Pan-Enterovirus Amplicon-Based High-Throughput Sequencing Detects the Complete Capsid of a EVA71 Genotype C1 Variant via Wastewater-Based Epidemiology in Arizona
Abstract
1. Introduction
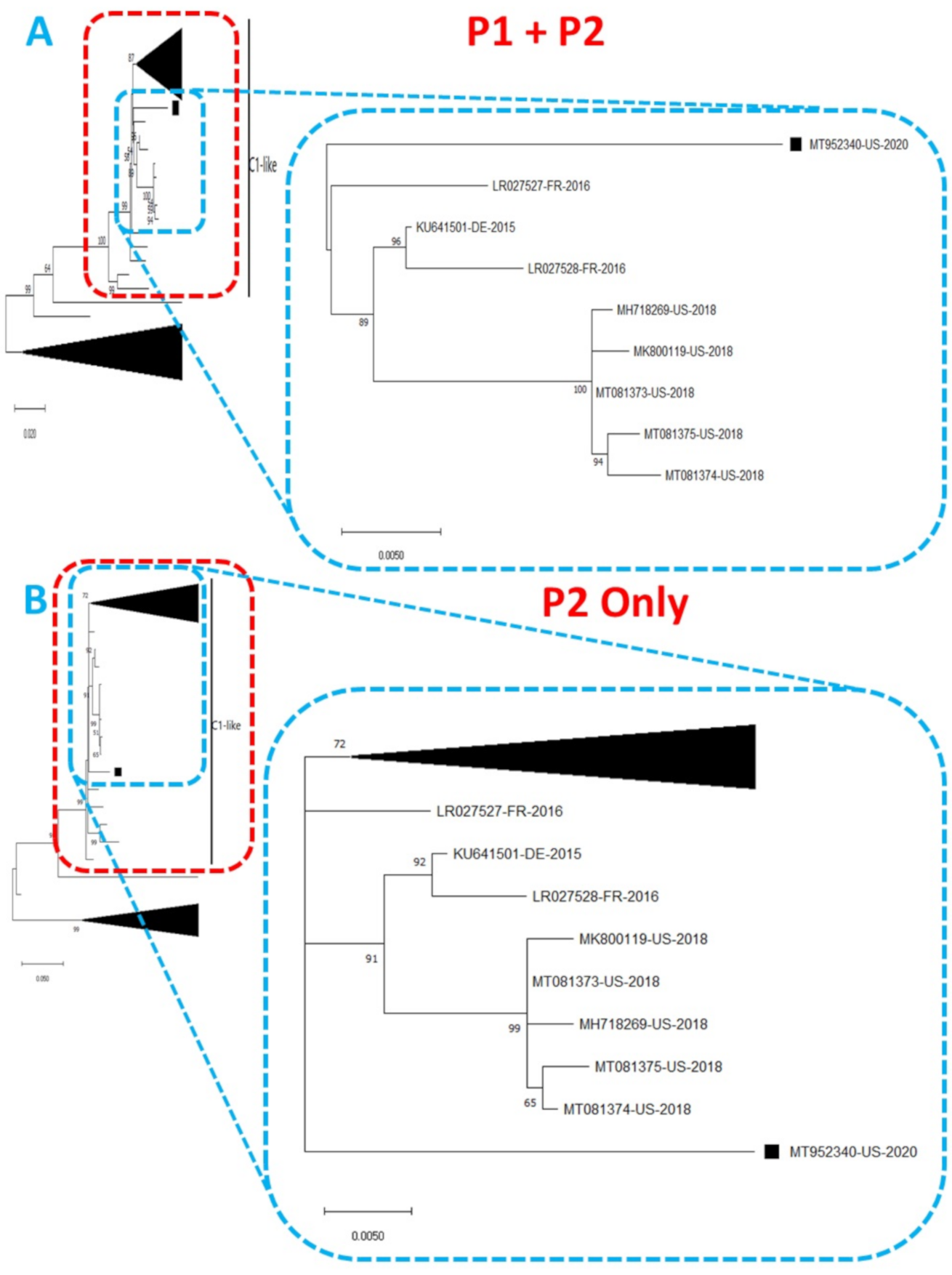
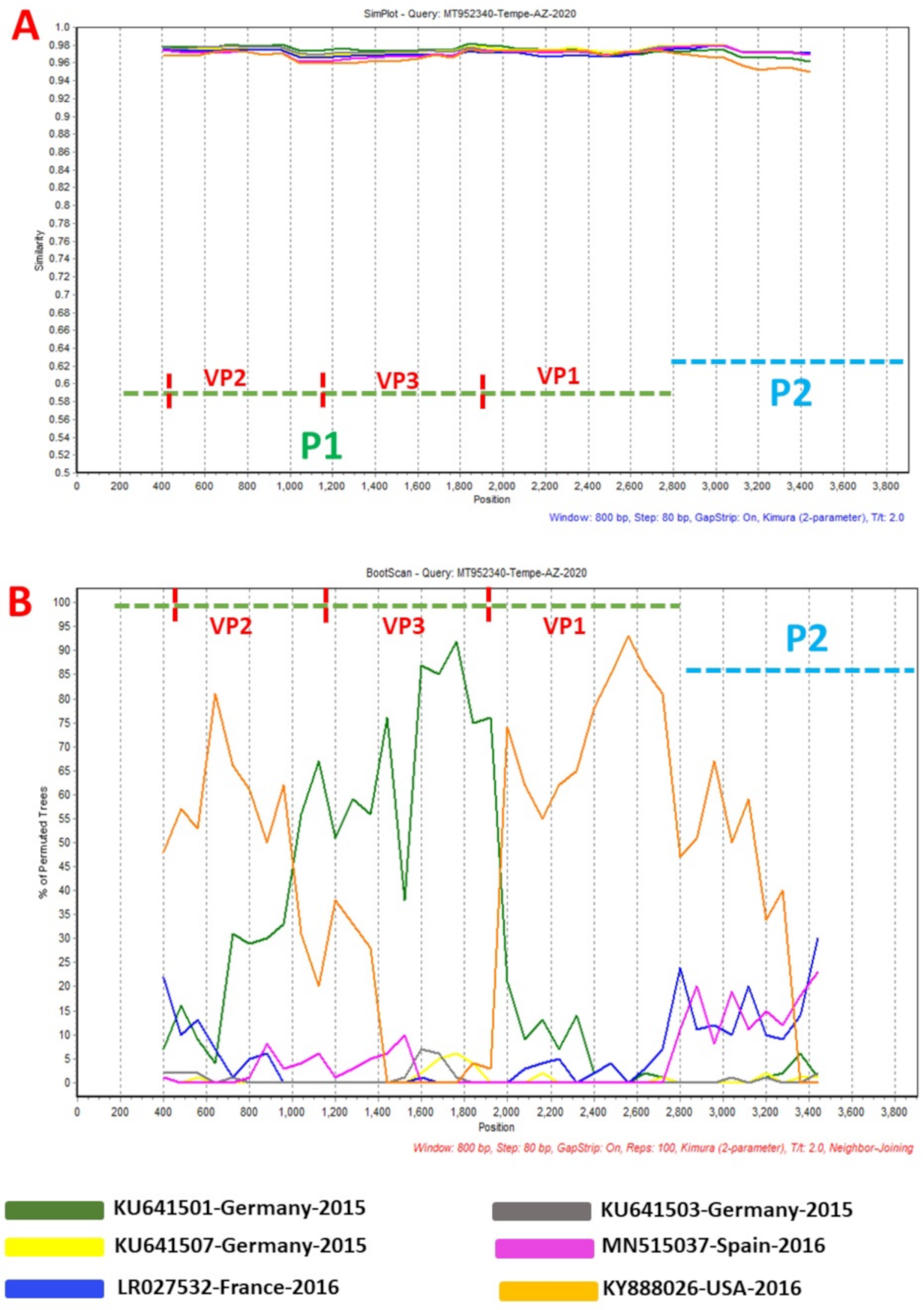
2. Results
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AFM | acute flaccid myelitis |
| ASU | Arizona State University |
| COVID-19 | Coronavirus Disease 2019 |
| EGT | enterovirus genotyping tool |
| EV | enteroviruses |
| EVA71 | enterovirus A71 |
| HFMD | hand, foot, and mouth disease |
| HHO | Human Health Observatory |
| HTS | high-throughput sequencing |
| ML | maximum-likelihood |
| MSA | multiple sequence alignment |
| ORF | open reading frame |
| PEG | polyethylene glycol |
| ppORF | polyprotein open reading frame |
| RT-PCR | reverse transcription polymerase chain reaction |
| SARS-CoV-2 | severe acute respiratory syndrome coronavirus 2 |
| uORF | upstream open reading frame |
| USA | United States of America |
| WBE | wastewater-based epidemiology |
