Prolonged Gut Dysbiosis and Fecal Excretion of Hepatitis A Virus in Patients Infected with Human Immunodeficiency Virus
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Subjects and Sample Collection
2.2. Measuring HAV RNA Load in Fecal Samples
2.3. DNA Extraction
2.4. Construction of DNA Library and Sequencing
2.5. Sequence Data Analyses
2.6. Measurement of the Cytokine Profile
2.7. RNA Purification and qRT-PCR Analysis
3. Results
3.1. General Characteristics of Participants
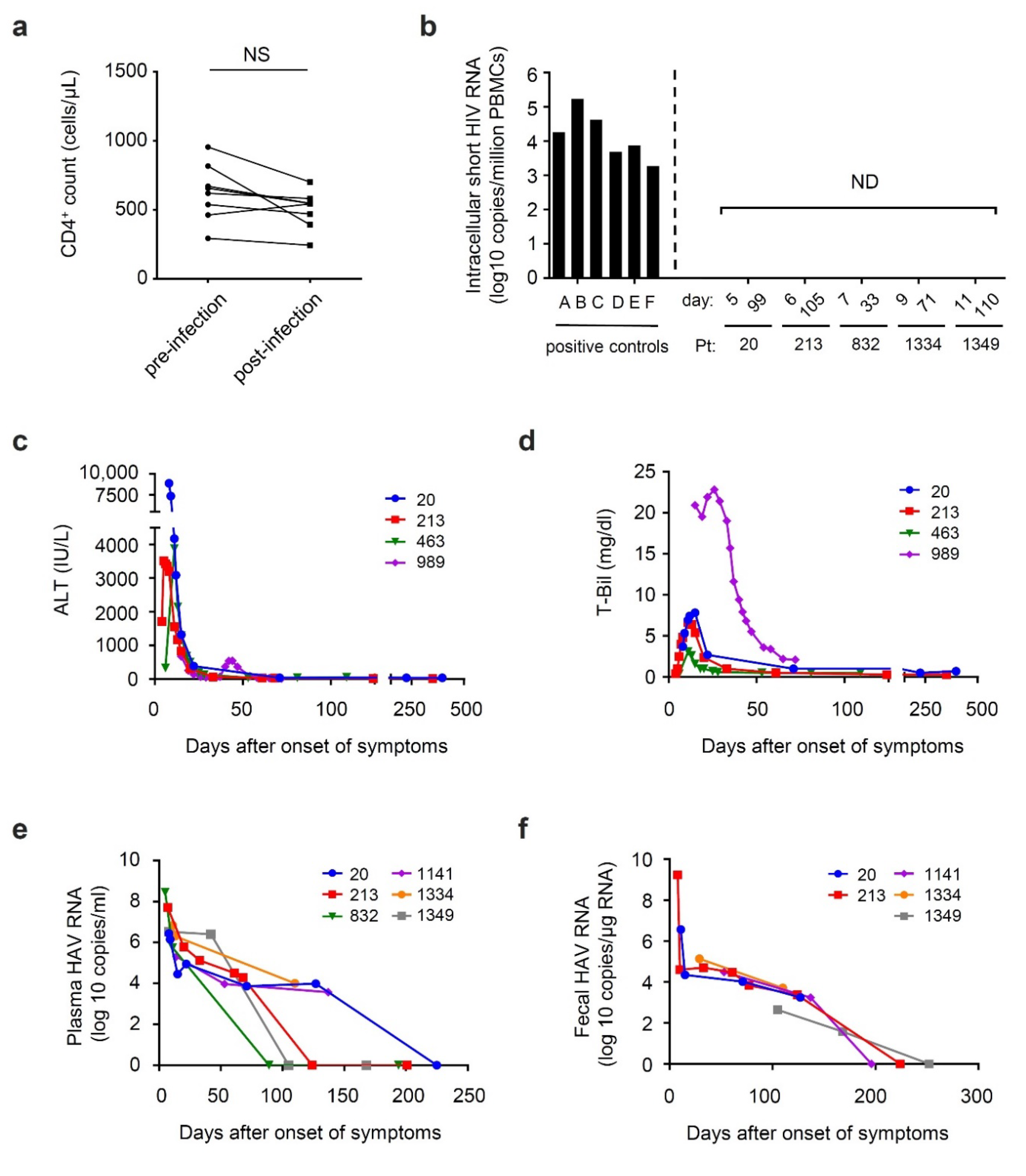
3.2. Time Course of Viral Shedding and Clinical Markers during HAV Co-Infection among Patients with Chronic HIV Infection
3.3. Time Course of Chemokine Expression during Acute HAV Infection
3.4. Prolonged Fecal Dysbiosis after HAV Infection
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lemon, S.M.; Ott, J.J.; Van Damme, P.; Shouval, D. Type A viral hepatitis: A summary and update on the molecular virology, epidemiology, pathogenesis and prevention. J. Hepatol. 2018, 68, 167–184. [Google Scholar] [CrossRef] [PubMed]
- Franco, E.; Meleleo, C.; Serino, L.; Sorbara, D.; Zaratti, L. Hepatitis A: Epidemiology and prevention in developing countries. World J. Hepatol. 2012, 4, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Villano, S.A.; Nelson, K.E.; Vlahov, D.; Purcell, R.H.; Saah, A.J.; Thomas, D.L. Hepatitis A among homosexual men and injection drug users: More evidence for vaccination. Clin. Infect. Dis. 1997, 25, 726–728. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mitchell, H.; Hughes, G. Recent epidemiology of sexually transmissible enteric infections in men who have sex with men. Curr. Opin. Infect. Dis. 2018, 31, 50–56. [Google Scholar] [CrossRef]
- Tanaka, S.; Kishi, T.; Ishihara, A.; Watanabe, D.; Uehira, T.; Ishida, H.; Shirasaka, T.; Mita, E. Outbreak of hepatitis A linked to European outbreaks among men who have sex with men in Osaka, Japan, from March to July 2018. Hepatol. Res. 2019, 49, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Koga, M.; Lim, L.A.; Ogishi, M.; Satoh, H.; Kikuchi, T.; Adachi, E.; Sugiyama, R.; Kiyohara, T.; Suzuki, R.; Muramatsu, M.; et al. Comparison of the Clinical Features of Hepatitis A in People Living with HIV between Pandemics in 1999–2000 and 2017–2018 in the Metropolitan Area of Japan. Jpn. J. Infect. Dis. 2020, 73, 89–95. [Google Scholar] [CrossRef]
- Castaneda, D.; Gonzalez, A.J.; Alomari, M.; Tandon, K.; Zervos, X.B. From hepatitis A to E: A critical review of viral hepatitis. World J. Gastroenterol. 2021, 27, 1691–1715. [Google Scholar] [CrossRef]
