Neutralization of Dengue Virus Serotypes by Sera from Dengue-Infected Individuals Is Preferentially Directed to Heterologous Serotypes and Not against the Autologous Serotype Present in Acute Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Viruses
2.3. Serum Specimens
2.4. Foci Reduction Neutralization Test
2.5. Maltose Binding Protein–ED3 Fusion Proteins
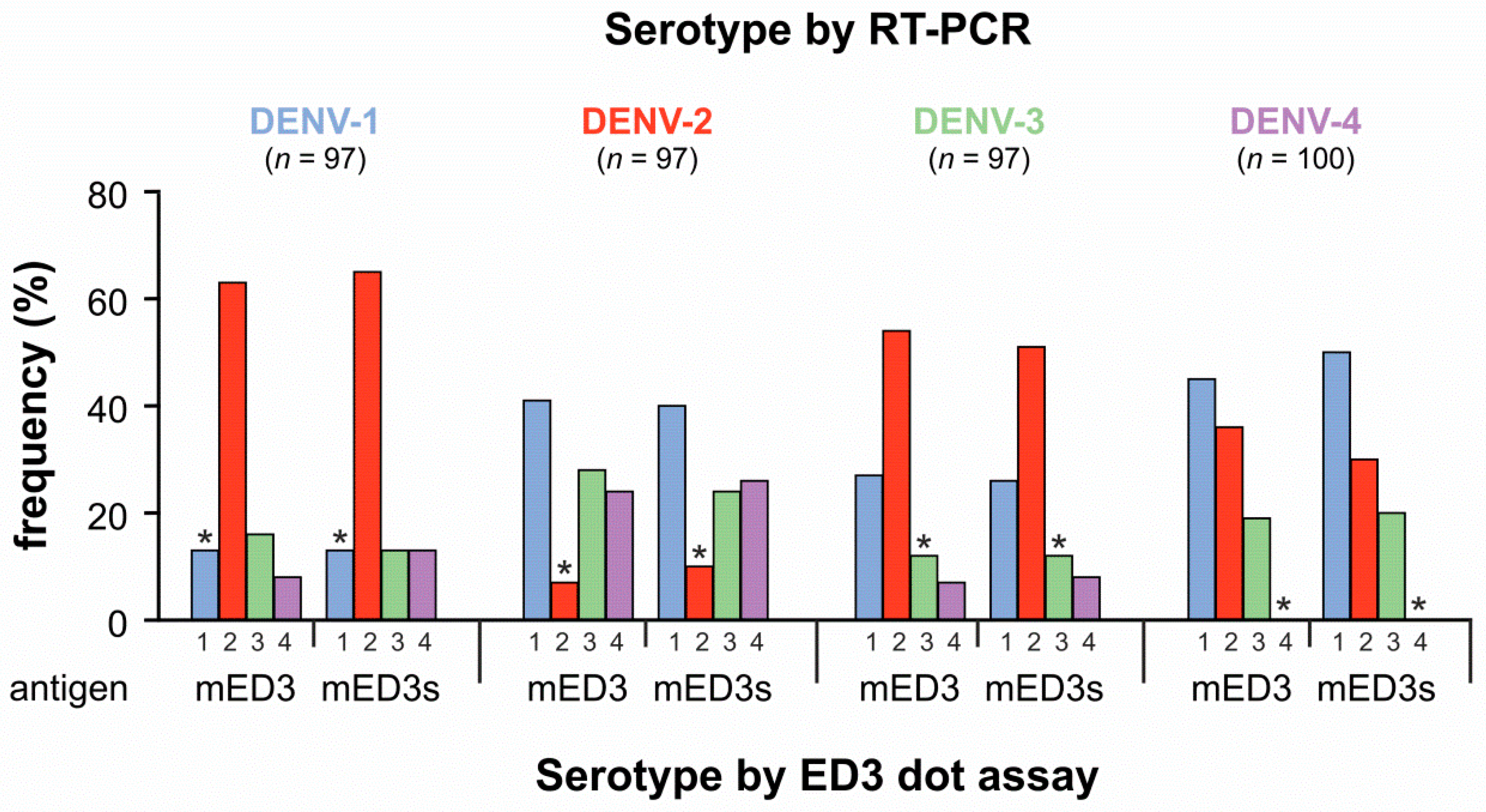
2.6. ED3 Dot Assay
2.7. ED3 ELISA
3. Results
3.1. Comparison of Direct DENV Detection and Serotyping Using the ED3 Dot Assay
3.2. Neutralization of DENV by Sera from RT-PCR Diagnosed DENV Cases
3.3. Neutralization of Heterologous and Homologous Serotypes Using DENV-1 Positive Sera
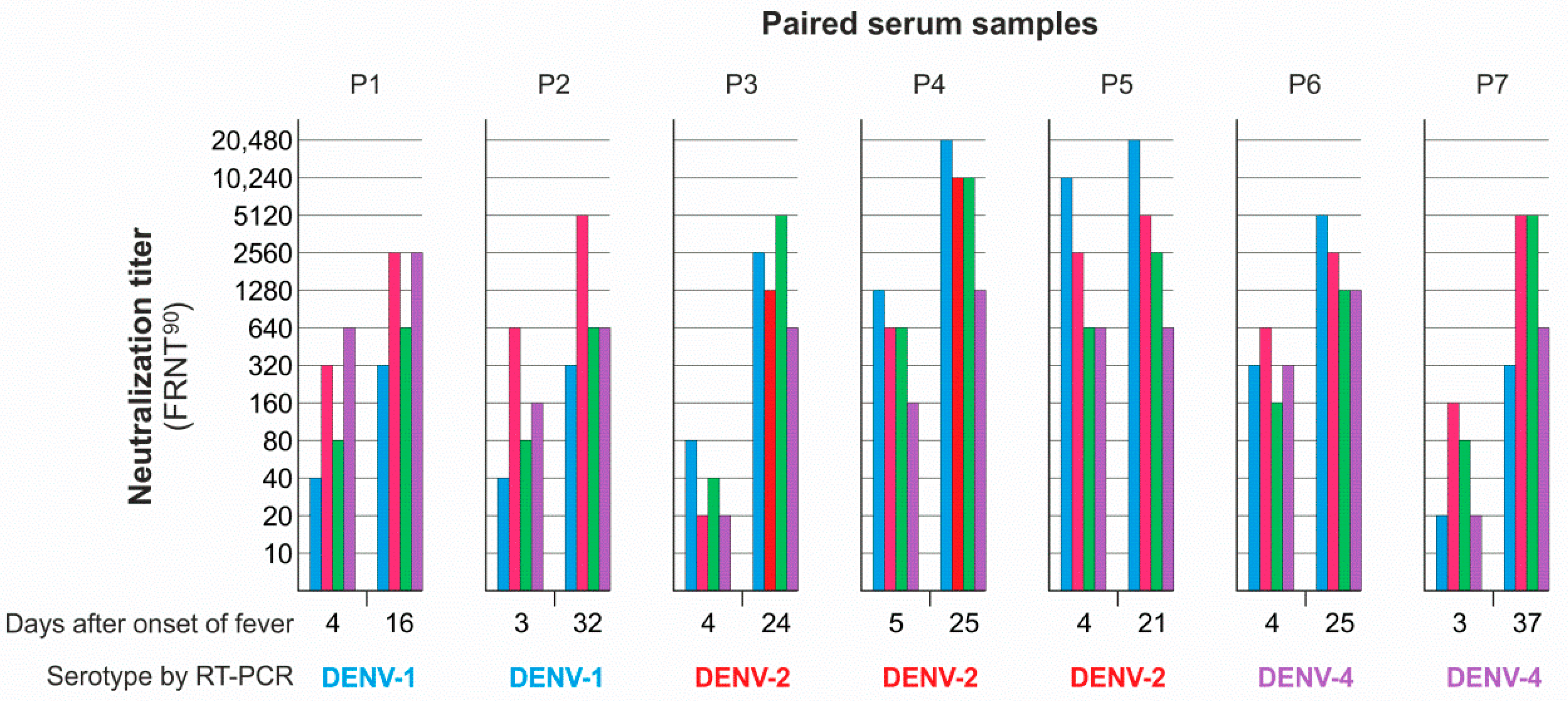
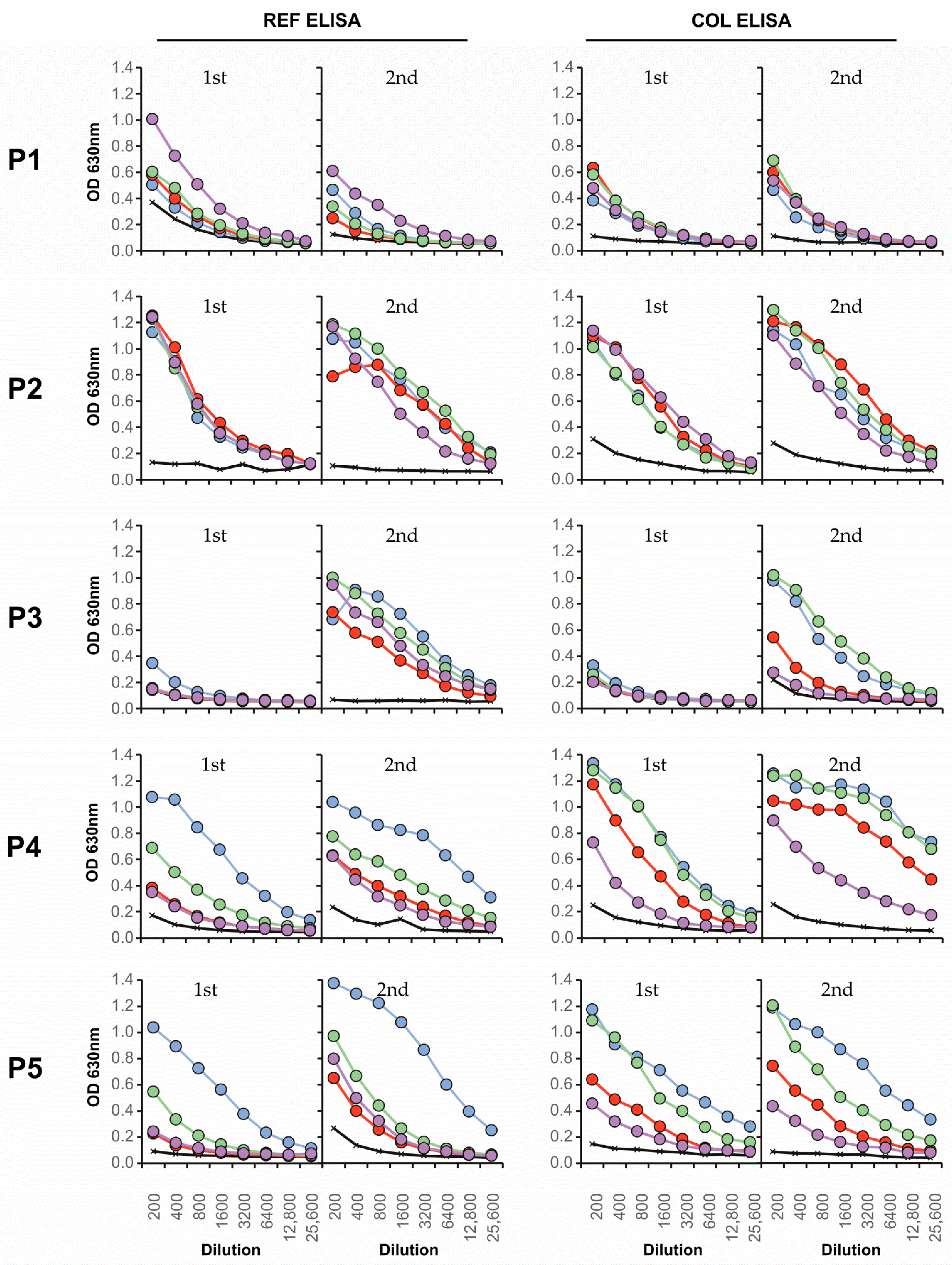
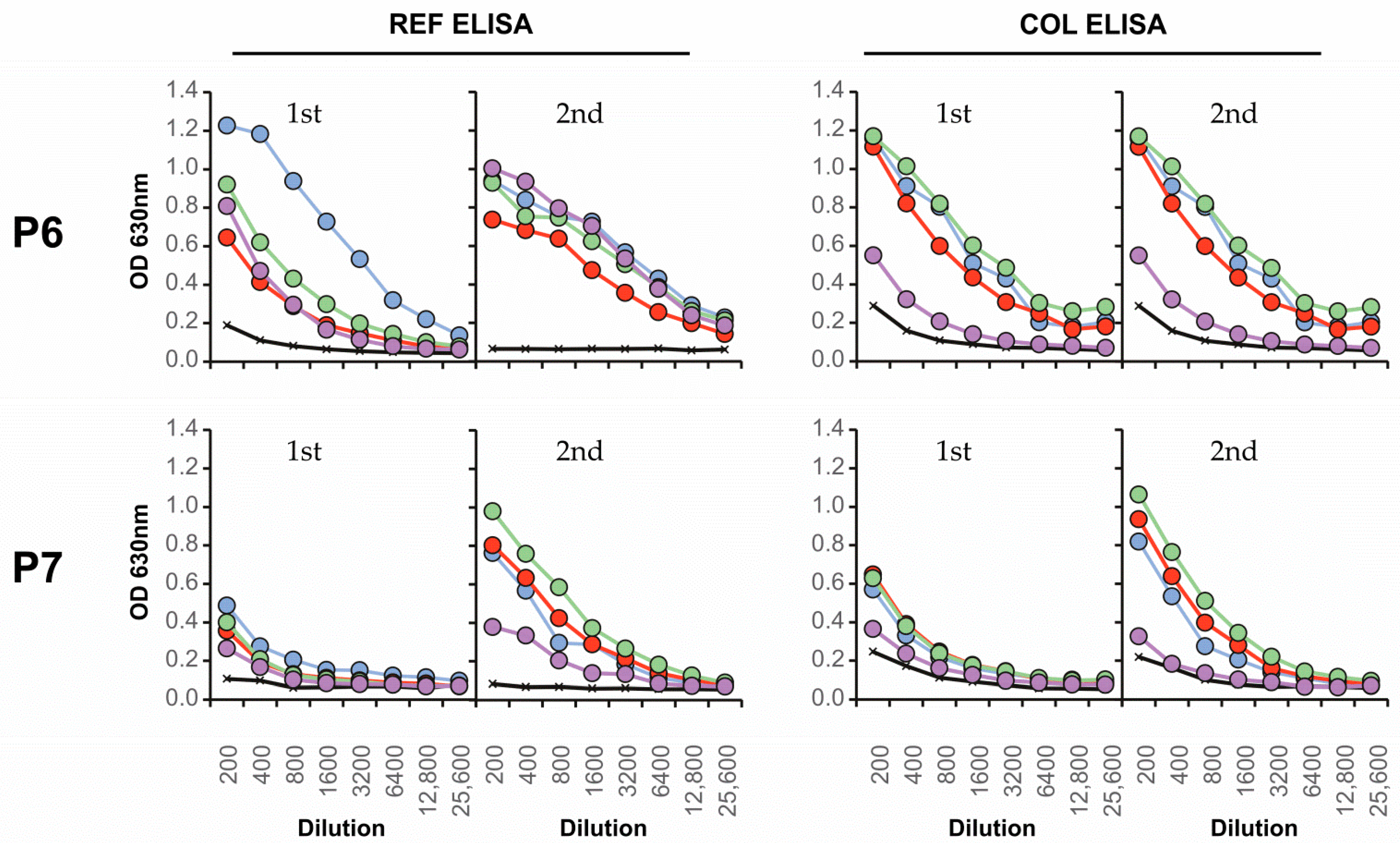
3.4. Neutralizing Antibody Responses and Serotype Specificity in Late Serum Samples
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| DENV-1 | |
| Oligo-1 | 5′-AAG GGG ATG TCA TAT GTG ATG TGC ACA GGC TCA TTT AAG CTA GAG AAG GAA GTG GCT GAG AC-3′ |
| Oligo-2 | 5′-CGC GTC TGT TCC TTC GTA TTT GAC CTG CAC TAG AAC AGT TCC ATG CTG GGT CTC AGC CAC TTC CTT CTC T-3′ |
| Oligo-3 | 5′-ACG AAG GAA CAG ACG CGC CAT GCA AGA TCC CCT TCT CGA CCC AAG ATG AGA AAG GAG TGA CCC AGA ATG-3‘ |
| Oligo-4 | 5′-GAC TGA TTT TTC TTT GTC AGT AAC TAT GGG ATT GGC TGT TAT CAA TCT CCC ATT CTG GGT CAC TCC TTT C-3′ |
| Oligo-5 | 5′-TTA CTG ACA AAG AAA AAT CAG TCA ACA TTG AGA CAG AAC CAC CTT TTG GTG AGA GCT ACA TCG TGG TAG GGG CAG-3′ |
| Oligo-6 | 5′-GCT TCC TTT CTT GAA CCA GCT TAG TTT CAA GGC TTT TTC ACC TGC CCC TAC CAC GAT-3′ |
| Oligo-7 BamHI-for | 5′-CAC GGA TCC AAG GGG ATG TCA TAT GTG ATG TG-3′ |
| Oligo-8 HindIII-rev | 5′-CAC AAG CTT GCT TCC TTT CTT GAA CCA GCT-3′ |
| DENV-2 | |
| Oligo-1 | 5′-TCA TAC TCT ATG TGT ACA GGA AAG TTT AAA ATT GTG AGA GAA ATA GCA GAA ACA CAA CAT G-3′ |
| Oligo-2 | 5′-CAT GGA GAA CCG TCC CCT TCA TAT TGT ATT CTG ATA ACT ATT GTT CCA TGT TGT GTT TCT GCT ATT TCT C-3′ |
| Oligo-3 | 5′-GGG GAC GGT TCT CCA TGT AAG ATC CCT TTT GAA ATA ACA GAC TTG GAA AAA AGA CAC GTC TTA GGT CGC-3′ |
| Oligo-4 | 5′-CTA TGT TGA CTG GGC TAT CTT TTT CTA TTA CGA TTG GGT TAA CTG TAA TCG GCG ACC TAA GAC GTG TCT TTT TT-3′ |
| Oligo-5 | 5′-AAT AGA AAA AGA TAG CCC AGT CAA CAT AGA AGC AGA ACC TCC ATT CGG AGA CAG CTA CAT CAT CAT AGG AGT AGA G-3′ |
| Oligo-6 | 5′-ACT TCC CTT CTT AAA CCA ATT GAG TTT CAA TTG TCC CGG CTC TAC TCC TAT GAT GAT GTA GCT GT-3′ |
| Oligo-7 BamHI-for | 5′-CAC GGA TCC TCA TAC TCT ATG TGT ACA GGA AAG TTT AAA-3′ |
| Oligo-8 HindIII-rev | 5′-GTG AAG CTT ACT TCC CCT CTT AAA CCA ATT GAG T-3′ |
| DENV-3 | |
| Oligo-1 | 5′-AAG GGG ATG AGC TAT GCA ATG TGC ACG AGT ACC TTT GTG TTG-3′ |
| Oligo-2 | 5′-ATG AGT ATT GTC CCA TGT TGC GTT TCT GAG ACT TCT TTC TTC AAC ACA AAG GTA CTC GTG C-3′ |
| Oligo-3 | 5′-CGC AAC ATG GGA CAA TAC TCA TCA AGG TCG AGT ACA AAG GGG AAG ATG TAC CTT GCA AG-3′ |
