Abstract
2A is an oligopeptide sequence that mediates a ribosome “skipping” effect and can mediate a co-translation cleavage of polyproteins. These sequences are widely distributed from insect to mammalian viruses and could act by accelerating adaptive capacity. These sequences have been used in many heterologous co-expression systems because they are versatile tools for cleaving proteins of biotechnological interest. In this work, we review and update the occurrence of 2A/2A-like sequences in different groups of viruses by screening the sequences available in the National Center for Biotechnology Information database. Interestingly, we reported the occurrence of 2A-like for the first time in 69 sequences. Among these, 62 corresponded to positive single-stranded RNA species, six to double stranded RNA viruses, and one to a negative-sense single-stranded RNA virus. The importance of these sequences for viral evolution and their potential in biotechnological applications are also discussed.
1. Introduction
2A and 2A-like sequences are oligopeptides with approximately 18–25 amino acids and can mediate a co-translation “cleavage” of polyproteins in eukaryotic cells. The “core” sequence at the C-terminus of 2A, together with the N-terminal proline of the downstream protein, contains the canonical motif—(G/H)1D2(V/I)3E4X5N6P7G8P9—involved in a ribosome “skipping” effect during translation, which separates two proteins without needing a proteinase [1,2].
The 2A cleavage occurs between the G8 site at the upstream protein (P1) and the P9 site at the downstream protein (P2). During amino acid insertion into the protein, the 2A sequence can cause a structural modification at the ribosome peptidyl-transferase center (PTC), making the ribosome “skip” the proline codon. It inhibits the formation of a glycine-proline peptide bond because of the hydrolysis of the peptidyl (2A)-tRNAGly ester linkage, releasing the polypeptide from the translational complex [3,4]. In this way, the first amino acid, proline, of the downstream encoded protein, is specified by the third codon in the sequence of P7G8P9, and the C-terminal amino acid of the upstream encoded protein is a glycine encoded by the second codon in that sequence [5,6]. This ribosome “skipping” effect is also referred to as “Stop-Carry On” or “StopGo” translation [6]. Thus, the ribosome activity does not depend on structural elements within the mRNA but a peptide sequence, differentiating this mechanism from the other forms of non-canonical mRNA processing. Because of this activity, the 2A and 2A-like sequences can be named CHYSELs (cis-acting hydrolase elements) [7].
Originally, the term “2A” was assigned to define a specific region of the genome of the foot-and-mouth disease virus (FMDV), a positive-sense single-stranded RNA (pssRNA) virus and member of the Picornaviridae family [1,4,8,9,10]. Similar sequences discovered in other viruses were named “2A-like.” These sequences have been described in other Picornaviridae, such as Equine rhinitis A virus and Porcine teschovirus-1, in other viruses of the Dicistroviridae and Iflaviviridae families [2], and even in the infectious myonecrosis virus (IMNV), a double-stranded RNA (dsRNA) virus belonging to the Totiviridae family [11].
From these first discoveries, the 2A and 2A-like proteolytic cleavage activities have been demonstrated in several eukaryotic systems in vitro and in vivo [2,12]. Because of their mechanism of action, some authors also refer to 2A and 2A-like peptides as cis-acting hydrolase elements [7,13].
In 2017, Yang et al. reviewed the 2A sequence structures and functions of Picornaviridae members [14]. The latest works analyzing 2A and 2A-like sequences, including viruses from other families, were conducted by Luke et al. in 2008, 2009, and 2014 and by Luke and Ryan in 2013 [2,15,16,17]. With advances in sequencing technology, in recent years, there has been a significant increase in the number of viral sequences added to the National Center for Biotechnology Information (NCBI) database. Therefore, the goal of this article was to introduce a new screening of 2A and 2A-like sequences in viral genomes available from the NCBI database to revise the principal 2A and 2A-like sequences, describe their occurrence in different viral families, and discuss their potential applications in biotechnology.
2. Materials and Methods
The sequences used in this study were obtained from the viral databank (https://www.ncbi.nlm.nih.gov/genome/viruses/, accessed on 9 January 2021). To find 2A/2A-like sequences, the viral genomes were aligned against some of the 2A/2A-like classical motifs (GDVEENPGP; GDVESNPGP; HDIETNPGP; GDVELNPGP; GDIELNPGP; GDIESNPGP; HDVEMNPGP) using the Blastp tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 9 January 2021) and the non-redundant protein sequences database (nr) only including viruses (taxid:10239). Search parameters were set to return a maximum of 500 sequences for each query. Repeated viral sequences were excluded from the analysis.
An active search was performed on the publication linked to the sequence annotation in the NCBI database to identify whether the sequences found had already been reported in the literature after the initial screening. If no report was found, an active search was performed using the Google Scholar search tool, with each respective virus name plus the word “2A” as keywords. If no articles reported the presence of 2A/2A-like in the query virus, we considered this finding novel.
3. Results and Discussion
3.1. 2A/2A-Like Distribution on Viruses
Table 1 shows the principal 2A or 2A-like motifs that had their self-cleavage efficiencies tested in vitro, confirming that these sequences are widely distributed among the pssRNA and dsRNA viruses, ranging from insect to mammalian viruses. Luke et al. were the first to report this wide distribution and identified motifs similar to those found in the FMDV [2].

Table 1.
Principal 2A/2A-like motifs described in literature and their cleavage efficiency.
The search for these motifs in the viral genomes available in the NCBI database revealed 69 sequences containing 2A-like motifs that had not been identified. Among these, 62 corresponded to pssRNA, six to dsRNA, and one to a negative-sense single-stranded RNA (nssRNA) virus. Additionally, 2A-like motifs, previously described in 102 sequences, were confirmed. All 2A/2A-like motifs and their respective species resulting from the search are described in Table 2 and Table 3.

Table 2.
Positive-sense single-stranded RNA virus containing 2A-like motifs.

Table 3.
Double-stranded RNA viruses identified in this study containing 2A-like motifs.
3.2. pssRNA Viruses
Here, we registered 62 new 2A-like notifications in pssRNA viruses, as presented in Table 2 (underlined). The positions in each respective genome are shown in Figure 1.
