Detection of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by Mass Spectrometry
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Samples and Workflow
2.2. Ribonucleic Acid Extraction
2.3. Real-Time Reverse Transcriptase Polymerase Chain Reaction
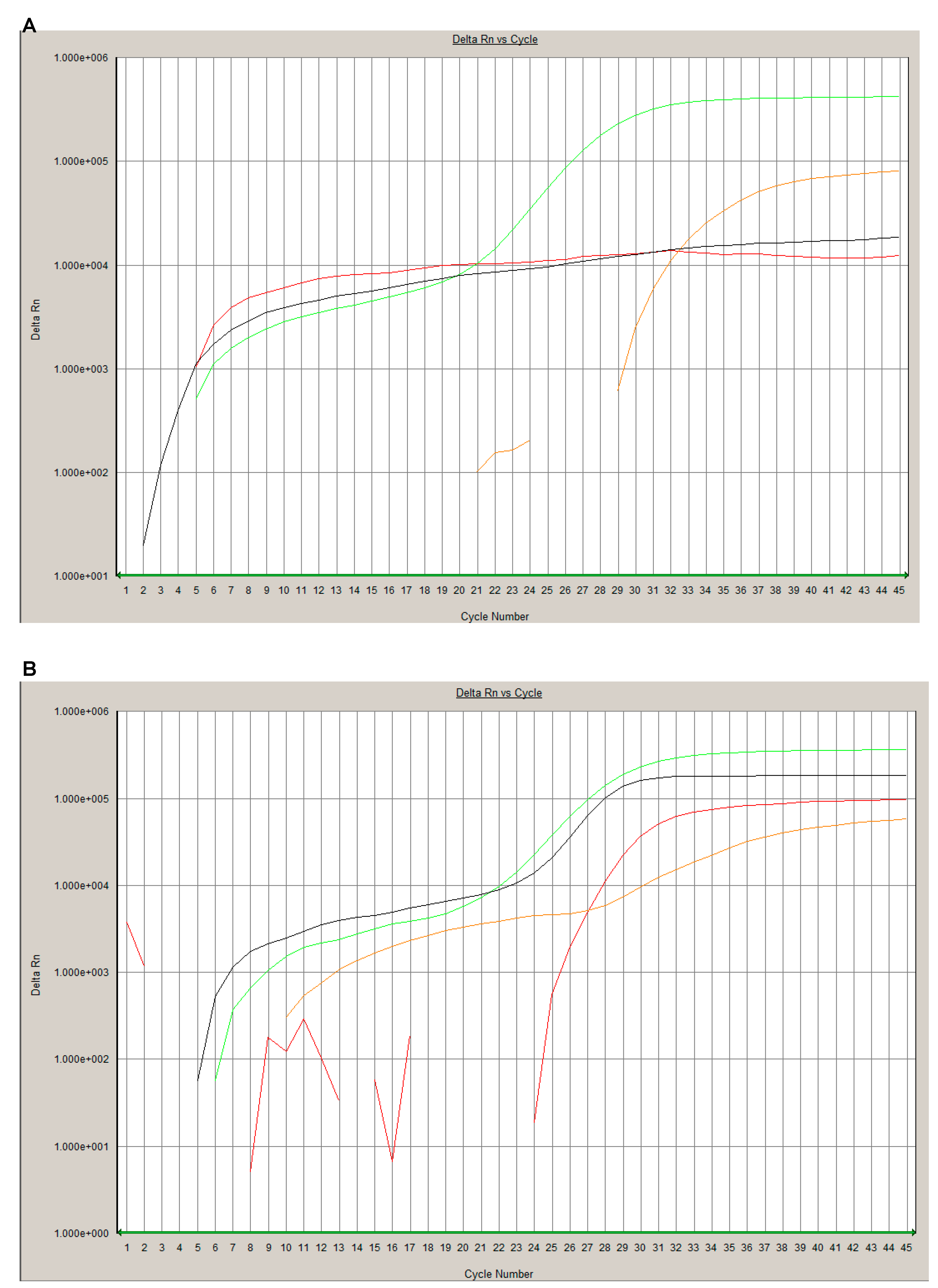
2.4. SARS-CoV-2 Mass Spectrometrical Assay
2.5. Descriptive Statistics
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Ou, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Studdert, D.M.; Hall, M.A. Disease Control, Civil Liberties, and Mass Testing—Calibrating Restrictions during the Covid-19 Pandemic. N. Engl. J. Med. 2020, 383, 102–104. [Google Scholar] [CrossRef]
- Nussbaumer-Streit, B.; Mayr, V.; Dobrescu, A.I.; Chapman, A.; Persad, E.; Klerings, I.; Wagner, G.; Siebert, U.; Christof, C.; Zachariah, C.; et al. Quarantine alone or in combination with other public health measures to control COVID-19: A rapid review. Cochrane Database Syst. Rev. 2020, 4, CD013574. [Google Scholar] [CrossRef]
- Long, C.; Xu, H.; Shen, Q.; Zhang, X.; Fan, B.; Wang, C.; Zeng, B.; Li, Z.; Li, X.; Li, H. Diagnosis of the Coronavirus disease (COVID-19): rRT-PCR or CT? Eur. J. Radiol. 2020, 126, 108961. [Google Scholar] [CrossRef]
- Zhu, J.; Zhong, Z.; Ji, P.; Li, H.; Li, B.; Pang, J.; Zhang, J.; Zhao, C. Clinicopathological characteristics of 8697 patients with COVID-19 in China: A meta-analysis. Fam. Med. Community Health 2020, 8, e000406. [Google Scholar] [CrossRef]