| Oligo-4 | 5′-TGT GAG CTT TCC CTT GTC CAT CCT CTG TGG AGA AAG GAA TCT TGC AAG GTA CAT CTT CCC C-3‘ |
| Oligo-5 | 5′-TGG ACA AGG GAA AGC TCA CAA TGG CAG ACT GAT TAC AGC CAA CCC AGT GGT GAC TAA GAG-3′ |
| Oligo-6 | 5′-CCA AAA GGA GGT TCA GCC TCA ATA TTG ACA GGC TCC TCC CTC TTA GTC ACC ACT GGG TTG-3′ |
| Oligo-7 | 5′-TGA GGC TGA ACC TCC TTT TGG GGA AAG TAA TAT AGT AAT TGG AAT TGG AGA CAA CGC CTT-3′ |
| Oligo-8 | 5′-GCT TCC CTT CTT GTA CCA GTT GAT TTT CAA GGC GTT GTC TCC AAT TCC-3′ |
| Oligo-9 BamHI-for | 5′-CACGGATCCA AGGGGATGAG CTATGCAATG-3′ |
| Oligo-10 HindIII-rev | 5′-GTGAAGCTTG CTTCCCTTCT TGTACCAGTT G-3′ |
| DENV-4 | |
| Oligo-1 | 5′-AAA GGT ATG TCT TAC ACG ATG TGT TCG GGC AAG TTT AGT ATT GAT AAA GAA ATG GCG GAA ACG-3′ |
| Oligo-2 | 5′-GGG GGC TCC CGC CCC TTC GTA TTT GAC TTT CAC CAC TGT CGT TCC GTG CTG CGT TTC CGC CAT TTC TTT ATC-3′ |
| Oligo-3 | 5′-CGA AGG GGC GGG AGC CCC CTG TAA AGT CCC CAT TGA AAT TCG GGA CGT GAA CAA GGA GAA AGT CGT AGG-3′ |
| Oligo-4 | 5′-CTA TAT TCG TGA CAC TGT TCG TAT TTT CTG CCA GAG GTG TTG CAG AGA TCA CAC GTC CTA CGA CTT TCT CCT TGT TC-3′ |
| Oligo-5 | 5′-CGA ACA GTG TCA CGA ATA TAG AAC TTG AAC CGC CCT TTG GCG ACA GCT ATA TAA TGA TAG GGG TGG GC-3′ |
| Oligo-6 | 5′-CGA TCC TTT TCT GAA CCA ATG AAG GGT TAA TGC AGA ATT G CC CAC CCC TAT CAT TAT ATA GC-3′ |
| Oligo-7 BamHI-for | 5′-CAC GGA TCC AAA GGT ATG TCT T-3′ |
| Oligo-8 HindIII-rev | 5′-CAC AAG CTT CGA TCC TTT TCT G-3′ |
References
- Calisher, C.H.; Karabatsos, N.; Dalrymple, J.M.; Shope, R.E.; Porterfield, J.S.; Westaway, E.G.; Brandt, W.E. Antigenic Relationships between Flaviviruses as Determined by Cross-neutralization Tests with Polyclonal Antisera. J. Gen. Virol. 1989, 70, 37–43. [Google Scholar] [CrossRef]
- Russell, P.K.; Nisalak, A. Dengue virus identification by the plaque reduction neutralization test. J. Immunol. 1967, 99, 291–296. [Google Scholar]
- Henchal, E.A.; Gentry, M.K.; McCown, J.M.; Brandt, W.E. Dengue virus-specific and flavivirus group determinants identified with monoclonal antibodies by indirect immunofluorescence. Am. J. Trop. Med. Hyg. 1982, 31, 830–836. [Google Scholar] [CrossRef]
- Henchal, E.A.; McCown, J.M.; Seguin, M.C.; Gentry, M.K.; Brandt, W.E. Rapid identification of dengue virus isolates by using monoclonal antibodies in an indirect immunofluorescence assay. Am. J. Trop. Med. Hyg. 1983, 32, 164–169. [Google Scholar] [CrossRef]
- Nawa, M.; Ichikawa, Y.; Inouye, S. Serotyping of dengue viruses by an enzyme-linked immunosorbent assay. Jpn. J. Med. Sci. Biol. 1985, 38, 217–221. [Google Scholar] [CrossRef]
- Santiago, G.A.; Vergne, E.; Quiles, Y.; Cosme, J.; Vazquez, J.; Medina, J.F.; Medina, F.; Colón, C.; Margolis, H.; Muñoz-Jordán, J.L. Analytical and Clinical Performance of the CDC Real Time RT-PCR Assay for Detection and Typing of Dengue Virus. PLoS Negl. Trop. Dis. 2013, 7, e2311. [Google Scholar] [CrossRef]