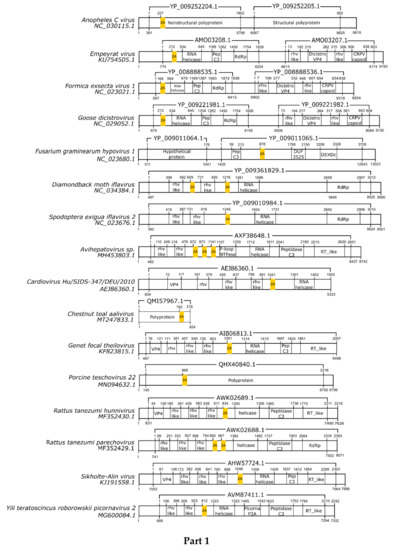
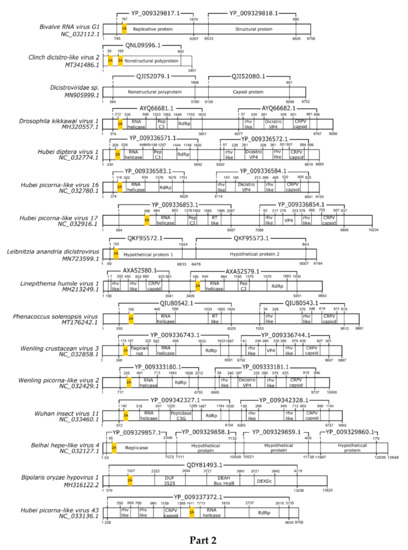
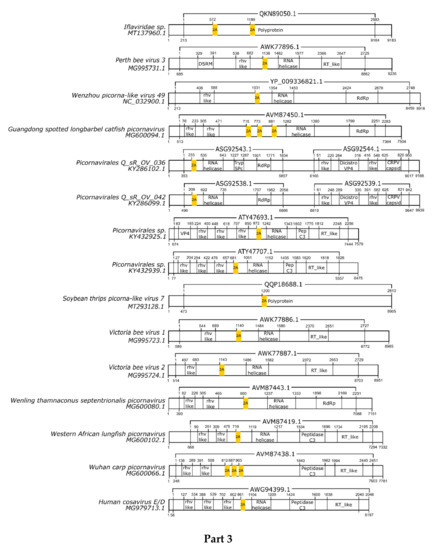
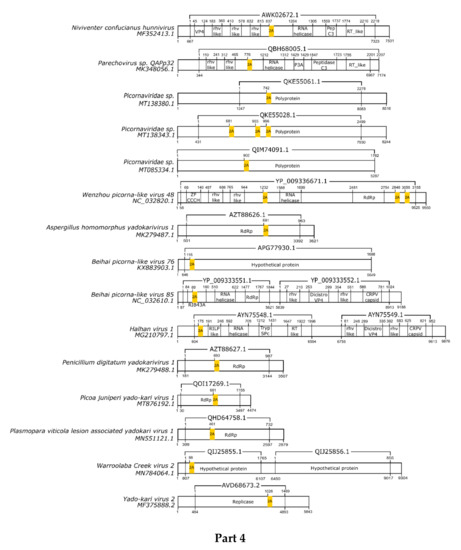
Figure 1.
Schematic representation of positive-sense single-strand RNA virus sequences. Schematic representations of pssRNA virus sequences showing the location of each respective 2A-like (yellow rectangles). The nucleotide positions and size of each predicted polypeptide are represented by the numbers below and above the bars, respectively. The annotations of each viral sequence were included according to the NCBI. The nucleotide and protein accession numbers are presented forward and above each scheme, respectively. Representations of each genome are not in scale. This figure is presented in four parts.
In most pssRNA viruses, 2A/2A-like segments are used in primary polypeptide processing. The pssRNA viruses commonly possess one 2A/2A-like sequence, but some viruses have two, three, or even four motifs (Table 2). Many of them are members of the order Picornavirales, such as Picornaviridae, Dicistroviridae, and Iflaviridae. Currently, the Picornaviridae family has 63 assigned genera [28], but 2A/2A-like sequences have been found in viruses assigned or tentatively assigned to 15 genera: Aphthovirus, Avihepatovirus, Cardiovirus, Cosavirus, Crohivirus, Erbovirus, Hunninvirus, Limnipivirus, Mischivirus, Mosavirus, Parechovirus, Pasivirus, Senecavirus, Teschovirus, and Torchivirus.
In aphthoviruses and cardioviruses, the 2A-like region self-cleaves at its own C-terminus, meaning that the 2A-like polypeptide remains as a C-terminal extension of the upstream polyprotein (P1) until it is removed by secondary proteinase cleavage [8,9]. However, in parechoviruses, the 2A-like region has no protease or protease-like activity, and its apparent function is to alter host cell metabolism because it possesses a high homology to cellular protein H-rev107 that regulates cell proliferation (H-box 2A) [29].
In insect Iflaviruses, the 2A-like sequence separates the capsid and replicative protein domains. The Dicistroviridae family is composed of the Aparavirus, Cripavirus, and Triatovirus genera, in which the 2A-like sequences occur at the N-terminal region of the replicative protein open reading frame (ORF) [2,14].
Members of the Permutotetraviridae and Carmotetraviridae families (previously Tetraviridae), Thosea asigna virus and Euprosterna elaeasa virus, encode a 2A-like sequence at the N-terminus of the structural ORF [1]. The Providence virus has three 2A-like sequences, 2A2 and 2A3, located in the capsid protein precursor (VCAP), and 2A1 at the N-terminus of the p130 ORF, which encodes the viral replicase [30].
3.3. dsRNA Viruses
Among the dsRNA viruses, 2A-like sequences not yet reported were found in six species. The new 2A-like sequences are underlined in Table 3, and their localization inside the genome is schematized in Figure 2.