- Tan, C.; Xiao, Y.; Meng, X.; Huang, X.; Li, C.; Wu, A. Asymptomatic SARS-CoV-2 infections: What we need to know? Infect. Control Hosp. Epidemiol. 2020, 1–2. [Google Scholar] [CrossRef]
- Seger, C.; Salzmann, L. After another decade: LC-MS/MS became routine in clinical diagnostics. Clin. Biochem. 2020, 82, 2–11. [Google Scholar] [CrossRef]
- Borden, S.A.; Palaty, J.; Termopoli, V.; Famiglini, G.; Cappiello, A.; Gill, C.G.; Palma, P. Mass Spectrometry Analysis of Drugs of Abuse: Challenges and Emerging Strategies. Mass Spectrom. Rev. 2020, 37, 258–280. [Google Scholar] [CrossRef]
- Fung, A.W.S.; Sugumar, V.; Ren, A.H.; Kulasingam, V. Emerging role of clinical mass spectrometry in pathology. J. Clin. Pathol. 2020, 73, 61–69. [Google Scholar] [CrossRef]
- Angeletti, S.; Ciccozzi, M. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry in clinical microbiology: An updating review. Infect. Genet. Evol. 2019, 76, 104063. [Google Scholar] [CrossRef]
- De Roock, W.; Claes, B.; Bernasconi, D.; De Schutter, J.; Biesmans, B.; Fountzilas, G.; Kalogeras, K.T.; Kotoula, V.; Papamichael, D.; Laurent-Puig, P.; et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: A retrospective consortium analysis. Lancet Oncol. 2010, 11, 753–762. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brunink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 2020, 25, 2000045. [Google Scholar] [CrossRef] [PubMed]
- Bioscience, A. Instructions for Use: SARS-CoV-2 Panel. Available online: https://agenabio.com/wp-content/uploads/2020/06/SC2_Panel_IFU-CUS-001_R02.pdf (accessed on 27 July 2020).
