Small RNA Response to Infection of the Insect-Specific Lammi Virus and Hanko Virus in an Aedes albopictus Cell Line
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Virus Stocks and Virus Titration
2.3. In Vitro Infection
2.4. RNA Extractions
2.5. qPCR
2.6. High-Throughput Sequencing of Small RNA
2.7. Small RNA Sequence Analysis
2.8. Data Availability
3. Results
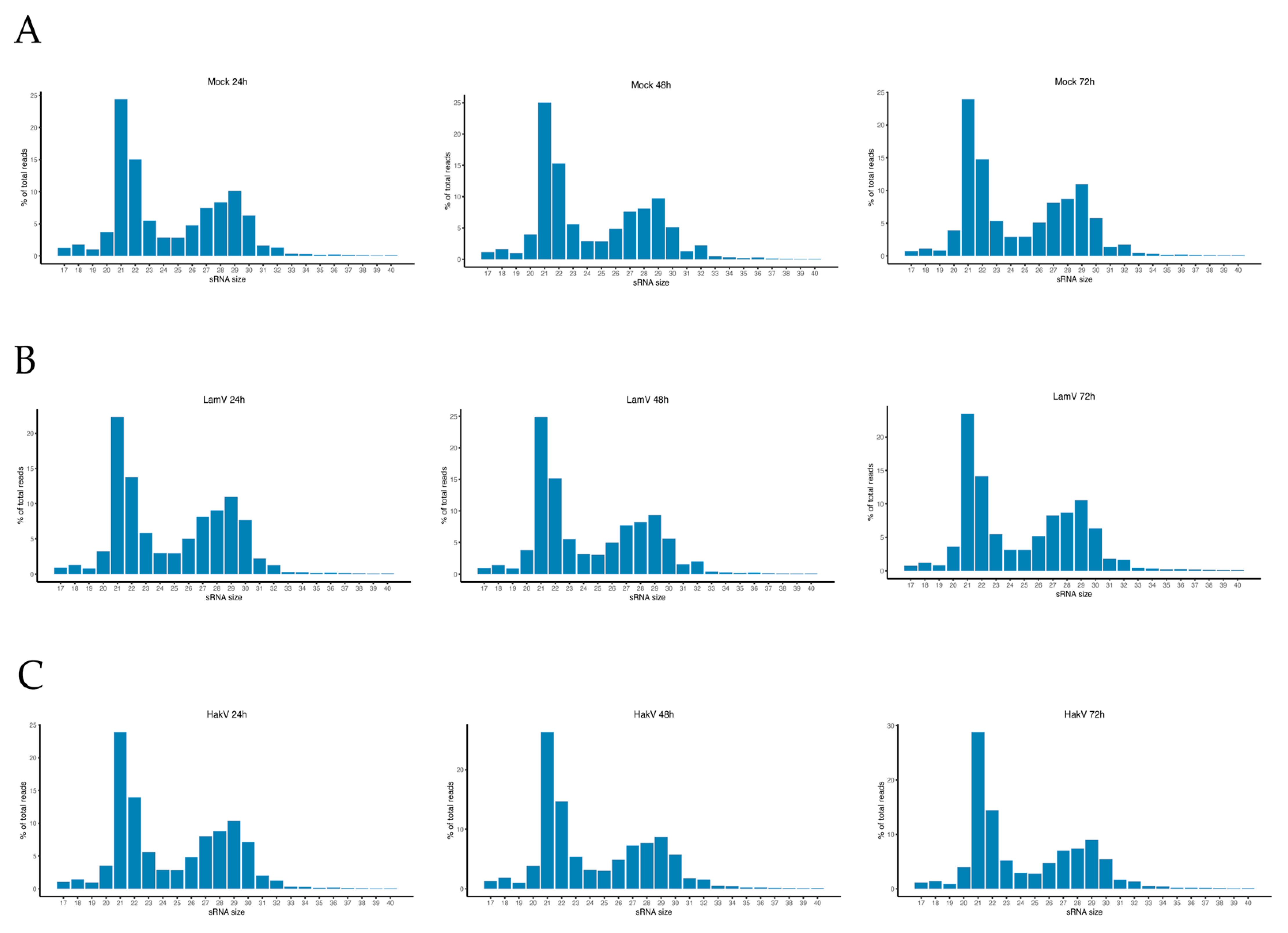
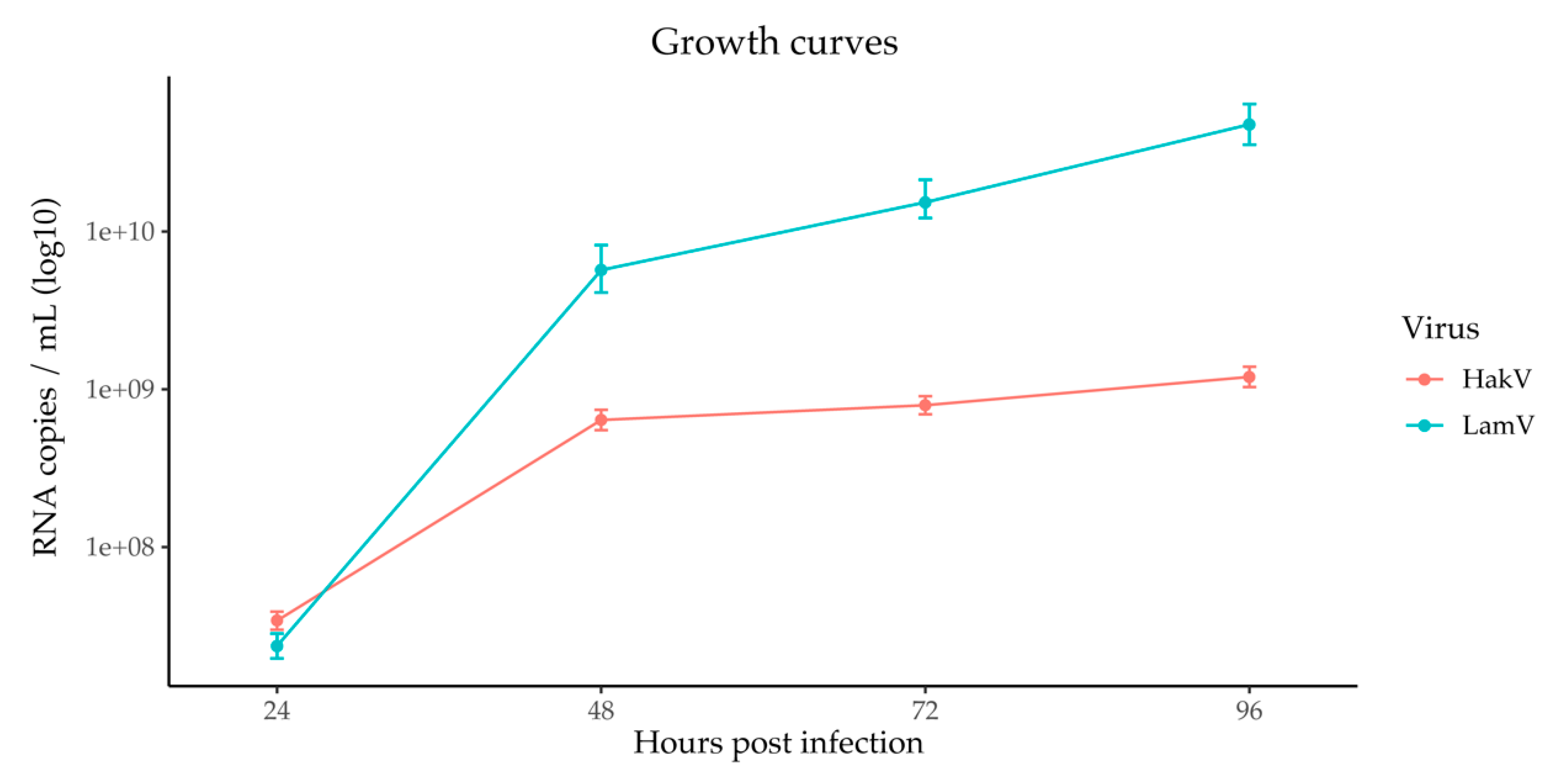
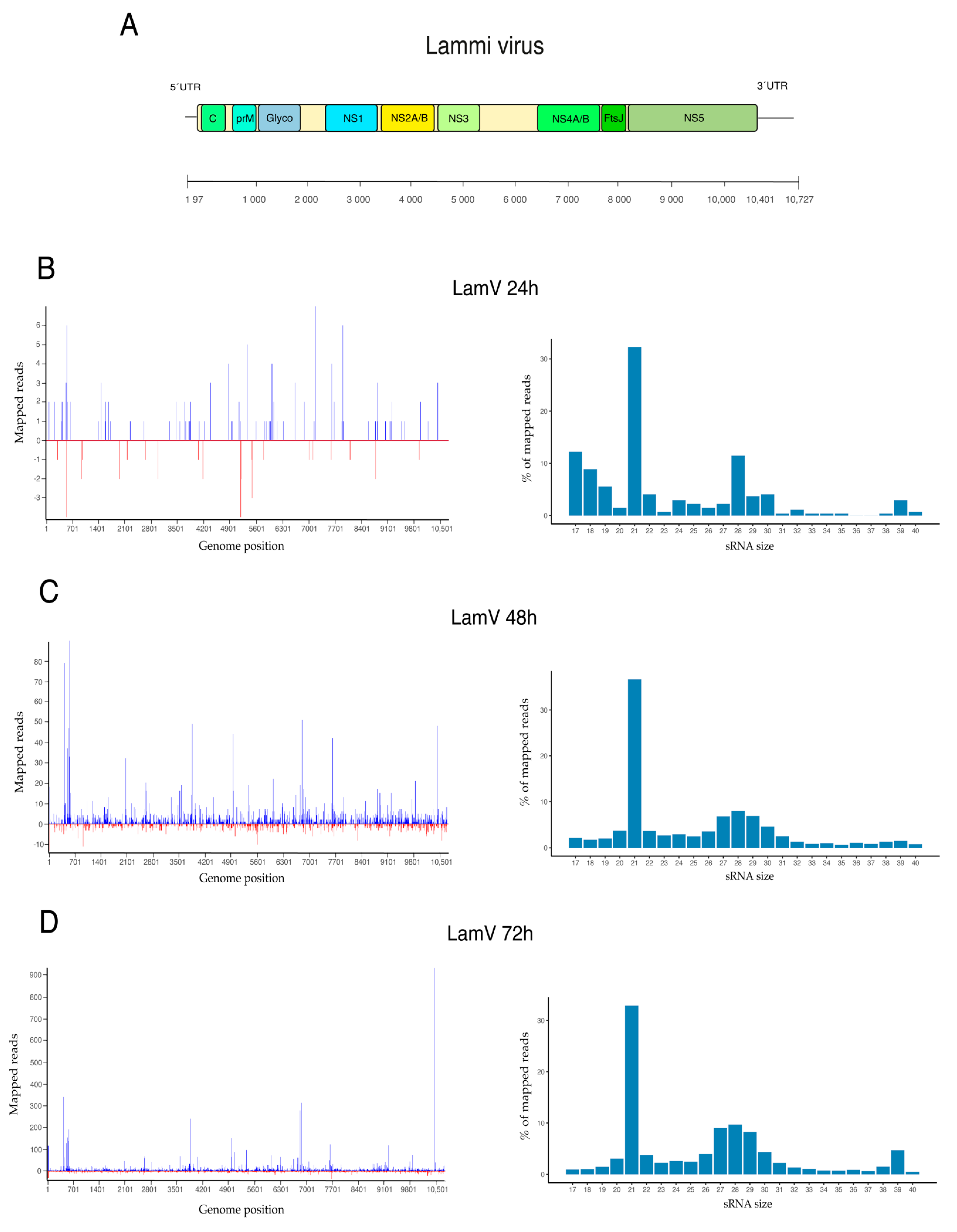
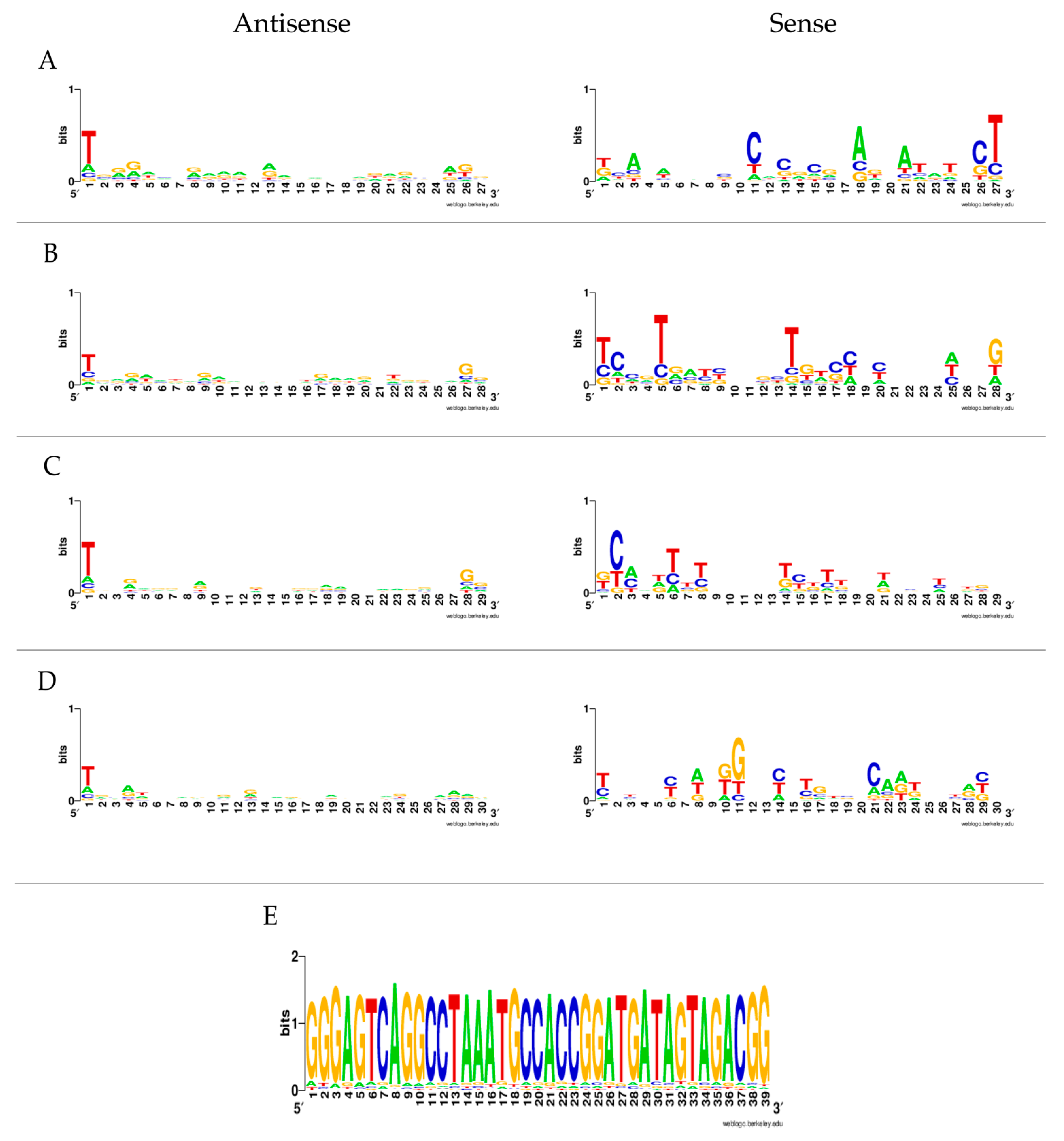
3.1. Viral Small RNA Profiles in U4.4 Infected with LamV
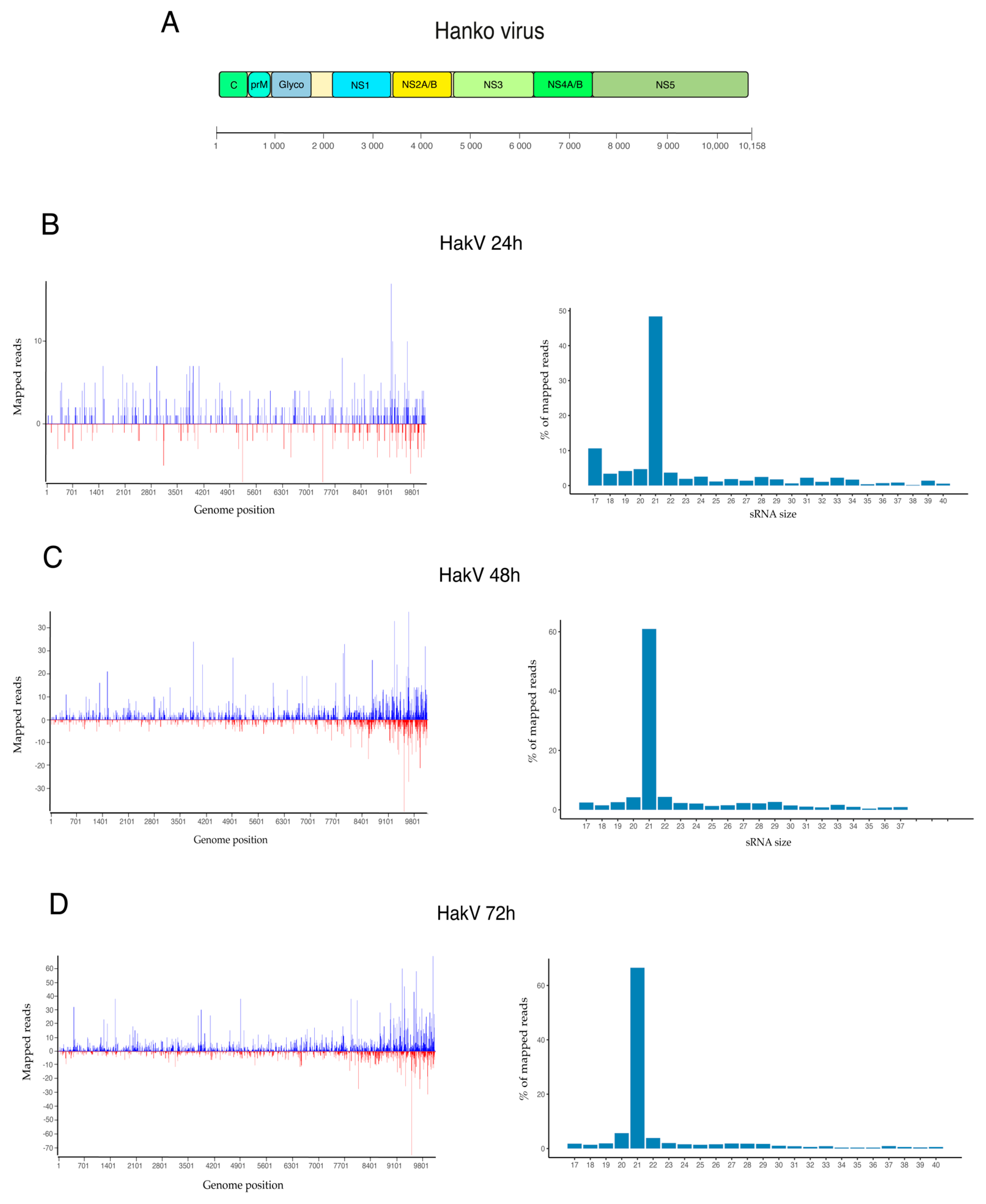
3.2. Viral Small RNA Profiles in U4.4 Infected with HakV
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nasar, F.; Gorchakov, R.V.; Tesh, R.B.; Weaver, S.C. Eilat virus host range restriction is present at multiple levels of the virus life cycle. J. Virol. 2015, 89, 1404–1418. [Google Scholar] [CrossRef] [Green Version]
- Saiyasombat, R.; Bolling, B.G.; Brault, A.C.; Bartholomay, L.C.; Blitvich, B.J. Evidence of efficient transovarial transmission of Culex flavivirus by Culex pipiens (Diptera: Culicidae). J. Med. Entomol. 2011, 48, 1031–1038. [Google Scholar] [CrossRef]
- Bolling, B.G.; Eisen, L.; Moore, C.G.; Blair, C.D. Insect-specific flaviviruses from Culex mosquitoes in Colorado, with evidence of vertical transmission. Am. J. Trop. Med. Hyg. 2011, 85, 169–177. [Google Scholar] [CrossRef] [Green Version]
- Haddow, A.D.; Guzman, H.; Popov, V.L.; Wood, T.G.; Widen, S.G.; Haddow, A.D.; Tesh, R.B.; Weaver, S.C. First isolation of Aedes flavivirus in the Western Hemisphere and evidence of vertical transmission in the mosquito Aedes (Stegomyia) albopictus (Diptera: Culicidae). Virology 2013, 