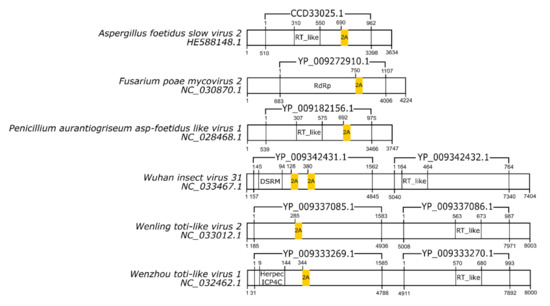
Figure 2.
Schematic representation of double-stranded RNA virus sequences. Schematic representations of dsRNA virus sequences showing the location of each respective 2A-like (yellow rectangles). The nucleotide positions and size of each predicted polypeptide are represented by the numbers below and above the bars, respectively. The annotations of each viral sequence were made according to the information available at the NCBI. The nucleotide and protein accession numbers are located forward and above each scheme, respectively. Representations of each genome are not in scale.
In double-stranded viruses, 2A-like sequences are present in two families: Totiviridae and Reoviridae. In Totiviridae, 2A-like sequences are distributed in all representatives of the IMNV-like group [31]. These viruses predominantly infect arthropods, such as penaeid shrimp [32], mosquitoes [33,34], and the fruit fly Drosophila melanogaster [35], except for the golden shiner Totivirus that infects the fish Notemigonus crysoleucas [36]. The genome of IMNV-like viruses is composed of two ORFs, and the 2A-like sequences separate an RNA-binding protein of other putative proteins in ORF1 [37].
In the Reoviridae family, 2A-like sequences are found in cypoviruses and rotaviruses with 2A-like sequences in one of the segments encoding a non-structural protein. In Operophtera brumata cypovirus 18 and Bombyx mori cypovirus 1, 2A-like sequences occur within segment 5. In type C rotaviruses, 2A-like sequences link the ssRNA-binding protein NSP3 to dsRNA-binding protein (dsRBP). In porcine and human rotavirus C, the 2A-like sequences are present at segment 6, although in the adult diarrhea virus, the sequence appears in segment 5 [1,2]. All cypoviruses and rotaviruses possess only one 2A-like sequence (Table 3).
3.4. nssRNA Virus
Surprisingly, one 2A-like motif (GDIEQNPGP) was found in a tentatively assigned virus of the Bunyaviridae family (Accession number: APG79245.1). This motif is located in the RNA-dependent RNA polymerase (RdRp) sequence (Figure 3). This is the first report of a 2A-like sequence in a nssRNA virus.
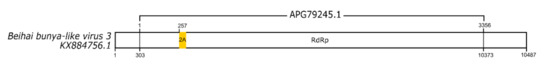
Figure 3.
Schematic representation of a negative-sense single-strand RNA virus sequence. Schematic representations of nssRNA virus sequence showing the location of its respective 2A-like sequence (yellow rectangle). The nucleotide positions and size of the predicted polypeptide are represented by the numbers below and above the bars, respectively. The annotations of the viral sequence were made according to NCBI. The nucleotide and protein accession numbers are located forward and above the scheme, respectively. Representation of the genome are not to scale.
3.5. 2A/2A-Likes Sequences and Viral Evolution
Previous studies concerning RNA viruses and 2A-like peptides have reported that these sequences emerged independently during the evolution of viral families [2,14]. However, in a previous study [31], we showed sequences very similar to functional 2A-like sequences in some RNA viruses that could be the precursors of 2A sequences.
In particular, RNA viruses depend on the activity of RNA-dependent RNA polymerases. These enzymes have a significant error rate (10−3 to 10−5 mutations per inserted nucleotide) because they do not have exonucleotide review activity [38]. This results in a high degree of genetic heterogeneity in populations of RNA viruses, which are believed to favor adaptability to different environments and hosts [39]. Considering this, the 2A/2A-like sequences could have emerged by subsequent mutation events that ended in a cleavage function, providing the advantage of releasing more than one protein from the same ORF. Therefore, this could directly impact viral adaptation potential and viral infection mechanisms to favor their fitness in complex multicellular systems [31].
Yang et al. also suggested that picornaviruses with more complex infection mechanisms than other viruses of the same family have more than one 2A-like sequence in their genomes [14]. Taking this evidence into account, it seems that 2A/2A-like sequences may be a key element in viral genome evolution and, once acquired, its loss of function may impact virus effectivity.
3.6. Biotechnology Applications
Various approaches have been employed to co-express multiple proteins in cells, including the use of internal ribosomal entry site (IRES) elements [40,41], dual promoter systems [42,43], and transfection of multiple vectors [44]. Each of these is associated with several limitations, such as uneven or unreliable protein expression levels, silencing of some promoters [45,46], and increased toxicity to cells (with multiple transfections) [47].
Co-expression systems, including 2A/2A-like peptides, could be an alternative strategy for expressing multiple genes under the control of a single promoter. These constructs could have the additional advantage of producing proteins at near-stoichiometric levels, unlike IRES-mediated polycistronic expression, where ribosomes are independently recruited at distinct regions with the mRNA [1,4,48,49]. This necessitates the optimization of the system by testing several combinations of promoters and/or IRES and the order of genes within the expression cassette [46]. Furthermore, IRES activity can be affected by cell type, and variable expression can be observed in the downstream coding sequence [50].
2A/2A-like sequences have been used in a range of heterologous expression systems because of their cleavage capacity. These systems include viruses [51], yeasts [52,53], fungi [54,55,56], insect cells [57,58], plants [59], human HTK-143 cells [9], rabbit reticulocytes [60], HeLa cells [61], CHO cells [62], HEK293 cells [63], algae [64], and other animals [65,66,67].
In yeasts, more than two 2A sequences have been used to co-express proteins from the same vector. As seen in [68] and [69], three proteins were produced using this strategy in S. cerevisiae. Surprisingly, up to nine proteins have been linked and successfully co-translated and separated with 2A sequences in the yeast Pichia pastoris [70].
Researchers have also attempted to use 2A for multi-gene transformation in staple crops [71,72]. They can also be used for gene fusion, as seen in tomatoes, potatoes, and others [73,74].