- Cui, J.; Li, F.; Shi, Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Wu, J.; Liu, J.; Li, S.; Peng, Z.; Xiao, Z.; Wang, X.; Yan, R.; Luo, J. Detection and analysis of nucleic acid in various biological samples of COVID-19 patients. Travel Med. Infect. Dis. 2020, 101673. [Google Scholar] [CrossRef]
- Cheng, P.K.; Wong, D.A.; Tong, L.K.; Ip, S.M.; Lo, A.C.; Lau, C.S.; Yeung, E.Y.; Lim, W.W. Viral shedding patterns of coronavirus in patients with probable severe acute respiratory syndrome. Lancet 2004, 363, 1699–1700. [Google Scholar] [CrossRef]
- Yan, Y.; Chang, L.; Wang, L. Laboratory testing of SARS-CoV, MERS-CoV, and SARS-CoV-2 (2019-nCoV): Current status, challenges, and countermeasures. Rev. Med. Virol. 2020, 30, e2106. [Google Scholar] [CrossRef]
- Li, Y.; Yao, L.; Li, J.; Chen, L.; Song, Y.; Cai, Z.; Yang, C. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J. Med. Virol. 2020. [Google Scholar] [CrossRef]
- Tahamtan, A.; Ardebili, A. Real-time RT-PCR in COVID-19 detection: Issues affecting the results. Expert Rev. Mol. Diagn. 2020, 20, 453–454. [Google Scholar] [CrossRef]
- Xiao, A.T.; Tong, Y.X.; Zhang, S. False-negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: Rather than recurrence. J. Med. Virol. 2020. [Google Scholar] [CrossRef]
- West, C.P.; Montori, V.M.; Sampathkumar, P. COVID-19 Testing: The Threat of False-Negative Results. Mayo Clin. Proc. 2020, 95, 1127–1129. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Long, L.; Zhang, D.; Yan, T.; Cui, S.; Yang, P.; Wang, Q.; Ren, S. Potential false-negative nucleic acid testing results for Severe Acute Respiratory Syndrome Coronavirus 2 from thermal inactivation of samples with low viral loads. Clin. Chem. 2020, 66, 794–801. [Google Scholar] [CrossRef] [PubMed]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef] [PubMed]
- Xiu, L.; Zhang, C.; Wu, Z.; Peng, J. Establishment and Application of a Universal Coronavirus Screening Method Using MALDI-TOF Mass Spectrometry. Front. Microbiol. 2017, 8, 1510. [Google Scholar] [CrossRef]
- Siuzdak, G. Probing viruses with mass spectrometry. J. Mass Spectrom. 1998, 33, 203–211. [Google Scholar] [CrossRef]
- Kriegsmann, M.; Wandernoth, P.; Lisenko, K.; Casadonte, R.; Longuespee, R.; Arens, N.; Kriegsmann, J. Detection of HPV subtypes by mass spectrometry in FFPE tissue specimens: A reliable tool for routine diagnostics. J. Clin. Pathol. 2017, 70, 417–423. [Google Scholar] [CrossRef]