440, 134–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marklewitz, M.; Zirkel, F.; Kurth, A.; Drosten, C.; Junglen, S. Evolutionary and phenotypic analysis of live virus isolates suggests arthropod origin of a pathogenic RNA virus family. Proc. Natl. Acad. Sci. USA 2015, 112, 7536–7541. [Google Scholar] [CrossRef] [Green Version]
- Li, C.X.; Shi, M.; Tian, J.H.; Lin, X.D.; Kang, Y.J.; Chen, L.J.; Qin, X.C.; Xu, J.; Holmes, E.C.; Zhang, Y.Z. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. eLife 2015, 4. [Google Scholar] [CrossRef]
- Blitvich, B.J.; Firth, A.E. Insect-specific flaviviruses: A systematic review of their discovery, host range, mode of transmission, superinfection exclusion potential and genomic organization. Viruses 2015, 7, 1927–1959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huhtamo, E.; Putkuri, N.; Kurkela, S.; Manni, T.; Vaheri, A.; Vapalahti, O.; Uzcategui, N.Y. Characterization of a novel flavivirus from mosquitoes in northern europe that is related to mosquito-borne flaviviruses of the tropics. J. Virol. 2009, 83, 9532–9540. [Google Scholar] [CrossRef] [Green Version]
- Calzolari, M.; Ze-Ze, L.; Ruzek, D.; Vazquez, A.; Jeffries, C.; Defilippo, F.; Osorio, H.C.; Kilian, P.; Ruiz, S.; Fooks, A.R.; et al. Detection of mosquito-only flaviviruses in Europe. J. Gen. Virol. 2012, 93, 1215–1225. [Google Scholar] [CrossRef] [Green Version]
- Blair, C.D. Mosquito RNAi is the major innate immune pathway controlling arbovirus infection and transmission. Future Microbiol. 2011, 6, 265–277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galiana-Arnoux, D.; Dostert, C.; Schneemann, A.; Hoffmann, J.A.; Imler, J.L. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat. Immunol. 2006, 7, 590–597. [Google Scholar] [CrossRef]