- Zolla-Pazner, S.; Gorny, M.K.; Nyambi, P.N.; VanCott, T.C.; Nádas, A. Immunotyping of human immunodeficiency virus type 1 (HIV): An approach to immunologic classification of HIV. J. Virol. 1999, 73, 4042–4051. [Google Scholar] [CrossRef]
- Nyambi, P.N.; Nádas, A.; Mbah, H.A.; Burda, S.; Williams, C.; Gorny, M.K.; Zolla-Pazner, S. Immunoreactivity of intact virions of human immunodeficiency virus type 1 (HIV-1) reveals the existence of fewer HIV-1 immunotypes than genotypes. J. Virol. 2000, 74, 10670–10680. [Google Scholar] [CrossRef][Green Version]
- Katzelnick, L.C.; Fonville, J.M.; Gromowski, G.D.; Arriaga, J.B.; Green, A.; James, S.L.; Lau, L.; Montoya, M.; Wang, C.; VanBlargan, L.A.; et al. Dengue viruses cluster antigenically but not as discrete serotypes. Science 2015, 349, 1338–1343. [Google Scholar] [CrossRef]
- Tsai, W.-Y.; Durbin, A.; Tsai, J.-J.; Hsieh, S.-C.; Whitehead, S.; Wang, W.-K. Complexity of Neutralizing Antibodies against Multiple Dengue Virus Serotypes after Heterotypic Immunization and Secondary Infection Revealed by In-Depth Analysis of Cross-Reactive Antibodies. J. Virol. 2015, 89, 7348–7362. [Google Scholar] [CrossRef]
- Morens, D.M.; Halstead, S.B.; Repik, P.M.; Putvatana, R.; Raybourne, N. Simplified plaque reduction neutralization assay for dengue viruses by semimicro methods in BHK-21 cells: Comparison of the BHK suspension test with standard plaque reduction neutralization. J. Clin. Microbiol. 1985, 22, 250–254. [Google Scholar] [CrossRef]
- Patel, B.; Longo, P.; Miley, M.J.; Montoya, M.; Harris, E.; de Silva, A.M. Dissecting the human serum antibody response to secondary dengue virus infections. PLoS Negl. Trop. Dis. 2017, 11, e0005554. [Google Scholar] [CrossRef]
- Guzman, M.G.; Alvarez, M.; Rodriguez-Roche, R.; Bernardo, L.; Montes, T.; Vazquez, S.; Morier, L.; Alvarez, A.; Gould, E.A.; Kourí, G.; et al. Neutralizing Antibodies after Infection with Dengue 1 Virus. Emerg. Infect. Dis. 2007, 13, 282–286. [Google Scholar] [CrossRef]
- Imrie, A.; Meeks, J.; Gurary, A.; Sukhbaatar, M.; Truong, T.T.; Cropp, C.B.; Effler, P. Antibody to dengue 1 detected more than 60 years after infection. Viral Immunol. 2007, 20, 672–675. [Google Scholar] [CrossRef]
- Katzelnick, L.C.; Montoya, M.; Gresh, L.; Balmaseda, A.; Harris, E. Neutralizing antibody titers against dengue virus correlate with protection from symptomatic infection in a longitudinal cohort. Proc. Natl. Acad. Sci. USA 2016, 113, 728–733. [Google Scholar] [CrossRef] [PubMed]
- Sabin, A.B. Research on dengue during World War II. Am. J. Trop. Med. Hyg. 1952, 1, 30–50. [Google Scholar] [CrossRef] [PubMed]
- Auerswald, H.; de Jesus, A.; Seixas, G.; Nazareth, T.; In, S.; Mao, S.; Duong, V.; Clara Silva, A.; Paul, R.; Dussart, P.; et al. First dengue virus seroprevalence study on Madeira Island after the 2012 outbreak indicates unreported dengue circulation. Parasit. Vectors 2019, 12, 103. [Google Scholar] [CrossRef]