To construct the co-expression vectors, the 2A/2A-like sequences are usually incorporated into an adenovirus [75], adeno-associated virus (AAV) [12], retrovirus [76], lentivirus [77,78], or plasmid vector [79,80]. Many other biotechnological applications that depend on the co-expression of multiple genes use 2A/2A-like sequences, e.g., the production of antibodies and antigens that can be used in vaccine production [80,81,82,83,84,85], observation of chromatin dynamics and genome (DNA and RNA) editing in the application of cell/gene therapies [78,79,86,87,88,89,90], and development of optogenetic tools [91,92,93]. More examples of viral 2As applications can be found in [94].
4. Conclusions
In this article, we reviewed the 2A/2A-like sequence distribution of viruses and described the occurrence of these motifs in viral species where these sequences have not been previously reported. These findings need to be confirmed through in vitro tests to verify they are active 2A-like sequences.
Because of its cleavage function, the 2A/2A-like sequences appear to directly affect the complexity of the viral genome, which plays a decisive role in viral evolution. Additionally, they are excellent alternatives for developing new biotechnological tools that depend on the expression of multiple products, such as vaccines, transgenic approaches, cell/gene therapy, and optogenetic tools.
Author Contributions
Conceptualization, J.G.S.d.L. and D.C.F.L.; methodology, J.G.S.d.L.; writing—original draft preparation, J.G.S.d.L. and D.C.F.L.; writing—review and editing, J.G.S.d.L. and D.C.F.L.; visualization, J.G.S.d.L. and D.C.F.L. All authors have read and agreed to the published version of the manuscript.
Funding
We would like to thank Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for financial support.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Donnelly, M.L.L.; Hughes, L.E.; Luke, G.; Mendoza, H.; Ten Dam, E.; Gani, D.; Ryan, M.D. The “cleavage” activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring “2A-like” sequences. J. Gen. Virol. 2001, 82, 1027–1041. [Google Scholar] [CrossRef]
- Luke, G.A.; de Felipe, P.; Lukashev, A.; Kallioinen, S.E.; Bruno, E.A.; Ryan, M.D. Occurrence, function and evolutionary origins of ‘2A-like’ sequences in virus genomes. J. Gen. Virol. 2008, 89, 1036–1042. [Google Scholar] [CrossRef]
- Ryan, M.D.; Donnelly, M.; Lewis, A.; Mehrotra, A.P.; Wilkie, J.; Gani, D. A model for nonstoichiometric, cotranslational protein scission in eukaryotic ribosomes. Bioorg. Chem. 1999, 27, 55–79. [Google Scholar] [CrossRef] [Green Version]
- Donnelly, M.L.L.; Luke, G.; Mehrotra, A.; Li, X.; Hughes, L.E.; Gani, D.; Ryan, M.D. Analysis of the aphthovirus 2A/2B polyprotein “cleavage” mechanism indicates not a proteolytic reaction, but a novel translational effect: A putative ribosomal “skip”. J. Gen. Virol. 2001, 82, 1013–1025. [Google Scholar] [CrossRef]
- Atkins, J.F.; Wills, N.M.; Loughran, G.; Wu, C.Y.; Parsawar, K.; Ryan, M.D.; Wang, C.H.; Nelson, C.C. A case for “StopGo”: Reprogramming translation to augment codon meaning of GGN by promoting unconventional termination (Stop) after addition of glycine and then allowing continued translation (Go). RNA 2007, 13, 803–810. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.D.; Ryan, M.D. Ribosome “Skipping”: “Stop-Carry On” or “StopGo” Translation. In Recoding: Expansion of Decoding Rules Enriches Gene Expression; Atkins, J.F., Gesteland, R.F., Eds.; Springer: New York, NY, USA, 2010; pp. 101–121. [Google Scholar]
- de Felipe, P. Skipping the co-expression problem: The new 2A “CHYSEL” technology. Genet. Vaccines Ther. 2004, 2, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryan, M.D.; King, A.M.Q.; Thomas, G.P. Cleavage of foot-and-mouth disease virus polyprotein is mediated by residues located within a 19 amino acid sequence. J. Gen. Virol. 1991, 72, 2727–2732. [Google Scholar] [CrossRef]
- Ryan, M.D.; Drew, J. Foot-and-mouth disease virus 2A oligopeptide mediated cleavage of an artificial polyprotein. EMBO J. 1994, 13, 928–933. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, M.L.L.; Gani, D.; Flint, M.; Monaghan, S.; Ryan, M.D. The cleavage activities of aphthovirus and cardiovirus 2A proteins. J. Gen. Virol. 1997, 78, 13–21. [Google Scholar] [CrossRef]
- Nibert, M.L. “2A-like” and “shifty heptamer” motifs in penaeid shrimp infectious myonecrosis virus, a monosegmented double-stranded RNA virus. J. Gen. Virol. 2007, 88, 1315–1318. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.E.; Brameld, J.M.; Hill, P.; Barrett, P.; Ebling, F.J.P.; Jethwa, P.H. The use of a viral 2A sequence for the simultaneous over-expression of both the vgf gene and enhanced green fluorescent protein (eGFP) in vitro and in vivo. J. Neurosci. Methods 2015, 256, 22–29. [Google Scholar] [CrossRef]