- Counihan, N.A.; Anderson, D.A. Specific IgA Enhances the Transcytosis and Excretion of Hepatitis A Virus. Sci Rep. 2016, 6, 21855. [Google Scholar] [CrossRef]
- Feng, Z.; Hensley, L.; McKnight, K.L.; Hu, F.; Madden, V.; Ping, L.; Jeong, S.H.; Walker, C.; Lanford, R.E.; Lemon, S.M. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 2013, 496, 367–371. [Google Scholar] [CrossRef]
- Wang, X.; Ren, J.; Gao, Q.; Hu, Z.; Sun, Y.; Li, X.; Rowlands, D.J.; Yin, W.; Wang, J.; Stuart, D.I.; et al. Hepatitis A virus and the origins of picornaviruses. Nature 2015, 517, 85–88. [Google Scholar] [CrossRef]
- Hirai-Yuki, A.; Hensley, L.; Whitmire, J.K.; Lemon, S.M. Biliary Secretion of Quasi-Enveloped Human Hepatitis A Virus. mBio 2016, 7, e01998-16. [Google Scholar] [CrossRef]
- Locarnini, S.A.; Coulepis, A.G.; Kaldor, J.; Gust, I.D. Coproantibodies in hepatitis A: Detection by enzyme-linked immunosorbent assay and immune electron microscopy. J. Clin. Microbiol. 1980, 11, 710–716. [Google Scholar] [CrossRef]
- Blank, C.A.; Anderson, D.A.; Beard, M.; Lemon, S.M. Infection of polarized cultures of human intestinal epithelial cells with hepatitis A virus: Vectorial release of progeny virions through apical cellular membranes. J. Virol. 2000, 74, 6476–6484. [Google Scholar] [CrossRef]
- Dotzauer, A.; Brenner, M.; Gebhardt, U.; Vallbracht, A. IgA-coated particles of Hepatitis A virus are translocalized antivectorially from the apical to the basolateral site of polarized epithelial cells via the polymeric immunoglobulin receptor. J. Gen. Virol. 2005, 86 Pt 10, 2747–2751. [Google Scholar] [CrossRef] [PubMed]
- Dotzauer, A.; Heitmann, A.; Laue, T.; Kraemer, L.; Schwabe, K.; Paulmann, D.; Flehmig, B.; Vallbracht, A. The role of immunoglobulin A in prolonged and relapsing hepatitis A virus infections. J. Gen. Virol. 2012, 93 Pt 4, 754–760. [Google Scholar] [CrossRef] [PubMed]
- Tjon, G.M.; Coutinho, R.A.; van den Hoek, A.; Esman, S.; Wijkmans, C.J.; Hoebe, C.J.; Wolters, B.; Swaan, C.; Geskus, R.B.; Dukers, N.; et al. High and persistent excretion of hepatitis A virus in immunocompetent patients. J. Med. Virol. 2006, 78, 1398–1405. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yotsuyanagi, H.; Koike, K.; Yasuda, K.; Moriya, K.; Shintani, Y.; Fujie, H.; Kurokawa, K.; Iino, S. Prolonged fecal excretion of hepatitis A virus in adult patients with hepatitis A as determined by polymerase chain reaction. Hepatology 1996, 24, 10–13. [Google Scholar] [CrossRef]
