Assessment of Metagenomic Sequencing and qPCR for Detection of Influenza D Virus in Bovine Respiratory Tract Samples
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Sample Preparation
2.3. qPCR Confirmation and Quantification
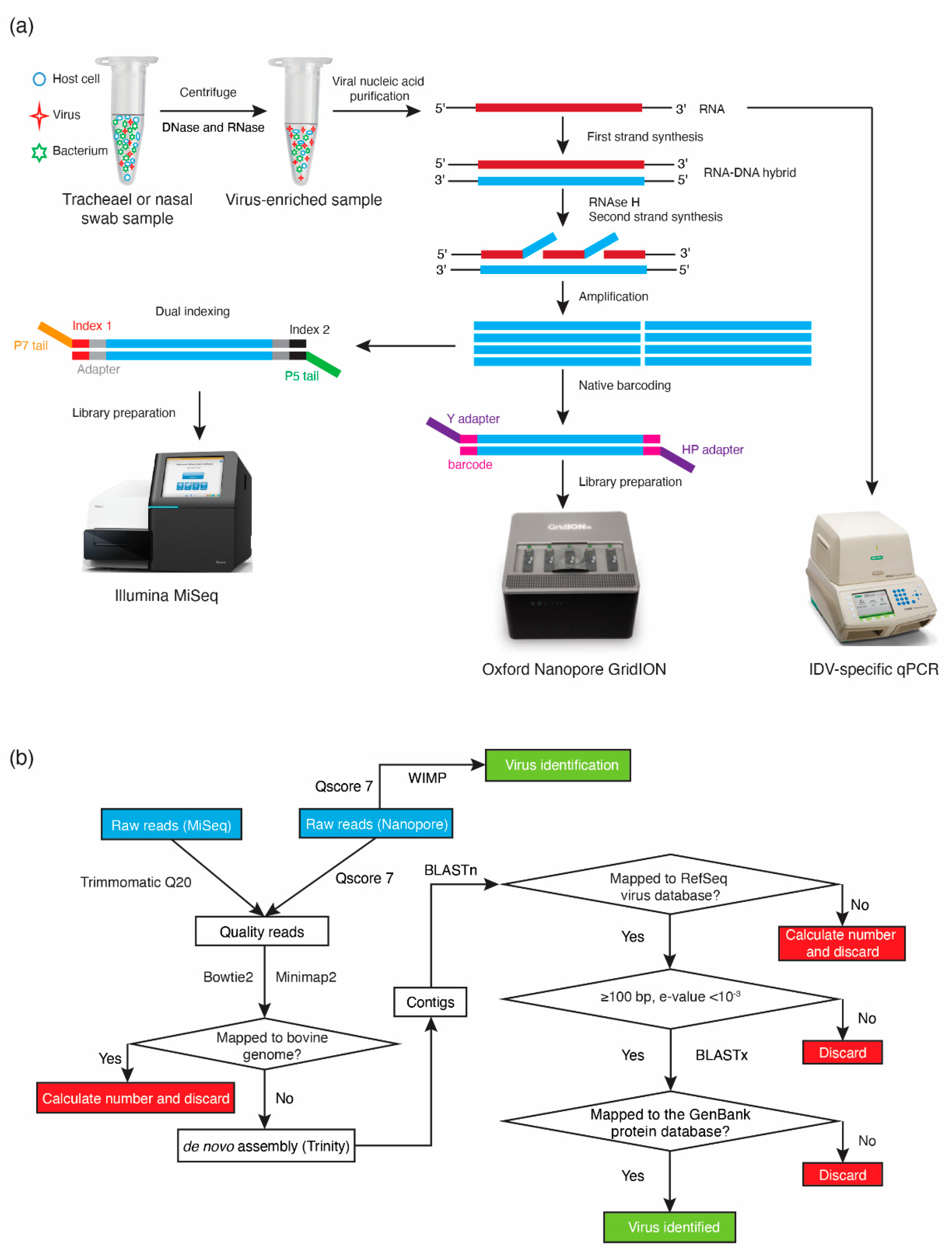
2.4. GridION Library Preparation and Sequencing
2.5. Bioinformatic Analysis
3. Results
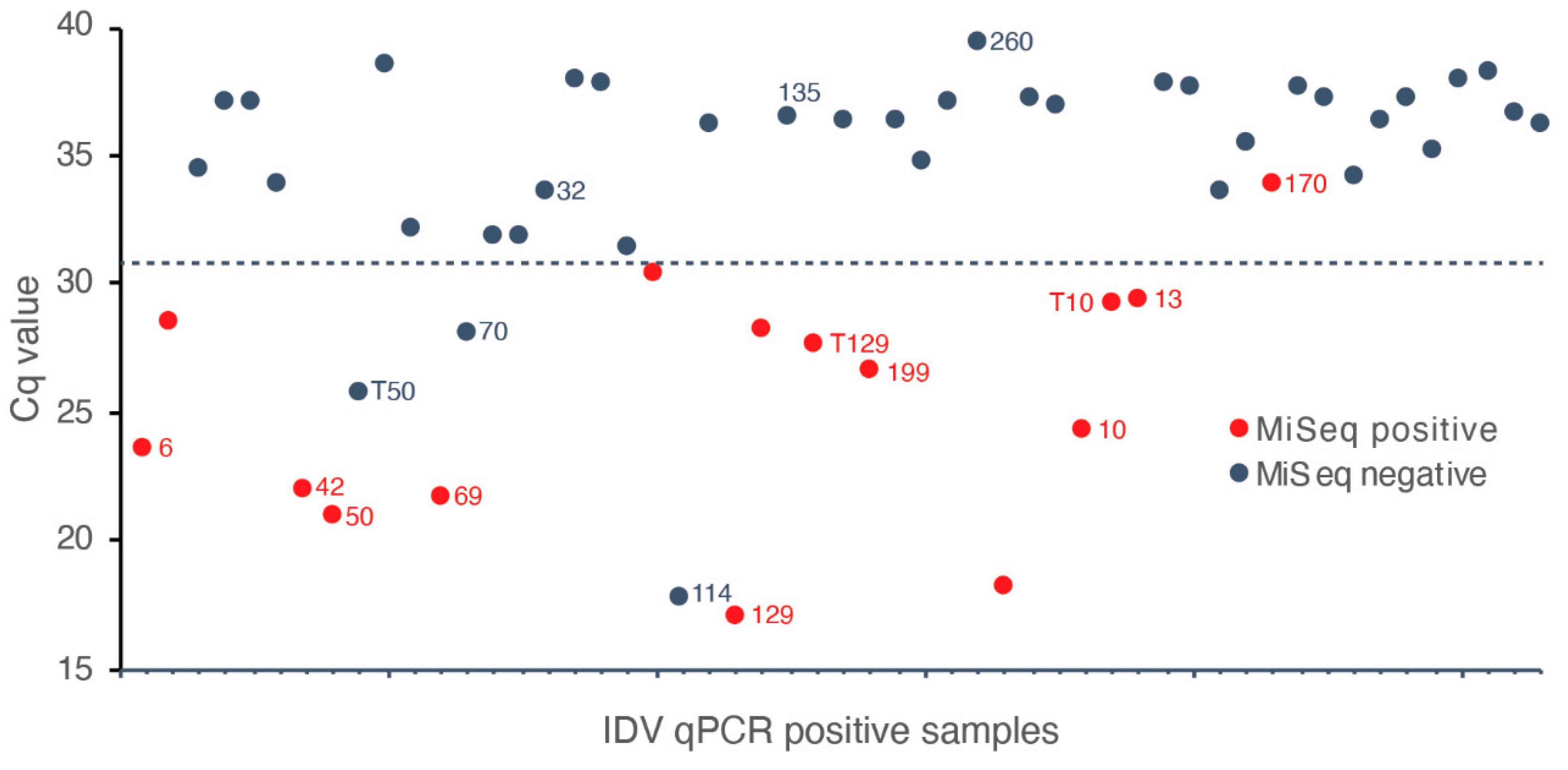
3.1. Comparison of IDV Detection by MiSeq and qPCR
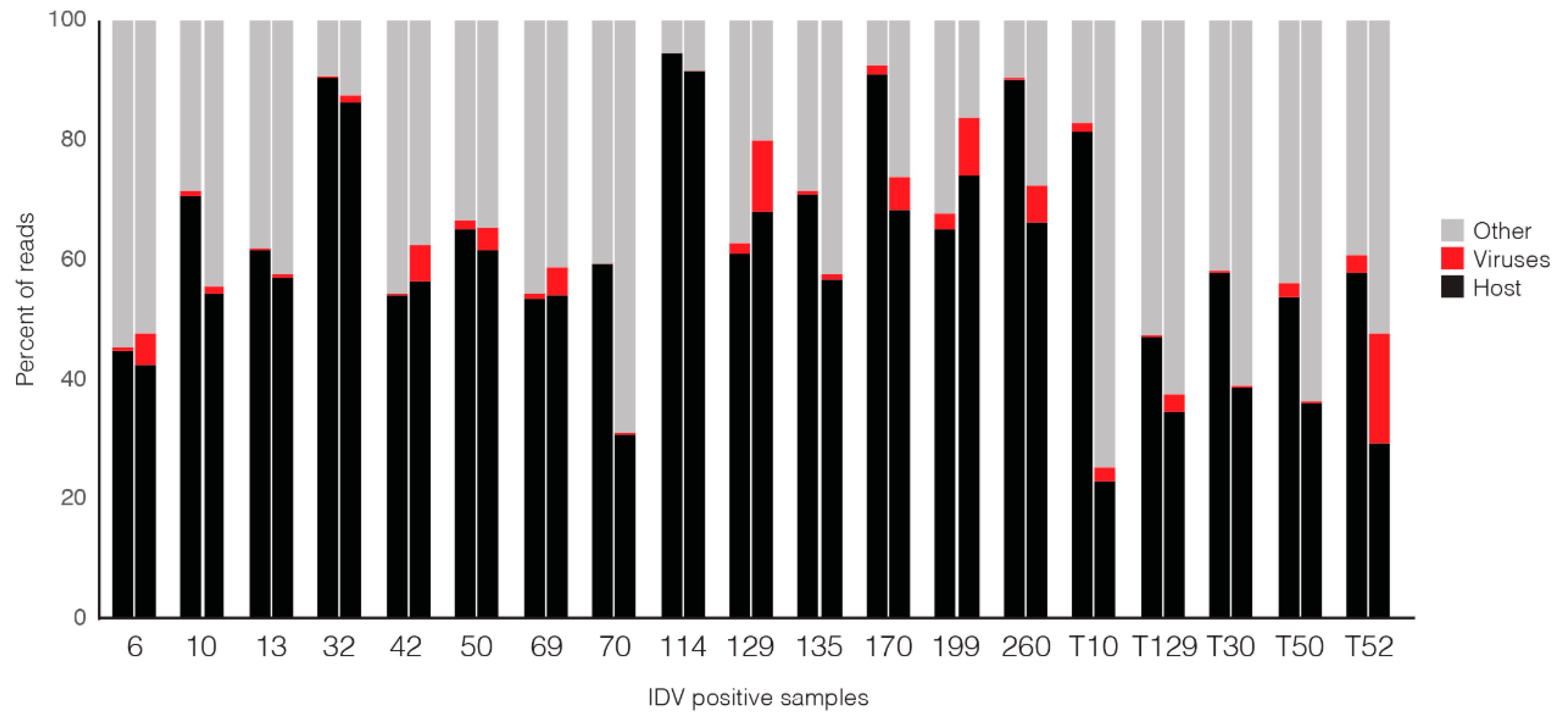
3.2. Comparison of Nanopore Sequencing Results with Previously Determined MiSeq Data
3.3. Comparison of IDV Detection by qPCR, MiSeq and Nanopore Sequencing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kafetzopoulou, L.E.; Efthymiadis, K.; Lewandowski, K.; Crook, A.; Carter, D.; Osborne, J.; Aarons, E.; Hewson, R.; Hiscox, J.A.; Carroll, M.W.; et al. Assessment of metagenomic Nanopore and Illumina sequencing for recovering whole genome sequences of chikungunya and dengue viruses directly from clinical samples. Euro Surveill. 2018, 23, 100228. [Google Scholar] [CrossRef]
- Wamaitha, M.J.; Nigam, D.; Maina, S.; Stomeo, F.; Wangai, A.; Njuguna, J.N.; Holton, T.A.; Wanjala, B.W.; Wamalwa, M.; Lucas, T.; et al. Metagenomic analysis of viruses associated with maize lethal necrosis in Kenya. Virol. J. 2018, 15, 90. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.; Chen, J. Application of next generation sequencing for the detection of human viral pathogens in clinical specimens. J. Clin. Virol. 2017, 86, 20–26. [Google Scholar] [CrossRef]
- Tyler, A.D.; Mataseje, L.; Urfano, C.J.; Schmidt, L.; Antonation, K.S.; Mulvey, M.R.; Corbett, C.R. Evaluation of Oxford Nanopore’s MinION Sequencing Device for Microbial Whole Genome Sequencing Applications. Sci. Rep. 2018, 8, 10931. [Google Scholar] [CrossRef]
- Andersen, K.G.; Shapiro, B.J.; Matranga, C.B.; Sealfon, R.; Lin, A.E.; Moses, L.M.; Folarin, O.A.; Goba, A.; Odia, I.; Ehiane, P.E.; et al. Clinical Sequencing Uncovers Origins and Evolution of Lassa Virus. Cell 2015, 162, 738–750. [Google Scholar] [CrossRef]