- Doronina, V.A.; de Felipe, P.; Wu, C.; Sharma, P.; Sachs, M.S.; Ryan, M.D.; Brown, J.D. Dissection of a co-translational nascent chain separation event. Biochem. Soc. Trans. 2008, 36, 712–716. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Cheng, A.; Wang, M.; Jia, R.; Sun, K.; Pan, K.; Yang, Q.; Wu, Y.; Zhu, D.; Chen, S.; et al. Structures and corresponding functions of five types of picornaviral 2A proteins. Front. Microbiol. 2017, 8, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Luke, G.A.; Escuin, H.; De Felipe, P.; Ryan, M. 2A to the Fore–Research, Technology and Applications 2A to the Fore–Research, Technology and Applications. Biotechnol. Genet. Eng. Rev. 2009, 26, 223–260. [Google Scholar] [CrossRef] [PubMed]
- Luke, G.A.; Pathania, U.S.; Roulston, C.; De Felipe, P.; Ryan, M.D. DxExNPGP-Motives for the motif. In Recent Research Developments in Virology; Research Signpost: Kerala, India, 2014; Volume 9, pp. 25–42. [Google Scholar]
- Luke, G.A.; Ryan, M.D. The protein coexpression problem in biotechnology and biomedicine: Virus 2A and 2A-like sequences provide a solution. Future Virol. 2013, 8, 983–996. [Google Scholar] [CrossRef] [Green Version]
- Gorbalenya, A.E.; Pringle, F.M.; Zeddam, J.L.; Luke, B.T.; Cameron, C.E.; Kalmakoff, J.; Hanzlik, T.N.; Gordon, K.H.J.; Ward, V.K. The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J. Mol. Biol. 2002, 324, 47–62. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, J.; Lu, J.; Yi, F.; Liu, C.; Hu, Y. Sequence analysis and genomic organization of a new insect picorna-like virus, Ectropis obliqua picorna-like virus, isolated from Ectropis obliqua. J. Gen. Virol. 2004, 85, 1145–1151. [Google Scholar] [CrossRef]
- Wu, C.Y.; Lo, C.F.; Huang, C.J.; Yu, H.T.; Wang, C.H. The complete genome sequence of Perina nuda picorna-like virus, an insect-infecting RNA virus with a genome organization similar to that of the mammalian picornaviruses. Virology 2002, 294, 312–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.; Browning, G.F.; Studdert, M.J.; Crabb, B.S. Equine rhinovirus 1 is more closely related to foot-and-mouth disease virus than to other picornaviruses. Proc. Natl. Acad. Sci. USA 1996, 93, 990–995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wutz, G.; Auer, H.; Nowotny, N.; Grosse, B.; Skern, T.; Kuechler, E. Equine rhinovirus serotypes 1 and 2: Relationship to each other and to aphthoviruses and cardioviruses. J. Gen. Virol. 1996, 77, 1719–1730. [Google Scholar] [CrossRef] [PubMed]
- Lindberg, A.M.; Johansson, S. Phylogenetic analysis of Ljungan virus and A-2 plaque virus, new members of the Picornaviridae. Virus Res. 2002, 85, 61–70. [Google Scholar] [CrossRef]
- Doherty, M.; Todd, D.; McFerran, N.; Hoey, E.M. Sequence analysis of a porcine enterovirus serotype 1 isolate: Relationships with other picornaviruses. J. Gen. Virol. 1999, 80, 1929–1941. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.S.; Lukashov, V.V.; Ganac, R.D.; Schnurr, D.P. Discovery of a novel human picornavirus in a stool sample from a pediatric patient presenting with fever of unknown origin. J. Clin. Microbiol. 2007, 45, 2144–2150. [Google Scholar] [CrossRef] [Green Version]
- Hagiwara, K.; Kobayashi, J.; Tomita, M.; Yoshimura, T. Nucleotide sequence of genome segment 5 from Bombyx mori cypovirus 1. Arch. Virol. 2001, 146, 181–187. [Google Scholar] [CrossRef]
- Yang, H.; Makeyev, E.V.; Kang, Z.; Ji, S.; Bamford, D.H.; Van Dijk, A.A. Cloning and sequence analysis of dsRNA segments 5, 6 and 7 of a novel non-group A, B, C adult rotavirus that caused an outbreak of gastroenteritis in China. Virus Res. 2004, 106, 15–26. [Google Scholar] [CrossRef]
- Zell, R.; Delwart, E.; Gorbalenya, A.E.; Hovi, T.; King, A.M.Q.; Knowles, N.J.; Lindberg, A.M.; Pallansch, M.A.; Palmenberg, A.C.; Reuter, G.; et al. ICTV virus taxonomy profile: Picornaviridae. J. Gen. Virol. 2017, 98, 2421–2422. [Google Scholar] [CrossRef] [PubMed]
- Hughes, P.J.; Stanway, G. The 2A proteins of three diverse picornaviruses are related to each other and to the H-rev107 family of proteins involved in the control of cell proliferation. J. Gen. Virol. 2000, 81, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Walter, C.T.; Pringle, F.M.; Nakayinga, R.; De Felipe, P.; Ryan, M.D.; Ball, L.A.; Dorrington, R.A. Genome organization and translation products of Providence virus: Insight into a unique tetravirus. J. Gen. Virol. 2010, 91, 2826–2835. [Google Scholar] [CrossRef] [PubMed]
- de Lima, J.G.S.; Teixeira, D.G.; Freitas, T.T.; Lima, J.P.M.S.; Lanza, D.C.F. Evolutionary origin of 2A-like sequences in Totiviridae genomes. Virus Res. 2019, 259, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Poulos, B.T.; Tang, K.F.J.; Pantoja, C.R.; Bonami, J.R.; Lightner, D.V. Purification and characterization of infectious myonecrosis virus of penaeid shrimp. J. Gen. Virol. 2006, 87, 987–996. [Google Scholar] [CrossRef]
- Zhai, Y.; Attoui, H.; Mohd Jaafar, F.; Wang, H.-Q.; Cao, Y.-X.; Fan, S.-P.; Sun, Y.-X.; Liu, L.-D.; Mertens, P.P.C.; Meng, W.-S.; et al. Isolation and full-length sequence analysis of Armigeres subalbatus totivirus, the first totivirus isolate from mosquitoes representing a proposed novel genus (Artivirus) of the family Totiviridae. J. Gen. Virol. 2010, 91, 2836–2845. [Google Scholar] [CrossRef]
- Isawa, H.; Kuwata, R.; Hoshino, K.; Tsuda, Y.; Sakai, K.; Watanabe, S.; Nishimura, M.; Satho, T.; Kataoka, M.; Nagata, N.; et al. Identification and molecular characterization of a new nonsegmented double-stranded RNA virus isolated from Culex mosquitoes in Japan. Virus Res. 2011, 155, 147–155. [Google Scholar] [CrossRef]