- Fan, H.Z.; Zhang, R.; Tian, T.; Zhong, Y.L.; Wu, M.P.; Xie, C.N.; Yang, J.J.; Huang, P.; Yu, R.B.; Zhang, Y.; et al. CYP24A1 genetic variants in the vitamin D metabolic pathway are involved in the outcomes of hepatitis C virus infection among high-risk Chinese population. Int. J. Infect. Dis. 2019, 84, 80–88. [Google Scholar] [CrossRef]
- Kriegsmann, M.; Arens, N.; Endris, V.; Weichert, W.; Kriegsmann, J. Detection of KRAS, NRAS and BRAF by mass spectrometry—A sensitive, reliable, fast and cost-effective technique. Diagn. Pathol. 2015, 10, 132. [Google Scholar] [CrossRef]
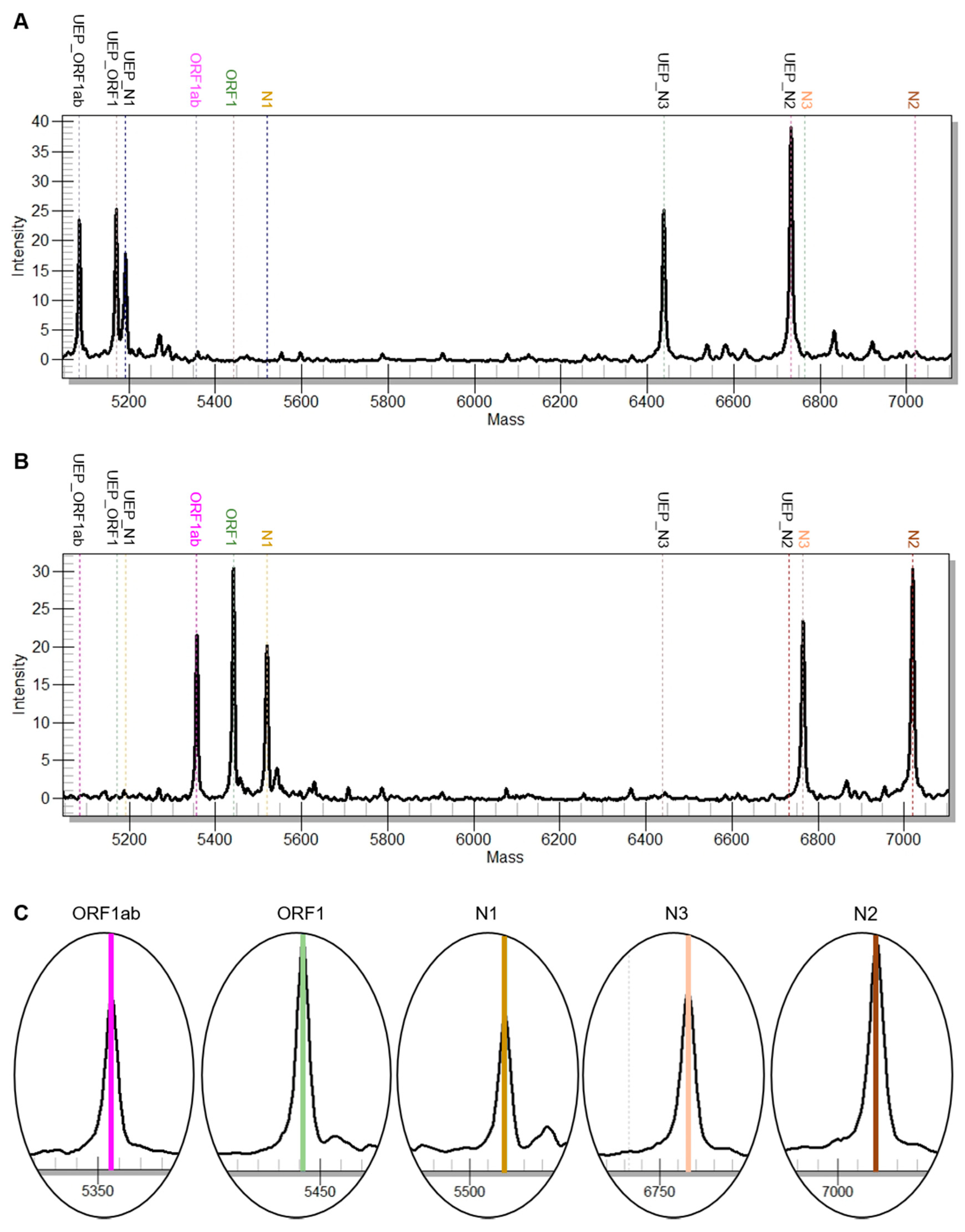
| MS Assay | ||
|---|---|---|
| Target | Genomic Region | Mass EA (Da) |
| SC2_N1 | N1 | 5519.50 |
| SC2_N2 | N2 | 7019.60 |
| SC2_N3 | N3 | 6765.40 |
| SC2_ORF1 | ORF1ab/nsp3 | 5441.60 |
| SC2_ORF1ab | ORF1ab/nsp10 | 5356.50 |
| Steps | rRT-PCR | MS Assay | ||
|---|---|---|---|---|
| Run Time (min) | Hands-on-Time (min) | Run Time (min) | Hands-on-Time (min) | |
| rRT-PCR | 75 | 30 | 150 | 15 |
| SAP | / | / | 50 | 5 |
| Extensions-PCR | / | / | 120 | 10 |
| CPM process | / | / | 90 | 5 |
| Data acquisition and analysis | 5 | 10 | 30 | 5 |
| Overall | 120 | 480 | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wandernoth, P.; Kriegsmann, K.; Groh-Mohanu, C.; Daeumer, M.; Gohl, P.; Harzer, O.; Kriegsmann, M.; Kriegsmann, J. Detection of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by Mass Spectrometry. Viruses 2020, 12, 849. https://doi.org/10.3390/v12080849
Wandernoth P, Kriegsmann K, Groh-Mohanu C, Daeumer M, Gohl P, Harzer O, Kriegsmann M, Kriegsmann J. Detection of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by Mass Spectrometry. Viruses. 2020; 12(8):849. https://doi.org/10.3390/v12080849
Chicago/Turabian StyleWandernoth, Petra, Katharina Kriegsmann, Cristina Groh-Mohanu, Martin Daeumer, Peter Gohl, Oliver Harzer, Mark Kriegsmann, and Joerg Kriegsmann. 2020. "Detection of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by Mass Spectrometry" Viruses 12, no. 8: 849. https://doi.org/10.3390/v12080849
APA StyleWandernoth, P., Kriegsmann, K., Groh-Mohanu, C., Daeumer, M., Gohl, P., Harzer, O., Kriegsmann, M., & Kriegsmann, J. (2020). Detection of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by Mass Spectrometry. Viruses, 12(8), 849. https://doi.org/10.3390/v12080849