References
- Lulla, V.; Dinan, A.M.; Hosmillo, M.; Chaudhry, Y.; Sherry, L.; Irigoyen, N.; Nayak, K.M.; Stonehouse, N.J.; Zilbauer, M.; Goodfellow, I.; et al. An upstream protein-coding region in enteroviruses modulates virus infection in gut epithelial cells. Nat. Microbiol. 2019, 4, 280–292. [Google Scholar] [CrossRef] [PubMed]
- Bessaud, M.; Razafindratsimandresy, R.; Nougairede, A.; Joffret, M.L.; Deshpande, J.M.; Dubot-Peres, A.; Heraud, J.M.; de Lamballerie, X.; Delpeyroux, F.; Bailly, J.L. Molecular comparison and evolutionary analyses of VP1 nucleotide sequences of new African human enterovirus 71 isolates reveal a wide genetic diversity. PLoS ONE 2014, 9, e90624. [Google Scholar] [CrossRef] [PubMed]
- Uprety, P.; Graf, E.H. Enterovirus infection and acute flaccid myelitis. Curr. Opin. Virol. 2020, 40, 55–60. [Google Scholar] [CrossRef] [PubMed]
- CDC. AFM Cases and Outbreaks. 2021. Available online: https://www.cdc.gov/acute-flaccid-myelitis/cases-in-us.html (accessed on 5 January 2021).