- Costa-Mattioli, M.; Allavena, C.; Poirier, A.S.; Billaudel, S.; Raffi, F.; Ferre, V. Prolonged hepatitis A infection in an HIV-1 seropositive patient. J. Med. Virol. 2002, 68, 7–11. [Google Scholar] [CrossRef]
- Shalimar. Gut microbiome and liver diseases. J. Clin. Exp. Hepatol. 2014, 4, 267–268. [Google Scholar] [CrossRef]
- Ishizaka, A.; Koga, M.; Mizutani, T.; Parbie, P.K.; Prawisuda, D.; Yusa, N.; Sedohara, A.; Kikuchi, T.; Ikeuchi, K.; Adachi, E.; et al. Unique Gut Microbiome in HIV Patients on Antiretroviral Therapy (ART) Suggests Association with Chronic Inflammation. Microbiol. Spectr. 2021, 9, e0070821. [Google Scholar] [CrossRef]
- Vujkovic-Cvijin, I.; Sortino, O.; Verheij, E.; Sklar, J.; Wit, F.W.; Kootstra, N.A.; Sellers, B.; Brenchley, J.M.; Ananworanich, J.; Loeff, M.S.V.; et al. HIV-associated gut dysbiosis is independent of sexual practice and correlates with noncommunicable diseases. Nat. Commun. 2020, 11, 2448. [Google Scholar] [CrossRef] [PubMed]
- Parbie, P.K.; Mizutani, T.; Ishizaka, A.; Kawana-Tachikawa, A.; Runtuwene, L.R.; Seki, S.; Abana, C.Z.; Kushitor, D.; Bonney, E.Y.; Ofori, S.B.; et al. Dysbiotic Fecal Microbiome in HIV-1 Infected Individuals in Ghana. Front. Cell. Infect. Microbiol. 2021, 11, 646467. [Google Scholar] [CrossRef] [PubMed]
- Pinto-Cardoso, S.; Klatt, N.R.; Reyes-Teran, G. Impact of antiretroviral drugs on the microbiome: Unknown answers to important questions. Curr. Opin. HIV AIDS 2018, 13, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, A.J.S.; Shaffer, M.; Nusbacher, N.M.; Griesmer, C.; Fiorillo, S.; Schneider, J.M.; Preston Neff, C.; Li, S.X.; Fontenot, A.P.; Campbell, T.; et al. An exploration of Prevotella-rich microbiomes in HIV and men who have sex with men. Microbiome 2018, 6, 198. [Google Scholar] [CrossRef]
- Noguera-Julian, M.; Rocafort, M.; Guillen, Y.; Rivera, J.; Casadella, M.; Nowak, P.; Hildebrand, F.; Zeller, G.; Parera, M.; Bellido, R.; et al. Gut Microbiota Linked to Sexual Preference and HIV Infection. Ebiomedicine 2016, 5, 135–146. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Ishizaka, A.; Sato, H.; Nakamura, H.; Koga, M.; Kikuchi, T.; Hosoya, N.; Koibuchi, T.; Nomoto, A.; Kawana-Tachikawa, A.; Mizutani, T. Short Intracellular HIV-1 Transcripts as Biomarkers of Residual Immune Activation in Patients on Antiretroviral Therapy. J. Virol. 2016, 90, 5665–5676. [Google Scholar] [CrossRef]