- Libraty, D.H.; Zhang, L.; Obcena, A.; Brion, J.D.; Capeding, R.Z. Anti-dengue virus envelope protein domain III IgG ELISA among infants with primary dengue virus infections. Acta Trop. 2015, 142, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Sirivichayakul, C.; Sabchareon, A.; Limkittikul, K.; Yoksan, S. Plaque reduction neutralization antibody test does not accurately predict protection against dengue infection in Ratchaburi cohort, Thailand. Virol. J. 2014, 11, 48. [Google Scholar] [CrossRef] [PubMed]
- Dejnirattisai, W.; Jumnainsong, A.; Onsirisakul, N.; Fitton, P.; Vasanawathana, S.; Limpitikul, W.; Puttikhunt, C.; Edwards, C.; Duangchinda, T.; Supasa, S.; et al. Cross-reacting antibodies enhance dengue virus infection in humans. Science 2010, 328, 745–748. [Google Scholar] [CrossRef]
- Lai, C.-Y.; Tsai, W.-Y.; Lin, S.-R.; Kao, C.-L.; Hu, H.-P.; King, C.-C.; Wu, H.-C.; Chang, G.-J.; Wang, W.-K. Antibodies to Envelope Glycoprotein of Dengue Virus during the Natural Course of Infection Are Predominantly Cross-Reactive and Recognize Epitopes Containing Highly Conserved Residues at the Fusion Loop of Domain II. J. Virol. 2008, 82, 6631–6643. [Google Scholar] [CrossRef]
- Durbin, A.P.; Karron, R.A.; Sun, W.; Vaughn, D.W.; Reynolds, M.J.; Perreault, J.R.; Thumar, B.; Men, R.; Lai, C.-J.; Elkins, W.R.; et al. Attenuation and immunogenicity in humans of a live Dengue virus type-4 vaccine candidate with a 30 nucleotide deletion in its 3-untranslated region. Am. J. Trop. Med. Hyg 2001, 65, 405–413. [Google Scholar] [CrossRef]
- Halstead, S.B.; Casals, L.; Shotwell, H.; Palumboii, N. Studies on the immunization of monkeys against Dengue. Protection derived from single and sequential virus infections. Am. J. Trop. Med. Hyg. 1973, 22, 366–374. [Google Scholar]
- Auerswald, H.; Klepsch, L.; Schreiber, S.; Hülsemann, J.; Franzke, K.; Kann, S.; Y., B.; Duong, V.; Buchy, P.; Schreiber, M.; et al. The Dengue ED3 Dot Assay, a Novel Serological Test for the Detection of Denguevirus Type-Specific Antibodies and Its Application in a Retrospective Seroprevalence Study. Viruses 2019, 11, 304. [Google Scholar] [CrossRef]
- Halstead, S.B.; Rojanasuphot, S.; Sangkawibha, N. Original antigenic sin in dengue. Am. J. Trop. Med. Hyg. 1983, 32, 154–156. [Google Scholar] [CrossRef]
- Monto, A.S.; Malosh, R.E.; Petrie, J.G.; Martin, E.T. The Doctrine of Original Antigenic Sin: Separating Good from Evil. J. Infect. Dis. 2017, 215, 1782–1788. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Lin, X.-D.; Vasilakis, N.; Tian, J.-H.; Li, C.-X.; Chen, L.-J.; Eastwood, G.; Diao, X.-N.; Chen, M.-H.; Chen, X.; et al. Divergent viruses discovered in arthropods and vertebrates revise the evolutionary history of the Flaviviridae and related viruses. J. Virol. 2015, 90, 659–669. [Google Scholar] [CrossRef]
- Aguiar, M.; Stollenwerk, N.; Halstead, S. The impact of the newly licensed dengue vaccine in endemic countries. PLoS Negl. Trop. Dis. 2016, 10, e0005179. [Google Scholar] [CrossRef]