- Wu, Q.; Luo, Y.; Lu, R.; Lau, N.; Lai, E.C.; Li, W.X.; Ding, S.W. Virus discovery by deep sequencing and assembly of virus-derived small silencing RNAs. Proc. Natl. Acad. Sci. USA 2010, 107, 1606–1611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Virol, A.; Mor, S.K.; Benjamin, N.; Phelps, D. Molecular detection of a novel totivirus from golden shiner (Notemigonus crysoleucas) baitfish in the USA. Arch. Virol. 2016. [Google Scholar] [CrossRef]
- Danielle, M.; Dantas, A.; Henrique, G.; Cavalcante, O.; Oliveira, R.A.C.C.; Lanza, D.C.F.F.; Dantas, M.D.A.; Cavalcante, G.H.O.; Oliveira, R.A.C.C.; Lanza, D.C.F.F. New insights about ORF1 coding regions support the proposition of a new genus comprising arthropod viruses in the family Totiviridae. Virus Res. 2016, 211, 159–164. [Google Scholar] [CrossRef] [Green Version]
- Modrow, S.; Falke, D.; Truyen, U.; Schätzl, H. How Do Mutations Lead to the Emergence of Novel Viruses? Springer: Berlin/Heidelberg, Germany, 2013; Volume 9783642207, ISBN 978-3-642-20717-4. [Google Scholar]
- Orton, R.J.; Wright, C.F.; King, D.P.; Haydon, D.T. Estimating viral bottleneck sizes for FMDV transmission within and between hosts and implications for the rate of viral evolution. Interface Focus 2019, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khodamoradi, S.; Shenagari, M.; Kheiri, M.T.; Sabahi, F.; Jamali, A.; Heidari, A.; Ashrafkhani, B. IRES-based co-expression of influenza virus conserved genes can promote synergistic antiviral effects both in vitro and in vivo. Arch. Virol. 2018, 163, 877–886. [Google Scholar] [CrossRef] [PubMed]
- Akiyoshi, S.; Ishii, T.; Bai, Z.; Mombaerts, P. Subpopulations of vomeronasal sensory neurons with coordinated coexpression of type 2 vomeronasal receptor genes are differentially dependent on Vmn2r1. Eur. J. Neurosci. 2018, 47, 887–900. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Su, L.; Duan, X.; Liu, L.; Wu, J. High-level extracellular protein production in Bacillus subtilis using an optimized dual-promoter expression system. Microb. Cell Fact. 2017, 16, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bayat, H.; Hossienzadeh, S.; Pourmaleki, E.; Ahani, R.; Rahimpour, A. Evaluation of different vector design strategies for the expression of recombinant monoclonal antibody in CHO cells. Prep. Biochem. Biotechnol. 2018, 48, 160–164. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, W.; Ehrlich, I.; Wolff, S.B.E.; Michalski, A.M.; Wölfl, S.; Hasan, M.T.; Lüthi, A.; Sprengel, R. Faithful expression of multiple proteins via 2A-peptide self-processing: A versatile and reliable method for manipulating brain circuits. J. Neurosci. 2009, 29, 8621–8629. [Google Scholar] [CrossRef] [PubMed]
- Radcliffe, P.A.; Mitrophanous, K.A. Multiple gene products from a single vector: “Self-cleaving” 2A peptides. Gene Ther. 2004, 11, 1673–1674. [Google Scholar] [CrossRef]
- Arbab, A.S.; Yocum, G.T.; Wilson, L.B.; Parwana, A.; Jordan, E.K.; Kalish, H.; Frank, J.A. Comparison of transfection agents in forming complexes with ferumoxides, cell labeling efficiency, and cellular viability. Mol. Imaging 2004, 3, 24–32. [Google Scholar] [CrossRef] [PubMed]
- de Felipe, P.; Luke, G.A.; Hughes, L.E.; Gani, D.; Halpin, C.; Ryan, M.D. E unum pluribus: Multiple proteins from a self-processing polyprotein. Trends Biotechnol. 2006, 24, 68–75. [Google Scholar] [CrossRef]
- Hellen, C.U.T.; Sarnow, P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001, 15, 1593–1612. [Google Scholar] [CrossRef] [Green Version]
- Bouabe, H.; Fässler, R.; Heesemann, J. Improvement of reporter activity by IRES-mediated polycistronic reporter system. Nucleic Acids Res. 2008, 36, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Momose, F.; Morikawa, Y. Polycistronic expression of the influenza A virus RNA-dependent RNA polymerase by using the Thosea asigna virus 2A-like self-processing sequence. Front. Microbiol. 2016, 7, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.F.; Lin, Y.; Zhang, J.H.; Zheng, S.P.; Ye, Y.R.; Liang, X.X.; Han, S.Y. Double Candida antarctica lipase B co-display on Pichia pastoris cell surface based on a self-processing foot-and-mouth disease virus 2A peptide. Appl. Microbiol. Biotechnol. 2012, 96, 1539–1550. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Yao, Q.; Tao, J.; Qiao, Y.; Zhang, Z. Co-ordinate expression of glycine betaine synthesis genes linked by the FMDV 2A region in a single open reading frame in Pichia pastoris. Appl. Microbiol. Biotechnol. 2007, 77, 891–899. [Google Scholar] [CrossRef]
- Subramanian, V.; Schuster, L.A.; Moore, K.T.; Ii, L.E.T.; Baker, J.O.; Wall, T.A.; Vander Linger, J.G.; Himmel, M.E.; Decker, S.R. Biotechnology for Biofuels A versatile 2A peptide - based bicistronic protein expressing platform for the industrial cellulase producing fungus, Trichoderma reesei. Biotechnol. Biofuels 2017, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Schuetze, T.; Meyer, V. Polycistronic gene expression in Aspergillus niger. Microb. Cell Fact. 2017, 16, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.; Liu, Q.; Li, X.; Zhang, C.; Li, J.; Sun, W.; Liu, D.; Xiao, D.; Tian, C. Construction of a new thermophilic fungus Myceliophthora thermophila platform for enzyme production using a versatile 2A peptide strategy combined with efficient CRISPR-Cas9 system. Biotechnol. Lett. 2020, 42, 1181–1191. [Google Scholar] [CrossRef]