- Filloux, D.; Fernandez, E.; Loire, E.; Claude, L.; Galzi, S.; Candresse, T.; Winter, S.; Jeeva, M.L.; Makeshkumar, T.; Martin, D.P.; et al. Nanopore-based detection and characterization of yam viruses. Sci. Rep. 2018, 8, 17879. [Google Scholar] [CrossRef]
- Quick, J.; Loman, N.J.; Duraffour, S.; Simpson, J.T.; Severi, E.; Cowley, L.; Bore, J.A.; Koundouno, R.; Dudas, G.; Mikhail, A.; et al. Real-time, portable genome sequencing for Ebola surveillance. Nature 2016, 530, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Schadt, E.E.; Turner, S.; Kasarskis, A. A window into third-generation sequencing. Hum. Mol. Genet. 2010, 19, R227–R240. [Google Scholar] [CrossRef] [PubMed]
- Greninger, A.L.; Naccache, S.N.; Federman, S.; Yu, G.; Mbala, P.; Bres, V.; Stryke, D.; Bouquet, J.; Somasekar, S.; Linnen, J.M.; et al. Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Med. 2015, 7, 99. [Google Scholar] [CrossRef]
- Buermans, H.P.; den Dunnen, J.T. Next generation sequencing technology: Advances and applications. Biochim. Biophys. Acta 2014, 1842, 1932–1941. [Google Scholar] [CrossRef] [PubMed]
- Mitra, N.; Cernicchiaro, N.; Torres, S.; Li, F.; Hause, B.M. Metagenomic characterization of the virome associated with bovine respiratory disease in feedlot cattle identified novel viruses and suggests an etiologic role for influenza D virus. J. Gen. Virol. 2016, 97, 1771–1784. [Google Scholar] [CrossRef] [PubMed]
- Ng, T.F.; Kondov, N.O.; Deng, X.; Van Eenennaam, A.; Neibergs, H.L.; Delwart, E. A metagenomics and case-control study to identify viruses associated with bovine respiratory disease. J. Virol. 2015, 89, 5340–5349. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Hill, J.E.; Fernando, C.; Alexander, T.W.; Timsit, E.; van der Meer, F.; Huang, Y. Respiratory viruses identified in western Canadian beef cattle by metagenomic sequencing and their association with bovine respiratory disease. Transbound. Emerg. Dis. 2019, 66, 1379–1386. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, L.; Eckard, L.; Epperson, W.B.; Long, L.P.; Smith, D.; Huston, C.; Genova, S.; Webby, R.; Wan, X.F. Influenza D virus infection in Mississippi beef cattle. Virology 2015, 486, 28–34. [Google Scholar] [CrossRef]
- Dane, H.; Duffy, C.; Guelbenzu, M.; Hause, B.; Fee, S.; Forster, F.; McMenamy, M.J.; Lemon, K. Detection of influenza D virus in bovine respiratory disease samples, UK. Transbound. Emerg. Dis. 2019, 66, 2184–2187. [Google Scholar] [CrossRef]
- Hause, B.M.; Collin, E.A.; Liu, R.; Huang, B.; Sheng, Z.; Lu, W.; Wang, D.; Nelson, E.A.; Li, F. Characterization of a novel influenza virus in cattle and Swine: Proposal for a new genus in the Orthomyxoviridae family. MBio 2014, 5, e00031-14. [Google Scholar] [CrossRef]
- Hause, B.M.; Ducatez, M.; Collin, E.A.; Ran, Z.; Liu, R.; Sheng, Z.; Armien, A.; Kaplan, B.; Chakravarty, S.; Hoppe, A.D.; et al. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog. 2013, 9, e1003176. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Fu, X.; Li, G.; Kerlin, F.; Veit, M. Novel Influenza D virus: Epidemiology, pathology, evolution and biological characteristics. Virulence 2017, 8, 1580–1591. [Google Scholar] [CrossRef]
- White, S.K.; Ma, W.; McDaniel, C.J.; Gray, G.C.; Lednicky, J.A. Serologic evidence of exposure to influenza D virus among persons with occupational contact with cattle. J. Clin. Virol. 2016, 81, 31–33. [Google Scholar] [CrossRef] [PubMed]
- Zhai, S.L.; Zhang, H.; Chen, S.N.; Zhou, X.; Lin, T.; Liu, R.; Lv, D.H.; Wen, X.H.; Wei, W.K.; Wang, D.; et al. Influenza D Virus in Animal Species in Guangdong Province, Southern China. Emerg. Infect. Dis. 2017, 23, 1392–1396. [Google Scholar] [CrossRef]
- Sreenivasan, C.; Thomas, M.; Sheng, Z.; Hause, B.M.; Collin, E.A.; Knudsen, D.E.; Pillatzki, A.; Nelson, E.; Wang, D.; Kaushik, R.S.; et al. Replication and Transmission of the Novel Bovine Influenza D Virus in a Guinea Pig Model. J. Virol. 2015, 89, 11990–12001. [Google Scholar] [CrossRef]