- Chalin, A.; Lefevre, B.; Devisme, C.; Pronier, C.; Carriere, V.; Thibault, V.; Amiot, L.; Samson, M. Serum CXCL10, CXCL11, CXCL12, and CXCL14 chemokine patterns in patients with acute liver injury. Cytokine 2018, 111, 500–504. [Google Scholar] [CrossRef]
- Wasmuth, H.E.; Lammert, F.; Zaldivar, M.M.; Weiskirchen, R.; Hellerbrand, C.; Scholten, D.; Berres, M.L.; Zimmermann, H.; Streetz, K.L.; Tacke, F.; et al. Antifibrotic effects of CXCL9 and its receptor CXCR3 in livers of mice and humans. Gastroenterology 2009, 137, 309–319.e3. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Shen, J.; Man, K.; Chu, E.S.; Yau, T.O.; Sung, J.C.; Go, M.Y.; Deng, J.; Lu, L.; Wong, V.W.; et al. CXCL10 plays a key role as an inflammatory mediator and a non-invasive biomarker of non-alcoholic steatohepatitis. J. Hepatol. 2014, 61, 1365–1375. [Google Scholar] [CrossRef] [PubMed]
- Chalin, A.; Lefevre, B.; Devisme, C.; Barget, N.; Amiot, L.; Samson, M. Circulating levels of CXCL11 and CXCL12 are biomarkers of cirrhosis in patients with chronic hepatitis C infection. Cytokine 2019, 117, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Flier, J.; Boorsma, D.M.; Bruynzeel, D.P.; Van Beek, P.J.; Stoof, T.J.; Scheper, R.J.; Willemze, R.; Tensen, C.P. The CXCR3 activating chemokines IP-10, Mig, and IP-9 are expressed in allergic but not in irritant patch test reactions. J. Investig. Dermatol. 1999, 113, 574–578. [Google Scholar] [CrossRef]
- Geissmann, F.; Cameron, T.O.; Sidobre, S.; Manlongat, N.; Kronenberg, M.; Briskin, M.J.; Dustin, M.L.; Littman, D.R. Intravascular immune surveillance by CXCR6+ NKT cells patrolling liver sinusoids. PLoS Biol. 2005, 3, e113. [Google Scholar] [CrossRef]
- Olson, T.S.; Ley, K. Chemokines and chemokine receptors in leukocyte trafficking. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2002, 283, R7–R28. [Google Scholar] [CrossRef]
- Ida, S.; Tachikawa, N.; Nakajima, A.; Daikoku, M.; Yano, M.; Kikuchi, Y.; Yasuoka, A.; Kimura, S.; Oka, S. Influence of human immunodeficiency virus type 1 infection on acute hepatitis A virus infection. Clin. Infect. Dis. 2002, 34, 379–385. [Google Scholar] [CrossRef]
- Zhou, Y.; Callendret, B.; Xu, D.; Brasky, K.M.; Feng, Z.; Hensley, L.L.; Guedj, J.; Perelson, A.S.; Lemon, S.M.; Lanford, R.E.; et al. Dominance of the CD4(+) T helper cell response during acute resolving hepatitis A virus infection. J. Exp. Med. 2012, 209, 1481–1492. [Google Scholar] [CrossRef]
- Fenwick, C.; Joo, V.; Jacquier, P.; Noto, A.; Banga, R.; Perreau, M.; Pantaleo, G. T-cell exhaustion in HIV infection. Immunol. Rev. 2019, 292, 149–163. [Google Scholar] [CrossRef]