- Matranga, C.; Tomari, Y.; Shin, C.; Bartel, D.P.; Zamore, P.D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 2005, 123, 607–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rand, T.A.; Petersen, S.; Du, F.; Wang, X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 2005, 123, 621–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell, C.L.; Keene, K.M.; Brackney, D.E.; Olson, K.E.; Blair, C.D.; Wilusz, J.; Foy, B.D. Aedes aegypti uses RNA interference in defense against Sindbis virus infection. BMC Microbiol. 2008, 8, 47. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Vargas, I.; Scott, J.C.; Poole-Smith, B.K.; Franz, A.W.; Barbosa-Solomieu, V.; Wilusz, J.; Olson, K.E.; Blair, C.D. Dengue virus type 2 infections of Aedes aegypti are modulated by the mosquito’s RNA interference pathway. PLoS Pathog 2009, 5, e1000299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keene, K.M.; Foy, B.D.; Sanchez-Vargas, I.; Beaty, B.J.; Blair, C.D.; Olson, K.E. RNA interference acts as a natural antiviral response to O’nyong-nyong virus (Alphavirus; Togaviridae) infection of Anopheles gambiae. Proc. Natl. Acad. Sci. USA 2004, 101, 17240–17245. [Google Scholar] [CrossRef] [Green Version]
- Franz, A.W.; Sanchez-Vargas, I.; Adelman, Z.N.; Blair, C.D.; Beaty, B.J.; James, A.A.; Olson, K.E. Engineering RNA interference-based resistance to dengue virus type 2 in genetically modified Aedes aegypti. Proc. Natl. Acad. Sci. USA 2006, 103, 4198–4203. [Google Scholar] [CrossRef] [Green Version]