- Aguiar, M.; Stollenwerk, N.; Halstead, S. The risks behind Dengvaxia recommendation. Lancet. Infect. Dis. 2016, 16, 882–883. [Google Scholar] [CrossRef]
- Aguiar, M.; Halstead, S.; Stollenwerk, N. Consider stopping Dengvaxia administration without immunological screening. Expert Rev. Vaccines 2017, 16, 301–302. [Google Scholar] [CrossRef]
- Aguiar, M.; Stollenwerk, N. Dengvaxia Efficacy Dependency on Serostatus: A Closer Look at More Recent Data. Clin. Infect. Dis. 2018, 66, 641–642. [Google Scholar] [CrossRef] [PubMed]
- Chatchen, S.; Sabchareon, A.; Sirivichayakul, C. Serodiagnosis of asymptomatic dengue infection. Asian Pac. J. Trop. Med. 2017, 10, 11–14. [Google Scholar] [CrossRef] [PubMed]
- Kyaw, A.K.; Ngwe Tun, M.M.; Naing, S.T.; Htwe, T.T.; Mar, T.T.; Khaing, T.M.; Aung, T.; Aye, K.S.; Thant, K.Z.; Morita, K.; et al. Inapparent dengue virus infection among students in Mandalay, Myanmar. Trans. R. Soc. Trop. Med. Hyg. 2019. [Google Scholar] [CrossRef] [PubMed]
- Olivero, R.M.; Hamer, D.H.; MacLeod, W.B.; Benoit, C.M.; Sanchez-Vegas, C.; Jentes, E.S.; Chen, L.H.; Wilson, M.E.; Marano, N.; Yanni, E.A.; et al. Dengue Virus Seroconversion in Travelers to Dengue-Endemic Areas. Am. J. Trop. Med. Hyg. 2016, 95, 1130–1136. [Google Scholar] [CrossRef] [PubMed]
- Grange, L.; Simon-Loriere, E.; Sakuntabhai, A.; Gresh, L.; Paul, R.; Harris, E. Epidemiological Risk Factors Associated with High Global Frequency of Inapparent Dengue Virus Infections. Front. Immunol. 2014, 5, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Liu, Q.; Huang, W.; Li, X.; Wang, Y. Current status on the development of pseudoviruses for enveloped viruses. Rev. Med. Virol. 2018, 28, e1963. [Google Scholar] [CrossRef]
- Mukherjee, S.; Pierson, T.C.; Dowd, K.A. Pseudo-infectious reporter virus particles for measuring antibody-mediated neutralization and enhancement of dengue virus infection. Methods Mol. Biol. 2014, 1138, 75–97. [Google Scholar]
- Kretschmer, M.; Kadlubowska, P.; Hoffmann, D.; Schwalbe, B.; Auerswald, H.; Schreiber, M. Zikavirus prME Envelope Pseudotyped Human Immunodeficiency Virus Type-1 as a Novel Tool for Glioblastoma-Directed Virotherapy. Cancers 2020, 12, 1000. [Google Scholar] [CrossRef] [PubMed]

| REF | ||||
| DENV-1 | 16007 | Thailand | 1964 | Genbank AF180817 |
| DENV-2 | 16681 | Thailand | 1984 | Genbank U87411.1 |
| DENV-3 | H87 | Philippines | 1956 | Genbank M93130 |
| DENV-4 | H241 | Philippines | 1956 | Genbank AB609591 |
| IPC A | ||||
| DENV-1 | KH/BID-V2004/2006 | Cambodia | 2006 | Genbank FJ639687 |
| DENV-2 | D2KH/06PHP | Cambodia | 2006 | IPC virus isolate |
| DENV-3 | KH/BID-V2058/2005 | Cambodia | 2005 | Genbank GQ868628 |
| DENV-4 | D4KH/98PHP | Cambodia | 1998 | IPC virus isolate |
| IPC B | ||||
| DENV-1 | KH/BID-V2011/2007 | Cambodia | 2007 | Genbank FJ639693 |
| DENV-2 | KH/BID-V2066/2007 | Cambodia | 2007 | Genbank FJ639717 |