- Vallbracht, A.; Maier, K.; Stierhof, Y.D.; Wiedmann, K.H.; Flehmig, B.; Fleischer, B. Liver-derived cytotoxic T cells in hepatitis A virus infection. J. Infect. Dis. 1989, 160, 209–217. [Google Scholar] [CrossRef]
- Walker, C.M.; Feng, Z.; Lemon, S.M. Reassessing immune control of hepatitis A virus. Curr. Opin. Virol. 2015, 11, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Fensterl, V.; Grotheer, D.; Berk, I.; Schlemminger, S.; Vallbracht, A.; Dotzauer, A. Hepatitis A virus suppresses RIG-I-mediated IRF-3 activation to block induction of beta interferon. J. Virol. 2005, 79, 10968–10977. [Google Scholar] [CrossRef] [PubMed]
- Qu, L.; Feng, Z.; Yamane, D.; Liang, Y.; Lanford, R.E.; Li, K.; Lemon, S.M. Disruption of TLR3 signaling due to cleavage of TRIF by the hepatitis A virus protease-polymerase processing intermediate, 3CD. PLoS Pathog. 2011, 7, e1002169. [Google Scholar] [CrossRef]
- Yang, Y.; Liang, Y.; Qu, L.; Chen, Z.; Yi, M.; Li, K.; Lemon, S.M. Disruption of innate immunity due to mitochondrial targeting of a picornaviral protease precursor. Proc. Natl. Acad. Sci. USA 2007, 104, 7253–7258. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Fang, L.; Wei, D.; Zhang, H.; Luo, R.; Chen, H.; Li, K.; Xiao, S. Hepatitis A virus 3C protease cleaves NEMO to impair induction of beta interferon. J. Virol. 2014, 88, 10252–10258. [Google Scholar] [CrossRef] [PubMed]
- Lanford, R.E.; Feng, Z.; Chavez, D.; Guerra, B.; Brasky, K.M.; Zhou, Y.; Yamane, D.; Perelson, A.S.; Walker, C.M.; Lemon, S.M. Acute hepatitis A virus infection is associated with a limited type I interferon response and persistence of intrahepatic viral RNA. Proc. Natl. Acad. Sci. USA 2011, 108, 11223–11228. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Wu, Z.; Xu, W.; Yang, J.; Chen, Y.; Li, L. Intestinal microbiota was assessed in cirrhotic patients with hepatitis B virus infection. Intestinal microbiota of HBV cirrhotic patients. Microb. Ecol. 2011, 61, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Wang, B.; Fu, Y.; Chen, Y.; Yang, F.; Lu, H.; Chen, Y.; Xu, J.; Li, L. Changes of fecal Bifidobacterium species in adult patients with hepatitis B virus-induced chronic liver disease. Microb. Ecol. 2012, 63, 304–313. [Google Scholar] [CrossRef]
- Wei, X.; Yan, X.; Zou, D.; Yang, Z.; Wang, X.; Liu, W.; Wang, S.; Li, X.; Han, J.; Huang, L.; et al. Abnormal fecal microbiota community and functions in patients with hepatitis B liver cirrhosis as revealed by a metagenomic approach. BMC Gastroenterol. 2013, 13, 175. [Google Scholar] [CrossRef]