- Osborn, M.J.; Panoskaltsis-Mortari, A.; McElmurry, R.T.; Bell, S.K.; Vignali, D.A.A.; Ryan, M.D.; Wilber, A.C.; McIvor, R.S.; Tolar, J.; Blazar, B.R. A picornaviral 2A-like sequence-based tricistronic vector allowing for high-level therapeutic gene expression coupled to a dual-reporter system. Mol. Ther. 2005, 12, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Schwirz, J.; Yan, Y.; Franta, Z.; Schetelig, M.F. Bicistronic expression and differential localization of proteins in insect cells and Drosophila suzukii using picornaviral 2A peptides. Insect Biochem. Mol. Biol. 2020, 119, 103324. [Google Scholar] [CrossRef]
- Zhang, B.; Rapolu, M.; Kumar, S.; Gupta, M.; Liang, Z.; Han, Z.; Williams, P.; Su, W.W. Coordinated protein co-expression in plants by harnessing the synergy between an intein and a viral 2A peptide. Plant Biotechnol. J. 2017, 15, 718–728. [Google Scholar] [CrossRef] [Green Version]
- de Felipe, P.; Hughes, L.E.; Ryan, M.D.; Brown, J.D. Co-translational, intraribosomal cleavage of polypeptides by the foot-and-mouth disease virus 2A peptide. J. Biol. Chem. 2003, 278, 11441–11448. [Google Scholar] [CrossRef] [Green Version]
- de Felipe, P.; Luke, G.A.; Brown, J.D.; Ryan, M.D. Inhibition of 2A-mediated “cleavage” of certain artificial polyproteins bearing N-terminal signal sequences. Biotechnol. J. 2010, 5, 213–223. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Wang, M.; Wang, T.; Wei, Y.; Guo, X.; Mi, C.; Zhao, C.; Cao, X.; Dou, Y. Effects of different 2A peptides on transgene expression mediated by tricistronic vectors in transfected CHO cells. Mol. Biol. Rep. 2020, 47, 469–475. [Google Scholar] [CrossRef]
- Shin, S.; Kim, S.H.; Shin, S.W.; Grav, L.M.; Pedersen, L.E.; Lee, J.S.; Lee, G.M. Comprehensive Analysis of Genomic Safe Harbors as Target Sites for Stable Expression of the Heterologous Gene in HEK293 Cells. ACS Synth. Biol. 2020, 9, 1263–1269. [Google Scholar] [CrossRef]
- Rasala, B.A.; Lee, P.A.; Shen, Z.; Briggs, S.P.; Mendez, M.; Mayfield, S.P. Robust expression and secretion of xylanase1 in Chlamydomonas reinhardtii by fusion to a selection gene and processing with the FMDV 2A peptide. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeong, I.; Kim, E.; Seong, J.Y.; Park, H.C. Overexpression of Spexin 1 in the Dorsal Habenula Reduces Anxiety in Zebrafish. Front. Neural Circuits 2019, 13, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Pontes-Quero, S.; Fernández-Chacón, M.; Luo, W.; Lunella, F.F.; Casquero-Garcia, V.; Garcia-Gonzalez, I.; Hermoso, A.; Rocha, S.F.; Bansal, M.; Benedito, R. High mitogenic stimulation arrests angiogenesis. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Wang, F.; Xu, S.; Wang, R.; Chen, W.; Hou, K.; Tian, C.; Wang, F.; Zhao, P.; Xia, Q. Optimization of a 2A self-cleaving peptide-based multigene expression system for efficient expression of upstream and downstream genes in silkworm. Mol. Genet. Genomics 2019, 294, 849–859. [Google Scholar] [CrossRef] [PubMed]
- Beekwilder, J.; Van Rossum, H.M.; Koopman, F.; Sonntag, F.; Buchhaupt, M.; Schrader, J.; Hall, R.D.; Bosch, D.; Pronk, J.T.; Van Maris, A.J.A.; et al. Polycistronic expression of a β-carotene biosynthetic pathway in Saccharomyces cerevisiae coupled to β-ionone production. J. Biotechnol. 2014, 192, 383–392. [Google Scholar] [CrossRef]
- Park, M.; Kang, K.; Park, S.; Kim, Y.S.; Ha, S.H.; Lee, S.W.; Ahn, M.J.; Bae, J.M.; Back, K. Expression of serotonin derivative synthetic genes on a single self-processing polypeptide and the production of serotonin derivatives in microbes. Appl. Microbiol. Biotechnol. 2008, 81, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Geier, M.; Fauland, P.; Vogl, T.; Glieder, A. Compact multi-enzyme pathways in P. pastoris. Chem. Commun. 2015, 51, 1643–1646. [Google Scholar] [CrossRef] [Green Version]
- Park, S.; Kang, K.; Kim, Y.S.; Back, K. Endosperm-specific expression of tyramine N-hydroxycinnamoyltransferase and tyrosine decarboxylase from a single self-processing polypeptide produces high levels of tyramine derivatives in rice seeds. Biotechnol. Lett. 2009, 31, 911–915. [Google Scholar] [CrossRef]
- Quilis, J.; López-García, B.; Meynard, D.; Guiderdoni, E.; San Segundo, B. Inducible expression of a fusion gene encoding two proteinase inhibitors leads to insect and pathogen resistance in transgenic rice. Plant Biotechnol. J. 2014, 12, 367–377. [Google Scholar] [CrossRef]
- Yeo, E.T.; Kwon, H.B.; Han, S.E.; Lee, J.T.; Ryu, J.C.; Byun, M.O. Genetic engineering of drought resistant potato plants by introduction of the trehalose-6-phosphate synthase (TPS1) gene from Saccharomyces cerevisiae. Mol. Cells 2000, 10, 263–268. [Google Scholar]
- Ralley, L.; Enfissi, E.M.A.; Misawa, N.; Schuch, W.; Bramley, P.M.; Fraser, P.D. Metabolic engineering of ketocarotenoid formation in higher plants. Plant J. 2004, 39, 477–486. [Google Scholar] [CrossRef]