- Timsit, E.; Workentine, M.; van der Meer, F.; Alexander, T. Distinct bacterial metacommunities inhabit the upper and lower respiratory tracts of healthy feedlot cattle and those diagnosed with bronchopneumonia. Vet. Microbiol. 2018, 221, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Allander, T.; Tammi, M.T.; Eriksson, M.; Bjerkner, A.; Tiveljung-Lindell, A.; Andersson, B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. USA 2005, 102, 12891–12896. [Google Scholar] [CrossRef]
- Faccini, S.; De Mattia, A.; Chiapponi, C.; Barbieri, I.; Boniotti, M.B.; Rosignoli, C.; Franzini, G.; Moreno, A.; Foni, E.; Nigrelli, A.D. Development and evaluation of a new Real-Time RT-PCR assay for detection of proposed influenza D virus. J. Virol. Methods 2017, 243, 31–34. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Brister, J.R.; Ako-Adjei, D.; Bao, Y.; Blinkova, O. NCBI viral genomes resource. Nucleic Acids Res. 2015, 43, D571–D577. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Shan, T.; Wang, C.; Côté, C.; Kolman, J.; Onions, D.; Gulland, F.M.D.; Delwart, E. The Fecal Viral Flora of California Sea Lions. J. Virol. 2011, 85, 9909–9917. [Google Scholar] [CrossRef]
- Shan, T.; Li, L.; Simmonds, P.; Wang, C.; Moeser, A.; Delwart, E. The Fecal Virome of Pigs on a High-Density Farm. J. Virol. 2011, 85, 11697–11708. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Lou, X.; Yan, H.; Pan, J.; Mao, H.; Tang, H.; Shu, Y.; Zhao, Y.; Liu, L.; Li, J.; et al. Metagenomic analysis of viral nucleic acid extraction methods in respiratory clinical samples. BMC Genom. 2018, 19, 773. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.J.; Wang, J.; Todd, A.K.; Bissielo, A.B.; Yen, S.; Strydom, H.; Moore, N.E.; Ren, X.; Huang, Q.S.; Carter, P.E.; et al. Evaluation of rapid and simple techniques for the enrichment of viruses prior to metagenomic virus discovery. J. Virol. Methods 2014, 195, 194–204. [Google Scholar] [CrossRef] [PubMed]