- Brackney, D.E.; Beane, J.E.; Ebel, G.D. RNAi targeting of West Nile virus in mosquito midguts promotes virus diversification. PLoS Pathog. 2009, 5, e1000502. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.S.; Nakahara, K.; Pham, J.W.; Kim, K.; He, Z.; Sontheimer, E.J.; Carthew, R.W. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 2004, 117, 69–81. [Google Scholar] [CrossRef] [Green Version]
- Puthiyakunnon, S.; Yao, Y.; Li, Y.; Gu, J.; Peng, H.; Chen, X. Functional characterization of three MicroRNAs of the Asian tiger mosquito, Aedes albopictus. Parasit Vectors 2013, 6, 230. [Google Scholar] [CrossRef] [Green Version]
- Slonchak, A.; Hussain, M.; Torres, S.; Asgari, S.; Khromykh, A.A. Expression of mosquito microRNA Aae-miR-2940-5p is downregulated in response to West Nile virus infection to restrict viral replication. J. Virol. 2014, 88, 8457–8467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saldana, M.A.; Etebari, K.; Hart, C.E.; Widen, S.G.; Wood, T.G.; Thangamani, S.; Asgari, S.; Hughes, G.L. Zika virus alters the microRNA expression profile and elicits an RNAi response in Aedes aegypti mosquitoes. PLoS Negl. Trop. Dis. 2017, 11, e0005760. [Google Scholar] [CrossRef] [PubMed]
- Campbell, C.L.; Harrison, T.; Hess, A.M.; Ebel, G.D. MicroRNA levels are modulated in Aedes aegypti after exposure to Dengue-2. Insect Mol. Biol. 2014, 23, 132–139. [Google Scholar] [CrossRef] [Green Version]
- Skalsky, R.L.; Vanlandingham, D.L.; Scholle, F.; Higgs, S.; Cullen, B.R. Identification of microRNAs expressed in two mosquito vectors, Aedes albopictus and Culex quinquefasciatus. BMC Genom. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rozhkov, N.V.; Hammell, M.; Hannon, G.J. Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev. 2013, 27, 400–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marconcini, M.; Hernandez, L.; Iovino, G.; Houe, V.; Valerio, F.; Palatini, U.; Pischedda, E.; Crawford, J.E.; White, B.J.; Lin, T.; et al. Polymorphism analyses and protein modelling inform on functional specialization of Piwi clade genes in the arboviral vector Aedes albopictus. PLoS Negl. Trop. Dis. 2019, 13, e0007919. [Google Scholar] [CrossRef] [Green Version]
- Miesen, P.; Girardi, E.; van Rij, R.P. Distinct sets of PIWI proteins produce arbovirus and transposon-derived piRNAs in Aedes aegypti mosquito cells. Nucleic Acids Res. 2015, 43, 6545–6556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schnettler, E.; Donald, C.L.; Human, S.; Watson, M.; Siu, R.W.C.; McFarlane, M.; Fazakerley, J.K.; Kohl, A.; Fragkoudis, R. Knockdown of piRNA pathway proteins results in enhanced Semliki Forest virus production in mosquito cells. J. Gen. Virol. 2013, 94, 1680–1689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varjak, M.; Maringer, K.; Watson, M.; Sreenu, V.B.; Fredericks, A.C.; Pondeville, E.; Donald, C.L.; Sterk, J.; Kean, J.; Vazeille, M.; et al. Aedes aegypti Piwi4 Is a Noncanonical PIWI Protein Involved in Antiviral Responses. mSphere 2017, 2, e00144-17. [Google Scholar] [CrossRef] [Green Version]
- Tassetto, M.; Kunitomi, M.; Whitfield, Z.J.; Dolan, P.T.; Sanchez-Vargas, I.; Garcia-Knight, M.; Ribiero, I.; Chen, T.; Olson, K.E.; Andino, R. Control of RNA viruses in mosquito cells through the acquisition of vDNA and endogenous viral elements. eLife 2019, 8, e41244. [Google Scholar] [CrossRef]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000, 132, 365–386. [Google Scholar]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [Green Version]