- Brown, B.A.; Oberste, M.S.; Alexander, J.P.J.; Kennett, M.L.; Pallansch, M.A. Molecular epidemiology and evolution of enterovirus 71 strains isolated from 1970 to 1998. J. Virol. 1999, 73, 9969–9975. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Sayers, E.W.; Cavanaugh, M.; Clark, K.; Ostell, J.; Pruitt, K.D.; Karsch-Mizrachi, I. GenBank. Nucleic Acids Res. 2020, 48, D84–D86. [Google Scholar] [CrossRef]
- Antona, D.; Kossorotoff, M.; Schuffenecker, I.; Mirand, A.; Leruez-Ville, M.; Bassi, C.; Aubart, M.; Moulin, F.; Levy-Bruhl, D.; Henquell, C.; et al. Severe paediatric conditions linked with EV-A71 and EV-D68, France, May to October 2016. Eurosurveillance 2016, 21. [Google Scholar] [CrossRef]
- Böttcher, S.; Obermeier, P.E.; Neubauer, K.; Diedrich, S. Laboratory Network for Enterovirus Diagnostics. Recombinant Enterovirus A71 Subgenogroup C1 Strains, Germany, 2015. Emerg. Infect. Dis. 2016, 22, 1843–1846. [Google Scholar] [CrossRef]
- Ngangas, S.T.; Lukashev, A.; Jugie, G.; Ivanova, O.; Mansuy, J.M.; Mengelle, C.; Izopet, J.; L’Honneur, A.S.; Rozenberg, F.; Leyssene, D.; et al. Multirecombinant Enterovirus A71 Subgenogroup C1 Isolates Associated with Neurologic Disease, France, 2016–2017. Emerg. Infect. Dis. 2019, 25, 1204–1208. [Google Scholar] [CrossRef]
- Lole, K.S.; Bollinger, R.C.; Paranjape, R.S.; Gadkari, D.; Kulkarni, S.S.; Novak, N.G.; Ingersoll, R.; Sheppard, H.W.; Ray, S.C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999, 73, 152–160. [Google Scholar] [CrossRef]
- Snider, C.J.; Diop, O.M.; Burns, C.C.; Tangermann, R.H.; Wassilak, S.G. Surveillance Systems to Track Progress Toward Polio Eradication–Worldwide, 2014–2015. MMWR Morb. Mortal. Wkly. Rep. 2016, 65, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Jorba, J.; Campagnoli, R.; De, L.; Kew, O. Calibration of multiple poliovirus molecular clocks covering an extended evolutionary range. J. Virol. 2008, 82, 4429–4440. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Garcia, M.D.; Kebe, O.; Fall, A.D.; Dia, H.; Diop, O.M.; Delpeyroux, F.; Ndiaye, K. Enterovirus A71 Genogroups C and E in Children with Acute Flaccid Paralysis, West Africa. Emerg. Infect. Dis. 2016, 22, 753–755. [Google Scholar] [CrossRef] [PubMed]
- Caine, E.A.; Moncla, L.H.; Ronderos, M.D.; Friedrich, T.C.; Osorio, J.E. A Single Mutation in the VP1 of Enterovirus 71 Is Responsible for Increased Virulence and Neurotropism in Adult Interferon-Deficient Mice. J. Virol. 2016, 90, 8592–8604. [Google Scholar] [CrossRef]
- Mandary, M.B.; Masomian, M.; Ong, S.K.; Poh, C.L. Characterization of Plaque Variants and the Involvement of Quasi-Species in a Population of EV-A71. Viruses 2020, 12, 651. [Google Scholar] [CrossRef]
- Lukashev, A.N.; Lashkevich, V.A.; Ivanova, O.E.; Koroleva, G.A.; Hinkkanen, A.E.; Ilonen, J. Recombination in circulating Human enterovirus B: Independent evolution of structural and non-structural genome regions. J. Gen. Virol. 2005, 86, 3281–3290. [Google Scholar] [CrossRef]
- Huang, S.W.; Hsu, Y.W.; Smith, D.J.; Kiang, D.; Tsai, H.P.; Lin, K.H.; Wang, S.M.; Liu, C.C.; Su, I.J.; Wang, J.R. Reemergence of enterovirus 71 in 2008 in taiwan: dynamics of genetic and antigenic evolution from 1998 to 2008. J. Clin. Microbiol. 2009, 47, 3653–3662. [Google Scholar] [CrossRef]
- Bessaud, M.; Joffret, M.L.; Holmblat, B.; Razafindratsimandresy, R.; Delpeyroux, F. Genetic relationship between cocirculating Human enteroviruses species C. PLoS ONE 2011, 6, e24823. [Google Scholar] [CrossRef]
