; Shadwell, J.; Richardson, R.; Lara, d.C.; Mettke, E.; Kurioka, A.; Hansen, T.H.; Klenerman, P.; et al. CD161++ CD8+ T cells, including the MAIT cell subset, are specifically activated by IL-12+IL-18 in a TCR-independent manner. Eur. J. Immunol. 2014, 44, 195–203. [Google Scholar] [CrossRef]
- Wilgenburg, v.B.; Scherwitzl, I.; Hutchinson, E.C.; Leng, T.; Kurioka, A.; Kulicke, C.; Lara, C.d.; Cole, S.; Vasanawathana, S.; Limpitikul, W.; et al. MAIT cells are activated during human viral infections. Nat. Commun. 2016, 7, 11653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boeijen, L.L.; Montanari, N.R.; Groen, d.R.A.; Oord, d.G.W.; Heide-Mulder, M.v.d.; Knegt, R.J.d.; Boonstra, A. Mucosal-Associated Invariant T Cells Are More Activated in Chronic Hepatitis B, but Not Depleted in Blood: Reversal by Antiviral Therapy. J. Infect. Dis. 2017, 216, 969–976. [Google Scholar] [CrossRef]
- Locy, H.; Mey, S.d.; Mey, W.d.; Ridder, M.D.; Thielemans, K.; Maenhout, S.K. Immunomodulation of the Tumor Microenvironment: Turn Foe into Friend. Front. Immunol. 2018, 9, 2909. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Cao, Q.; Xiong, Y.; Zhang, E.; Lu, M. Interaction between Hepatitis B Virus and Toll-Like Receptors: Current Status and Potential Therapeutic Use for Chronic Hepatitis, B. Vaccines 2018, 6, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, G.; Ge, J.; Liu, C.; Pang, J.; Huang, Z.; Peng, J.; Sun, J.; Hou, J.L.; Zhang, X.Y. Impaired expression and function of TLR8 in chronic HBV infection and its association with treatment responses during peg-IFN-alpha-2a antiviral therapy. Clin. Res. Hepatol. Gastroenterol. 2017, 41, 386–398. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ayithan, N.; Ghosh, A.; Dwivedi, A.; Wallin, J.J.; Tan, S.K.; Chen, D.; Kottilil, S.; Poonia, B. Oral Selective TLR8 Agonist Selgantolimod Induces Multiple Immune Cell Responses in Humans. Viruses 2021, 13, 2400. https://doi.org/10.3390/v13122400
Ayithan N, Ghosh A, Dwivedi A, Wallin JJ, Tan SK, Chen D, Kottilil S, Poonia B. Oral Selective TLR8 Agonist Selgantolimod Induces Multiple Immune Cell Responses in Humans. Viruses. 2021; 13(12):2400. https://doi.org/10.3390/v13122400
Chicago/Turabian StyleAyithan, Natarajan, Alip Ghosh, Ankit Dwivedi, Jeffrey J. Wallin, Susanna K. Tan, Diana Chen, Shyam Kottilil, and Bhawna Poonia. 2021. "Oral Selective TLR8 Agonist Selgantolimod Induces Multiple Immune Cell Responses in Humans" Viruses 13, no. 12: 2400. https://doi.org/10.3390/v13122400
APA StyleAyithan, N., Ghosh, A., Dwivedi, A., Wallin, J. J., Tan, S. K., Chen, D., Kottilil, S., & Poonia, B. (2021). Oral Selective TLR8 Agonist Selgantolimod Induces Multiple Immune Cell Responses in Humans. Viruses, 13(12), 2400. https://doi.org/10.3390/v13122400