- Seguin, J.; Otten, P.; Baerlocher, L.; Farinelli, L.; Pooggin, M.M. MISIS-2: A bioinformatics tool for in-depth analysis of small RNAs and representation of consensus master genome in viral quasispecies. J. Virol. Methods 2016, 233, 37–40. [Google Scholar] [CrossRef] [Green Version]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leger, P.; Lara, E.; Jagla, B.; Sismeiro, O.; Mansuroglu, Z.; Coppee, J.Y.; Bonnefoy, E.; Bouloy, M. Dicer-2- and Piwi-mediated RNA interference in Rift Valley fever virus-infected mosquito cells. J. Virol. 2013, 87, 1631–1648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morazzani, E.M.; Wiley, M.R.; Murreddu, M.G.; Adelman, Z.N.; Myles, K.M. Production of virus-derived ping-pong-dependent piRNA-like small RNAs in the mosquito soma. PLoS Pathog. 2012, 8, e1002470. [Google Scholar] [CrossRef]
- Siu, R.W.; Fragkoudis, R.; Simmonds, P.; Donald, C.L.; Chase-Topping, M.E.; Barry, G.; Attarzadeh-Yazdi, G.; Rodriguez-Andres, J.; Nash, A.A.; Merits, A.; et al. Antiviral RNA interference responses induced by Semliki Forest virus infection of mosquito cells: Characterization, origin, and frequency-dependent functions of virus-derived small interfering RNAs. J. Virol. 2011, 85, 2907–2917. [Google Scholar] [CrossRef] [Green Version]
- Hess, A.M.; Prasad, A.N.; Ptitsyn, A.; Ebel, G.D.; Olson, K.E.; Barbacioru, C.; Monighetti, C.; Campbell, C.L. Small RNA profiling of Dengue virus-mosquito interactions implicates the PIWI RNA pathway in anti-viral defense. BMC Microbiol. 2011, 11, 45. [Google Scholar] [CrossRef] [Green Version]
- Myles, K.M.; Morazzani, E.M.; Adelman, Z.N. Origins of alphavirus-derived small RNAs in mosquitoes. RNA Biol. 2009, 6, 387–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dietrich, I.; Shi, X.; McFarlane, M.; Watson, M.; Blomstrom, A.L.; Skelton, J.K.; Kohl, A.; Elliott, R.M.; Schnettler, E. The Antiviral RNAi Response in