- Chu, V.T.; Weber, T.; Wefers, B.; Wurst, W.; Sander, S.; Rajewsky, K.; Kühn, R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015, 33, 543–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Browne, E.P. An Interleukin-1 Beta-Encoding Retrovirus Exhibits Enhanced Replication In Vivo. J. Virol. 2015, 89, 155–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, G.; Choi, J.G.; Bharaj, P.; Abraham, S.; Dang, Y.; Kafri, T.; Alozie, O.; Manjunath, M.N.; Shankar, P. CCR5 gene editing of resting CD4+ T cells by transient ZFN expression from HIV envelope pseudotyped nonintegrating lentivirus confers HIV-1 resistance in humanized mice. Mol. Ther.-Nucleic Acids 2014, 3, 1–10. [Google Scholar] [CrossRef]
- Peng, Y.; Yang, T.; Tang, X.; Chen, F.; Wang, S. Construction of an Inducible CRISPR/Cas9 System for CXCR4 Gene and Demonstration of its Effects on MKN-45 Cells. Cell Biochem. Biophys. 2020, 78, 23–30. [Google Scholar] [CrossRef]
- Fang, Y.; Stroukov, W.; Cathomen, T.; Mussolino, C. Chimerization enables gene synthesis and lentiviral delivery of customizable tale-based effectors. Int. J. Mol. Sci. 2020, 21, 795. [Google Scholar] [CrossRef] [Green Version]
- Mizote, Y.; Masumi-Koizumi, K.; Katsuda, T.; Yamaji, H. Production of an antibody Fab fragment using 2A peptide in insect cells. J. Biosci. Bioeng. 2020, 130, 205–211. [Google Scholar] [CrossRef]
- Arevalo-Villalobos, J.I.; Govea-Alonso, D.O.; Bañuelos-Hernández, B.; González-Ortega, O.; Zarazúa, S.; Rosales-Mendoza, S. Inducible expression of antigens in plants: A study focused on peptides related to multiple sclerosis immunotherapy. J. Biotechnol. 2020, 318, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.H.; Hu, C.C.; Liao, J.T.; Lee, Y.L.; Huang, Y.W.; Lin, N.S.; Lin, Y.L.; Hsu, Y.H. Production of Japanese encephalitis virus antigens in plants using bamboo mosaic virus-based vector. Front. Microbiol. 2017, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Hung, C.Y.; Bhattacharya, C.; Nichols, S.; Rahimuddin, H.; Kittur, F.S.; Leung, T.C.; Xie, J. An effective way of producing fully assembled antibody in transgenic tobacco plants by linking heavy and light chains via a self-cleaving 2a peptide. Front. Plant Sci. 2018, 9, 1–11. [Google Scholar] [CrossRef]
- Chen, L.; Yang, X.; Luo, D.; Yu, W. Efficient Production of a Bioactive Bevacizumab monoclonal antibody using the 2A self-cleavage peptide in transgenic rice callus. Front. Plant Sci. 2016, 7, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Monreal-Escalante, E.; Bañuelos-Hernández, B.; Hernández, M.; Fragoso, G.; Garate, T.; Sciutto, E.; Rosales-Mendoza, S. Expression of Multiple Taenia Solium Immunogens in Plant Cells Through a Ribosomal Skip Mechanism. Mol. Biotechnol. 2015, 57, 635–643. [Google Scholar] [CrossRef]
- Jin, S.; Yang, C.; Huang, J.; Liu, L.; Zhang, Y.; Li, S.; Zhang, L.; Sun, Q.; Yang, P. Conditioned medium derived from FGF-2-modified GMSCs enhances migration and angiogenesis of human umbilical vein endothelial cells. Stem Cell Res. Ther. 2020, 11, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knoll, A.; Kankowski, S.; Schöllkopf, S.; Meier, J.C.; Seitz, O. Chemo-biological mRNA imaging with single nucleotide specificity. Chem. Commun. 2019, 55, 14817–14820. [Google Scholar] [CrossRef] [PubMed]
- Quenneville, S.; Labouèbe, G.; Basco, D.; Metref, S.; Viollet, B.; Foretz, M.; Thorens, B. Hypoglycemia Sensing Neurons of the Ventromedial Hypothalamus Require AMPK-Induced Txn2 Expression But Are Dispensable For Physiological Counterregulation. Diabetes 2020, db200577. [Google Scholar] [CrossRef]
- Shibuta, M.K.; Matsuoka, M.; Matsunaga, S. 2a Peptides Contribute To the Co-Expression of Proteins for Imaging and Genome Editing. Cytologia 2019, 84, 107–111. [Google Scholar] [CrossRef] [Green Version]
- Sinha, D.; Steyer, B.; Shahi, P.K.; Mueller, K.P.; Valiauga, R.; Edwards, K.L.; Bacig, C.; Steltzer, S.S.; Srinivasan, S.; Abdeen, A.; et al. Human iPSC Modeling Reveals Mutation-Specific Responses to Gene Therapy in a Genotypically Diverse Dominant Maculopathy. Am. J. Hum. Genet. 2020, 107, 278–292. [Google Scholar] [CrossRef] [PubMed]
- Kawano, F.; Okazaki, R.; Yazawa, M.; Sato, M. A photoactivatable Cre-loxP recombination system for optogenetic genome engineering. Nat. Chem. Biol. 2016, 12, 1059–1064. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, V.V.; Khamo, J.S.; Mei, W.; Turgeon, A.J.; Ashraf, H.M.; Mondal, P.; Patel, D.B.; Risner, N.; Cho, E.E.; Yang, J.; et al. Reversible optogenetic control of kinase activity during differentiation and embryonic development. Development 2016, 143, 4085–4094. [Google Scholar] [CrossRef] [Green Version]
- Taslimi, A.; Zoltowski, B.; Miranda, J.G.; Pathak, G.P.; Hughes, R.M.; Tucker, C.L. Optimized second-generation CRY2-CIB dimerizers and photoactivatable Cre recombinase. Nat. Chem. Biol. 2016, 12, 425–430. [Google Scholar] [CrossRef] [Green Version]
- Luke, G.A.; Ryan, M.D. Therapeutic applications of the ‘NPGP’ family of viral 2As. Rev. Med. Virol. 2018, 28, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).