| DENV-3 | KH/BID-V2051/2007 | Cambodia | 2006 | Genbank FJ639713 |
| DENV-4 | D4KH/07 | Cambodia | 2007 | IPC virus isolate |
| FRNT90, 1st Serum | FRNT90, 2nd Serum | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Serotype by | DENV | DENV | |||||||
| RT-PCR | 1 | 2 | 3 | 4 | 1 | 2 | 3 | 4 | |
| 01 | DENV-1 | 320 | 5120 | 640 | 2560 | 640 | 20,480 | 640 | 5120 |
| 02 | DENV-1 | 20 | 320 | 40 | 160 | 1280 | 20,480 | 5120 | 5120 |
| 03 | DENV-1 | 80 | 1280 | 80 | 640 | 320 | 5120 | 640 | 5120 |
| 04 | DENV-1 | 40 | 320 | 40 | 320 | 1280 | 20,480 | 320 | 2560 |
| 05 | DENV-1 | <20 | 80 | 20 | 80 | 320 | 2560 | 160 | 2560 |
| 06 | DENV-1 | 20 | 80 | 320 | 80 | 1280 | 2560 | 20,480 | 640 |
| 07 | DENV-1 | 320 | 160 | 5120 | 320 | 320 | 320 | 5120 | 320 |
| 08 | DENV-1 | <20 | <20 | <20 | <20 | 20 | <20 | <20 | 40 |
| 09 | DENV-1 | <20 | <20 | <20 | <20 | 20 | <20 | <20 | <20 |
| 10 | DENV-1 | 20 | 20 | 20 | 20 | 80 | 640 | 80 | 640 |
| 11 | DENV-2 | 5120 | 320 | 80 | 320 | 20,480 | 2560 | 1280 | 5120 |
| 12 | DENV-2 | 160 | 40 | 20 | 40 | 20,480 | 2560 | 1280 | 2560 |
| 13 | DENV-2 | 2560 | 640 | 80 | 320 | 10,240 | 5120 | 640 | 1280 |
| 14 | DENV-2 | 1280 | 640 | 160 | 640 | 2560 | 1280 | 160 | 1280 |
| 15 | DENV-2 | 40 | 2560 | 640 | 1280 | 80 | 2560 | 640 | 1280 |
| 16 | DENV-2 | 160 | 1280 | 2560 | 1280 | 320 | 2560 | 5120 | 2560 |
| 17 | DENV-2 | 160 | 1280 | 2560 | 1280 | 640 | 2560 | 20,480 | 2560 |
| 18 | DENV-2 | 160 | 640 | 1280 | 1280 | 640 | 2560 | 10,240 | 2560 |
| 19 | DENV-2 | <20 | 160 | <20 | 1280 | 640 | 160 | 40 | 320 |
| 20 | DENV-2 | 40 | 1280 | 20 | 2560 | 80 | 2560 | 40 | 5120 |
| 21 | DENV-3 | 5120 | 640 | 1280 | 320 | 5120 | 320 | 1280 | 320 |
| 22 | DENV-3 | 5120 | 1280 | 320 | 640 | 5120 | 2560 | 320 | 2560 |
| 23 | DENV-3 | 2560 | 640 | 320 | 320 | 10,240 | 2560 | 5120 | 5120 |
| 24 | DENV-3 | 5120 | 2560 | 80 | 1280 | 5120 | 10,240 | 640 | 5120 |
| 25 | DENV-3 | 160 | 2560 | 20 | 1280 | 1280 | 5120 | 40 | 2560 |
| 26 | DENV-3 | 320 | 5120 | 160 | 2560 | 5120 | 20,480 | 1280 | 20,480 |
| 27 | DENV-3 | 160 | 5120 | 160 | 2560 | 160 | 5120 | 320 | 5120 |
| 28 | DENV-3 | <20 | 320 | <20 | 40 | 40 | 160 | <20 | 40 |
| 29 | DENV-3 | 640 | 5120 | 40 | 5120 | 640 | 5120 | 80 | 5120 |
| 30 | DENV-3 | 320 | 10,240 | 320 | 5120 | 1280 | 5120 | 320 | 5120 |
| 31 | DENV-4 | 1280 | 80 | 20 | 40 | 10,240 | 640 | 640 | 1280 |
| 32 | DENV-4 | 320 | 40 | <20 | 40 | 10,240 | 10,240 | 2560 | 1280 |
| 33 | DENV-4 | 5120 | 160 | 160 | 80 | 20,480 | 1280 | 1280 | 640 |
| 34 | DENV-4 | 640 | 20 | 20 | 40 | 10,240 | 2560 | 1280 | 1280 |
| 35 | DENV-4 | 10,240 | 1280 | 640 | 1280 | 20,480 | 2560 | 1280 | 5120 |
| 36 | DENV-4 | 40 | 1280 | 160 | 640 | 320 | 5120 | 2560 | 2560 |
| 37 | DENV-4 | 640 | 20,480 | 320 | 5120 | 640 | 20,480 | 640 | 10,240 |
| 38 | DENV-4 | 640 | 320 | 1280 | 80 | 20,480 | 2560 | 20,480 | 1280 |
| 39 | DENV-4 | 160 | 160 | 5120 | 80 | 1280 | 640 | 20,480 | 640 |