Vector and Non-vector Cells against Orthobunyaviruses. PLoS Negl. Trop. Dis. 2017, 11, e0005272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Q.; Srivastav, S.P.; Gamez, S.; Dayama, G.; Feitosa-Suntheimer, F.; Patterson, E.I.; Johnson, R.M.; Matson, E.M.; Gold, A.S.; Brackney, D.E.; et al. A mosquito small RNA genomics resource reveals dynamic evolution and host responses to viruses and transposons. Genome Res. 2021, 31, 512–528. [Google Scholar] [CrossRef]
- Goertz, G.P.; Miesen, P.; Overheul, G.J.; van Rij, R.P.; van Oers, M.M.; Pijlman, G.P. Mosquito Small RNA Responses to West Nile and Insect-Specific Virus Infections in Aedes and Culex Mosquito Cells. Viruses 2019, 11, 271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goic, B.; Stapleford, K.A.; Frangeul, L.; Doucet, A.J.; Gausson, V.; Blanc, H.; Schemmel-Jofre, N.; Cristofari, G.; Lambrechts, L.; Vignuzzi, M.; et al. Virus-derived DNA drives mosquito vector tolerance to arboviral infection. Nat. Commun. 2016, 7, 12410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poirier, E.Z.; Goic, B.; Tome-Poderti, L.; Frangeul, L.; Boussier, J.; Gausson, V.; Blanc, H.; Vallet, T.; Loyd, H.; Levi, L.I.; et al. Dicer-2-Dependent Generation of Viral DNA from Defective Genomes of RNA Viruses Modulates Antiviral Immunity in Insects. Cell Host Microbe 2018, 23, 353–365 e8. [Google Scholar] [CrossRef] [Green Version]
- Miesen, P.; Ivens, A.; Buck, A.H.; van Rij, R.P. Small RNA Profiling in Dengue Virus 2-Infected Aedes Mosquito Cells Reveals Viral piRNAs and Novel Host miRNAs. PLoS Negl. Trop. Dis. 2016, 10, e0004452. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007, 128, 1089–1103. [Google Scholar] [CrossRef] [Green Version]