- Nathanson, N.; Kew, O.M. From emergence to eradication: The epidemiology of poliomyelitis deconstructed. Am. J. Epidemiol. 2010, 172, 1213–1229. [Google Scholar] [CrossRef]
- Lange, S.J.; Ritchey, M.D.; Goodman, A.B.; Dias, T.; Twentyman, E.; Fuld, J.; Schieve, L.A.; Imperatore, G.; Benoit, S.R.; Kite-Powell, A.; et al. Potential Indirect Effects of the COVID-19 Pandemic on Use of Emergency Departments for Acute Life-Threatening Conditions—United States, January-May 2020. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 795–800. [Google Scholar] [CrossRef]
- Venkatesan, A.K.; Done, H.Y.; Halden, R.U. United States National Sewage Sludge Repository at Arizona State University—A new resource and research tool for environmental scientists, engineers, and epidemiologists. Environ. Sci. Pollut. Res. Int. 2015, 22, 1577–1586. [Google Scholar] [CrossRef] [PubMed]
- Halden, R.U.; Terlinden, E.; Kraberger, S.; Scotch, M.; Steele, J.; Varsani, A. Tracking harmful chemicals and pathogens using the Human Health Observatory at ASU. Online J. Public Health Inform. 2019, 11, e369. [Google Scholar] [CrossRef]
- Majumdar, M.; Martin, J. Detection by Direct Next Generation Sequencing Analysis of Emerging Enterovirus D68 and C109 Strains in an Environmental Sample From Scotland. Front. Microbiol. 2018, 9, 1956. [Google Scholar] [CrossRef]
- Nix, W.A.; Oberste, M.S.; Pallansch, M.A. Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. J. Clin. Microbiol. 2006, 44, 2698–2704. [Google Scholar] [CrossRef] [PubMed]
- Arkin, A.P.; Cottingham, R.W.; Henry, C.S.; Harris, N.L.; Stevens, R.L.; Maslov, S.; Dehal, P.; Ware, D.; Perez, F.; Canon, S.; et al. KBase: The United States Department of Energy Systems Biology Knowledgebase. Nat. Biotechnol. 2018, 36, 566–569. [Google Scholar] [CrossRef]
- Kroneman, A.; Vennema, H.; Deforche, K.V.D.; Avoort, H.V.D.; Penaranda, S.; Oberste, M.S.; Vinje, J.; Koopmans, M. An automated genotyping tool for enteroviruses and noroviruses. J. Clin. Virol. 2011, 51, 121–125. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Faleye, T.O.C.; Driver, E.; Bowes, D.; Adhikari, S.; Adams, D.; Varsani, A.; Halden, R.U.; Scotch, M. Pan-Enterovirus Amplicon-Based High-Throughput Sequencing Detects the Complete Capsid of a EVA71 Genotype C1 Variant via Wastewater-Based Epidemiology in Arizona. Viruses 2021, 13, 74. https://doi.org/10.3390/v13010074
Faleye TOC, Driver E, Bowes D, Adhikari S, Adams D, Varsani A, Halden RU, Scotch M. Pan-Enterovirus Amplicon-Based High-Throughput Sequencing Detects the Complete Capsid of a EVA71 Genotype C1 Variant via Wastewater-Based Epidemiology in Arizona. Viruses. 2021; 13(1):74. https://doi.org/10.3390/v13010074
Chicago/Turabian StyleFaleye, Temitope O. C., Erin Driver, Devin Bowes, Sangeet Adhikari, Deborah Adams, Arvind Varsani, Rolf U. Halden, and Matthew Scotch. 2021. "Pan-Enterovirus Amplicon-Based High-Throughput Sequencing Detects the Complete Capsid of a EVA71 Genotype C1 Variant via Wastewater-Based Epidemiology in Arizona" Viruses 13, no. 1: 74. https://doi.org/10.3390/v13010074
APA StyleFaleye, T. O. C., Driver, E., Bowes, D., Adhikari, S., Adams, D., Varsani, A., Halden, R. U., & Scotch, M. (2021). Pan-Enterovirus Amplicon-Based High-Throughput Sequencing Detects the Complete Capsid of a EVA71 Genotype C1 Variant via Wastewater-Based Epidemiology in Arizona. Viruses, 13(1), 74. https://doi.org/10.3390/v13010074