| 40 | DENV-4 | 640 | 1280 | 5120 | 2560 | 640 | 1280 | 20,480 | 2560 |
| Serotype-Specific Neutralization | Number of Sera with Serotype by RT-PCR | |||
|---|---|---|---|---|
| DENV-1 | DENV-2 | DENV-3 | DENV-4 | |
| DENV-1 | 4 | 20 | 7 | 11 |
| DENV-2 | 10 | 3 | 9 | 7 |
| DENV-3 | 7 | 7 | - | 5 |
| DENV-4 | 3 | 4 | - | - |
| DENV-1 + DENV-2 | 1 | - | 1 | 1 |
| DENV-1 + DENV-3 | 2 | - | - | 1 |
| DENV-2 + DENV-3 | 1 | - | - | 1 |
| DENV-2 + DENV-4 | 6 | - | 4 | - |
| DENV-3 + DENV-4 | - | 1 | - | - |
| >2 serotypes | 1 | - | - | - |
| negative | 2 | - | - | |
| Total number of sera | 37 | 35 | 21 | 26 |
| Serum | FRNT90 * | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| REF | IPC A | IPC B | ||||||||||
| DENV | DENV | DENV | ||||||||||
| 1 | 2 | 3 | 4 | 1 | 2 | 3 | 4 | 1 | 2 | 3 | 4 | |
| 24 | 640 | 2560 | <20 | 160 | 80 | 1280 | 640 | <20 | 40 | 1280 | 640 | 20 |
| 14 | 80 | 2560 | 40 | 160 | 20 | 2560 | 320 | <20 | <20 | 2560 | 640 | <20 |
| 17 | 80 | 5120 | <20 | 320 | <20 | 5120 | 1280 | 20 | <20 | 5120 | 1280 | <20 |
| 25 | 2560 | 10,240 | 80 | 320 | 80 | 5120 | 2560 | <20 | 80 | 10,240 | 2560 | 20 |
| 22 | 160 | 5120 | 20 | 160 | <20 | 10,240 | 320 | 20 | <20 | 5120 | 640 | 40 |
| 16 | 80 | 640 | 2560 | 320 | 80 | 640 | 2560 | <20 | 40 | 80 | 5120 | <20 |
| 23 | 640 | 640 | 2560 | 640 | 40 | 640 | 5120 | <20 | 20 | 80 | 5120 | 40 |
| 30 | 640 | 320 | 10,240 | 160 | 40 | 320 | 10,240 | 20 | 20 | 40 | 10,240 | 40 |
| 15 | 1280 | 320 | 10,240 | 160 | 40 | 320 | 20,480 | 20 | 40 | 320 | 20,480 | 160 |
| 13 | <20 | 80 | <20 | 40 | <20 | 160 | 80 | <20 | <20 | 80 | 80 | 20 |
| 03 | 20 | 320 | <20 | 160 | <20 | 320 | 80 | <20 | <20 | 80 | 80 | <20 |
| 31 | 320 | 1280 | <20 | 320 | <20 | 1280 | 160 | 20 | <20 | 640 | 640 | 20 |
| 33 | 640 | 2560 | <20 | 320 | 20 | 2560 | 320 | <20 | 20 | 1280 | 1280 | 20 |
| 01 | <20 | 80 | <20 | 40 | <20 | 80 | 40 | <20 | <20 | <20 | 80 | 20 |
| 29 | 320 | 2560 | <20 | 160 | 20 | 2560 | 640 | <20 | <20 | 320 | 1280 | 20 |
| 21 | 160 | 640 | 160 | 320 | 40 | 640 | 1280 | <20 | 20 | 160 | 1280 | 20 |
| 04 | 320 | 640 | <20 | 80 | 40 | 640 | 640 | <20 | 40 | 160 | 640 | <20 |
| 08 | <20 | 160 | <20 | 160 | <20 | 320 | 160 | <20 | <20 | 80 | 160 | <20 |
| 20 | 20 | 80 | <20 | 1280 | <20 | 80 | 320 | 20 | <20 | 40 | 320 | 160 |
| 19 | 40 | 320 | <20 | 1280 | <20 | 320 | 320 | 40 | <20 | 40 | 320 | 160 |
| 09 | <20 | <20 | <20 | 20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 |
| 28 | 160 | 160 | <20 | 2560 | <20 | 160 | 160 | 80 | <20 | 40 | 320 | 640 |
| 12 | 20 | 80 | <20 | 640 | <20 | 80 | 160 | 20 | <20 | 40 | 160 | 160 |
| 05 | 2560 | 640 | 40 | 80 | 640 | 640 | 640 | <20 | 640 | 320 | 1280 | 20 |
| 26 | 640 | 640 | <20 | 80 | 40 | 640 | 320 | <20 | 40 | 320 | 640 | 20 |
| 18 | 640 | 640 | 640 | 320 | 20 | 640 | 5120 | <20 | 20 | 80 | 1240 | 80 |
| 07 | 2560 | 640 | 20 | 80 | 160 | 320 | 1280 | <20 | 320 | 320 | 1280 | 20 |