- Quick, J.; Grubaugh, N.D.; Pullan, S.T.; Claro, I.M.; Smith, A.D.; Gangavarapu, K.; Oliveira, G.; Robles-Sikisaka, R.; Rogers, T.F.; Beutler, N.A.; et al. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat. Protoc. 2017, 12, 1261. [Google Scholar] [CrossRef]
- Esling, P.; Lejzerowicz, F.; Pawlowski, J. Accurate multiplexing and filtering for high-throughput amplicon-sequencing. Nucleic Acids Res. 2015, 43, 2513–2524. [Google Scholar] [CrossRef]
- Kim, D.; Song, L.; Breitwieser, F.P.; Salzberg, S.L. Centrifuge: Rapid and sensitive classification of metagenomic sequences. Genome Res. 2016, 26, 1721–1729. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
| Sample * | Cq Value | Copy Number (Per Reaction) | Number (%) of IDV Reads | Largest IDV Contig (bp) | Total Input Reads | ||||
|---|---|---|---|---|---|---|---|---|---|
| Nanopore (WIMP) | Nanopore (In-House) | MiSeq | Nanopore (In-House) | MiSeq | Nanopore | MiSeq | |||
| 129 | 16.99 | 6.25 × 107 | 321,638 (14.69) | 606,932 (27.72) | 2182 (1.48) | 2030 | 951 | 2,188,805 | 147,341 |
| 114 | 17.53 | 1.31 × 107 | 8 (<0.01) | ND | ND | N/A | N/A | 335,559 | 136,961 |
| 50 | 20.71 | 1.25 × 106 | 1088 (5.74) | 1625 (8.57) | 162 (1.1) | 1161 | 498 | 18,966 | 14,719 |
| 69 | 21.69 | 4.48 × 105 | 944 (7.83) | 2287 (18.97) | 1812 (0.69) | 980 | 842 | 12,053 | 263,262 |
| 42 | 22.04 | 2.86 × 105 | 608 (0.34) | 584 (0.33) | 26 (0.07) | 986 | 499 | 179,559 | 37,560 |
| 6 | 23.33 | 2.16 × 105 | 281 (1.44) | 350 (1.8) | 183 (0.08) | 1034 | 522 | 19,512 | 228,875 |
| 10 | 24.09 | 1.31 × 105 | 1656 (1.17) | 1650 (1.11) | 87 (0.2) | 1359 | 485 | 148,381 | 44,433 |
| 199 | 26.01 | 4.64 × 104 | 4211 (10.99) | 10861 (28.34) | 3250 (1.49) | 2308 | 552 | 38,329 | 218,800 |
| T50 | 26.76 | 2.93 × 104 | 3 (<0.01) | ND | ND | N/A | N/A | 43,193 | 1,256,918 |