- Aly, A.M.; Adel, A.; El-Gendy, A.O.; Essam, T.M.; Aziz, R.K. Gut microbiome alterations in patients with stage 4 hepatitis C. Gut Pathog. 2016, 8, 42. [Google Scholar] [CrossRef]
- Ilhan, Z.E.; Marcus, A.K.; Kang, D.W.; Rittmann, B.E.; Krajmalnik-Brown, R. pH-Mediated Microbial and Metabolic Interactions in Fecal Enrichment Cultures. mSphere 2017, 2, e00047-17. [Google Scholar] [CrossRef] [PubMed]
- Duncan, S.H.; Louis, P.; Thomson, J.M.; Flint, H.J. The role of pH in determining the species composition of the human colonic microbiota. Environ. Microbiol. 2009, 11, 2112–2122. [Google Scholar] [CrossRef] [PubMed]
- Kakiyama, G.; Pandak, W.M.; Gillevet, P.M.; Hylemon, P.B.; Heuman, D.M.; Daita, K.; Takei, H.; Muto, A.; Nittono, H.; Ridlon, J.M.; et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J. Hepatol. 2013, 58, 949–955. [Google Scholar] [CrossRef]
- Li, M.; Liu, S.; Wang, M.; Hu, H.; Yin, J.; Liu, C.; Huang, Y. Gut Microbiota Dysbiosis Associated with Bile Acid Metabolism in Neonatal Cholestasis Disease. Sci. Rep. 2020, 10, 7686. [Google Scholar] [CrossRef] [PubMed]
- Pinto-Cardoso, S.; Lozupone, C.; Briceno, O.; Alva-Hernandez, S.; Tellez, N.; Adriana, A.; Murakami-Ogasawara, A.; Reyes-Teran, G. Fecal Bacterial Communities in treated HIV infected individuals on two antiretroviral regimens. Sci. Rep. 2017, 7, 43741. [Google Scholar] [CrossRef]


| HIV and Acute HAV | HIV Alone | Healthy Controls | |
|---|---|---|---|
| Total patients (No.) | 10 | 25 | 22 |
| Males (No.) | 10 (100%) | 25 (100%) | 22 (100%) |
| MSM (No.) | 10 (100%) | 24 (96%) | - |
| Age in years | 46 (36.8–52.3) | 47 (42–50.5) | 45 (34–50.3) |
| BMI | 23.6 (20.8–25.2) | 23.6 (20.8–26.1) | - |
| HIV Viral load < 20 copies/mL (No.) | 10 (100%) 1 | 25 (100%) | - |
| CD4 count (cells/ul) | 579 (483–707.5) 1 | 613 (517–731.5) | - |
| Duration of ART (years) | 8 (3.5–14) | 8 (6.5–13) | - |
| Peak serum ALT (IU/L) | 3540 (581.3–4001.3) | - | - |
| Peak serum AST (IU/L) | 1958 (1288–3673.8) | ||
| Peak serum T-Bil (mg/dL) | 6.4 (3.0–11.8) | - | - |
| Patient No. | Age | BMI | Years after HIV Diagnosis | CD4 Counts (/μL) 1 | CD8 Counts (/μL) 1 | CD4/CD8 Ratio 1 | HIV-RNA Load (Copies/mL) 1 | ART Regimen | Underlying Health Conditions |
|---|---|---|---|---|---|---|---|---|---|
| 20 | 40 | 24.2 | 9 | 508 | 656 | 0.8 | <20 | TAF/FTC RPV | Dyslipidemia |
| 213 | 56 | 24.8 | 24 | 538 | 1035 | 0.5 | <20 | TDF/FTC DTG | Insomnia |