- Varjak, M.; Donald, C.L.; Mottram, T.J.; Sreenu, V.B.; Merits, A.; Maringer, K.; Schnettler, E.; Kohl, A. Characterization of the Zika virus induced small RNA response in Aedes aegypti cells. PLoS Negl. Trop. Dis. 2017, 11, e0006010. [Google Scholar] [CrossRef] [Green Version]
- Mohn, F.; Handler, D.; Brennecke, J. Noncoding RNA. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science 2015, 348, 812–817. [Google Scholar] [CrossRef] [Green Version]
| Primers | Binding Site | Sequence (5′ → 3′) | Ref |
|---|---|---|---|
| LamV-F | 4659–4678 | TGGGTGTTACCGGGTTATGT | FJ606789 |
| LamV-R | 4845–4864 | ACGTTCCATTCAGTTTCCAT | |
| HakV-F | 4655–4674 | TGTGTTACGGTGGAAACTGG | NC030401 |
| HakV-R | 4842–4861 | CAACTGGTTCTCCGTTGACA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Öhlund, P.; Hayer, J.; Hesson, J.C.; Blomström, A.-L. Small RNA Response to Infection of the Insect-Specific Lammi Virus and Hanko Virus in an Aedes albopictus Cell Line. Viruses 2021, 13, 2181. https://doi.org/10.3390/v13112181
Öhlund P, Hayer J, Hesson JC, Blomström A-L. Small RNA Response to Infection of the Insect-Specific Lammi Virus and Hanko Virus in an Aedes albopictus Cell Line. Viruses. 2021; 13(11):2181. https://doi.org/10.3390/v13112181
Chicago/Turabian StyleÖhlund, Pontus, Juliette Hayer, Jenny C. Hesson, and Anne-Lie Blomström. 2021. "Small RNA Response to Infection of the Insect-Specific Lammi Virus and Hanko Virus in an Aedes albopictus Cell Line" Viruses 13, no. 11: 2181. https://doi.org/10.3390/v13112181
APA StyleÖhlund, P., Hayer, J., Hesson, J. C., & Blomström, A.-L. (2021). Small RNA Response to Infection of the Insect-Specific Lammi Virus and Hanko Virus in an Aedes albopictus Cell Line. Viruses, 13(11), 2181. https://doi.org/10.3390/v13112181