| T129 | 28.20 | 8.00 × 103 | 403 (0.23) | 502 (0.28) | 34 (<0.01) | 843 | 327 | 177,706 | 444,979 |
| 70 | 28.71 | 5.70 × 103 | 2 (0.02) | ND | ND | N/A | N/A | 12,211 | 160,704 |
| T10 | 29.15 | 5.50 × 103 | 29 (0.54) | ND | 455 (0.03) | N/A | 913 | 5,413 | 1,415,256 |
| 13 | 29.22 | 4.15 × 103 | 116 (0.10) | 152 (0.13) | 48 (0.04) | 626 | 470 | 119,073 | 132,847 |
| 32 | 33.60 | 2.18 × 102 | ND | ND | ND | N/A | N/A | 275,387 | 32,312 |
| 170 | 35.62 | 8.25 × 101 | 1080 (2.03) | 1991 (3.75) | 8167 (0.74) | 903 | 1,584 | 53,131 | 1,100,167 |
| 135 | 36.51 | 4.88 × 101 | 10 (<0.01) | ND | ND | N/A | N/A | 142,737 | 154,220 |
| 260 | 39.46 | 6.88 | 9 (<0.01) | ND | ND | N/A | N/A | 164,607 | 959,935 |
| T30 | ND | - | 3 (<0.01) | ND | 2 (<0.01) | N/A | 249 | 139,037 | 1,156,213 |
| T52 | ND | - | 51 (0.1) | 52 (0.1) | 497 (0.05) | 1616 | 1,341 | 51,180 | 930,628 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Huang, Y.; Godson, D.L.; Fernando, C.; Alexander, T.W.; Hill, J.E. Assessment of Metagenomic Sequencing and qPCR for Detection of Influenza D Virus in Bovine Respiratory Tract Samples. Viruses 2020, 12, 814. https://doi.org/10.3390/v12080814
Zhang M, Huang Y, Godson DL, Fernando C, Alexander TW, Hill JE. Assessment of Metagenomic Sequencing and qPCR for Detection of Influenza D Virus in Bovine Respiratory Tract Samples. Viruses. 2020; 12(8):814. https://doi.org/10.3390/v12080814
Chicago/Turabian StyleZhang, Maodong, Yanyun Huang, Dale L. Godson, Champika Fernando, Trevor W. Alexander, and Janet E. Hill. 2020. "Assessment of Metagenomic Sequencing and qPCR for Detection of Influenza D Virus in Bovine Respiratory Tract Samples" Viruses 12, no. 8: 814. https://doi.org/10.3390/v12080814
APA StyleZhang, M., Huang, Y., Godson, D. L., Fernando, C., Alexander, T. W., & Hill, J. E. (2020). Assessment of Metagenomic Sequencing and qPCR for Detection of Influenza D Virus in Bovine Respiratory Tract Samples. Viruses, 12(8), 814. https://doi.org/10.3390/v12080814