| 463 | 49 | 27.7 | 20 | 955 | 1124 | 0.8 | <20 | ABC/3TC/DTG | Hypertension, dyslipidemia |
| 708 | 44 | 23.1 | 12 | 490 | 378 | 1.3 | <20 | ABC/3TC DRV/c | - |
| 832 | 53 | 24.2 | 7 | 817 | 839 | 1 | <20 | TAF/FTC DTG | - |
| 989 | 36 | 21.3 | 10 | 620 | 589 | 1.1 | <20 | TAF/FTC/RPV | Dyslipidemia |
| 1141 | 48 | 26.4 | 6 | 671 | 447 | 1.5 | <20 | ABC/3TC RAL | - |
| 1292 | 52 | 19.2 | 4 | 293 | 605 | 0.5 | <20 | TAF/FTC DTG | - |
| 1334 | 27 | 17.7 | 2 | 656 | 525 | 1.2 | <20 | TAF/FTC DTG | - |
| 1349 | 37 | 22.0 | 1 | 462 | 539 | 0.9 | <20 | ABC/3TC/DTG | Atopic dermatitis |
| Patient No. | Initial HA-IgM (s/co) | ALT (IU/L) | Max AST (IU/L) | Max T-Bil (mg/dL) | |
|---|---|---|---|---|---|
| Before | Max | ||||
| 20 | 5.7 | 34 | 8877 | 6996 | 7.8 |
| 213 | 0.58 | 23 | 3515 | 3530 | 6.6 |
| 463 | 1.09 | 114 | 3880 | 1722 | 3.1 |
| 708 | 3.25 | 16 | 2589 | 2194 | 15.2 |
| 832 | 3.24 | 21 | 3985 | 4105 | 6.2 |
| 989 | 11.2 | 11 | 677 | 1400 | 22.8 |
| 1141 | 11.7 | 26 | 294 | 152 | 2.5 |
| 1292 | 2.04 | 10 | 174 | 952 | 2.2 |
| 1334 | 10.1 | 8 | 4050 | 2860 | 10.6 |
| 1349 | 11.6 | 22 | 3565 | 1663 | 4.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ishizaka, A.; Koga, M.; Mizutani, T.; Lim, L.A.; Adachi, E.; Ikeuchi, K.; Ueda, R.; Aoyagi, H.; Tanaka, S.; Kiyono, H.; et al. Prolonged Gut Dysbiosis and Fecal Excretion of Hepatitis A Virus in Patients Infected with Human Immunodeficiency Virus. Viruses 2021, 13, 2101. https://doi.org/10.3390/v13102101
Ishizaka A, Koga M, Mizutani T, Lim LA, Adachi E, Ikeuchi K, Ueda R, Aoyagi H, Tanaka S, Kiyono H, et al. Prolonged Gut Dysbiosis and Fecal Excretion of Hepatitis A Virus in Patients Infected with Human Immunodeficiency Virus. Viruses. 2021; 13(10):2101. https://doi.org/10.3390/v13102101
Chicago/Turabian StyleIshizaka, Aya, Michiko Koga, Taketoshi Mizutani, Lay Ahyoung Lim, Eisuke Adachi, Kazuhiko Ikeuchi, Ryuta Ueda, Haruyo Aoyagi, Satoshi Tanaka, Hiroshi Kiyono, and et al. 2021. "Prolonged Gut Dysbiosis and Fecal Excretion of Hepatitis A Virus in Patients Infected with Human Immunodeficiency Virus" Viruses 13, no. 10: 2101. https://doi.org/10.3390/v13102101
APA StyleIshizaka, A., Koga, M., Mizutani, T., Lim, L. A., Adachi, E., Ikeuchi, K., Ueda, R., Aoyagi, H., Tanaka, S., Kiyono, H., Matano, T., Aizaki, H., Yoshio, S., Mita, E., Muramatsu, M., Kanto, T., Tsutsumi, T., & Yotsuyanagi, H. (2021). Prolonged Gut Dysbiosis and Fecal Excretion of Hepatitis A Virus in Patients Infected with Human Immunodeficiency Virus. Viruses, 13(10), 2101. https://doi.org/10.3390/v13102101