| 11 | 640 | 160 | <20 | 80 | 20 | 160 | 320 | <20 | 20 | 80 | 320 | <20 |
| 32 | 2560 | 80 | 20 | 160 | 80 | 80 | 320 | <20 | 40 | 20 | 1280 | 40 |
| 06 | 640 | 160 | 20 | 80 | 160 | 160 | 640 | <20 | 160 | 40 | 1280 | 20 |
| 10 | 1280 | 80 | 20 | 80 | 20 | 40 | 1280 | <20 | 20 | 80 | 1280 | <20 |
| 02 | 640 | 320 | 40 | 640 | 40 | 320 | 1280 | 20 | 40 | 160 | 2560 | 40 |
| 27 | 640 | 320 | 320 | 640 | 40 | 160 | 2560 | 20 | 20 | 80 | 5120 | 20 |
| Serotype-Specific Neutralization | Number of Sera | ||
|---|---|---|---|
| REF | IPC A | IPC B | |
| DENV-1 | 6 | - | - |
| DENV-2 | 13 | 13 | 5 |
| DENV-3 | 4 | 15 | 21 |
| DENV-4 | 5 | - | 1 |
| DENV-1 + 2 | 1 | - | - |
| DENV-1 + 4 | 2 | - | - |
| DENV-2 + 3 | - | 3 | 4 |
| DENV-2 + 4 | 1 | - | - |
| DENV-3 + 4 | - | - | 1 |
| >2 serotypes | 1 | 1 | - |
| negative | - | 1 | 1 |
| Total number of sera | 33 | 33 | 33 |
| Colombian | Serotype | FRNT90 | ED3 Dot Assay | ED3 ELISA | Days after | ||
|---|---|---|---|---|---|---|---|
| 2nd Sera | RT-PCR | REF | REF | COL | REF | COL | Fever |
| P1 | 1 | 24 | 4 | 4 | 4 | - | 16 d |
| P2 | 1 | 2 | 2 | 2 | - | - | 32 d |
| P3 | 2 | 13 | 13 | 13 | - | 13 | 24 d |
| P4 | 2 | 123 | 123 | 123 | 1 | 123 | 25 d |
| P5 | 2 | 1 | 1 | 1 | 1 | 1 | 21 d |
| P6 | 4 | 12 | 123 | 12 | - | 123 | 25 d |
| P7 | 4 | 23 | - | 2 | 123 | 123 | 37 d |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Auerswald, H.; Kann, S.; Klepsch, L.; Hülsemann, J.; Rudnik, I.; Schreiber, S.; Buchy, P.; Schreiber, M. Neutralization of Dengue Virus Serotypes by Sera from Dengue-Infected Individuals Is Preferentially Directed to Heterologous Serotypes and Not against the Autologous Serotype Present in Acute Infection. Viruses 2021, 13, 1957. https://doi.org/10.3390/v13101957
Auerswald H, Kann S, Klepsch L, Hülsemann J, Rudnik I, Schreiber S, Buchy P, Schreiber M. Neutralization of Dengue Virus Serotypes by Sera from Dengue-Infected Individuals Is Preferentially Directed to Heterologous Serotypes and Not against the Autologous Serotype Present in Acute Infection. Viruses. 2021; 13(10):1957. https://doi.org/10.3390/v13101957
Chicago/Turabian StyleAuerswald, Heidi, Simone Kann, Leonard Klepsch, Janne Hülsemann, Ines Rudnik, Sebastian Schreiber, Philippe Buchy, and Michael Schreiber. 2021. "Neutralization of Dengue Virus Serotypes by Sera from Dengue-Infected Individuals Is Preferentially Directed to Heterologous Serotypes and Not against the Autologous Serotype Present in Acute Infection" Viruses 13, no. 10: 1957. https://doi.org/10.3390/v13101957
APA StyleAuerswald, H., Kann, S., Klepsch, L., Hülsemann, J., Rudnik, I., Schreiber, S., Buchy, P., & Schreiber, M. (2021). Neutralization of Dengue Virus Serotypes by Sera from Dengue-Infected Individuals Is Preferentially Directed to Heterologous Serotypes and Not against the Autologous Serotype Present in Acute Infection. Viruses, 13(10), 1957. https://doi.org/10.3390/v13101957






